Abstract
Aims
To determine if immunohistochemistry (IHC) could be used to monitor nuclear factor‐κB (NF‐κB) activity in oesophageal adenocarcinoma and pre‐malignant (Barrett's) oesophageal tissues, relative to normal oesophageal mucosa. The pro‐inflammatory cytokine interleukin‐8 (IL‐8), a transcriptional target of NF‐κB, was also studied to better understand NF‐κB functionality; its RNA and protein levels were assessed in oesophageal tissues.
Methods
IHC was employed using an antibody against the nuclear localisation sequence (NLS) of the p65 subunit as well as an antibody against IL‐8. To assess NF‐κB function, changes in gene expression of NF‐κB controlled genes (IL‐8 and I‐κB) were also assessed in the histological sequence using real‐time PCR. More global expression changes were also studied using membrane arrays.
Results
IHC was effective at monitoring overall NF‐κB activity and IL‐8 abundance. This method also allowed NF‐κB activity and IL‐8 abundance to be pinpointed in specific cell types. There were significant increases in nuclear NF‐κB activity and IL‐8 abundance across the histological series. Gene expression analysis also showed consistent up‐regulation of IL‐8, confirming the IHC data and showing enhanced transcriptional NF‐κB activity. I‐κB (another NF‐κB target) showed down‐regulation in dysplastic and adenocarcinoma tissues. Down‐regulation of I‐κB gene expression may partly explain increased NF‐κB activity.
Conclusion
IHC, using antibodies against the NLS of p65, may be useful in monitoring overall NF‐κB activity in oesophageal tissues. As IHC is amenable to high‐throughput screening (whereas traditional electrophoretic mobility shift assay methods are not), this may lead to the development of a better screening tool for early cancer risk.
Keywords: immunohistochemistry, NF‐kB, IL‐8, Barrett's oesophagus, adenocarcinoma, biomarker
Barrett's oesophagus is a pre‐malignant lesion occurring in approximately 10% of patients with chronic gastro‐oesophageal reflux disease.1,2 Approximately 1–3% of Western populations are thought to have acquired Barrett's oesophagus,3 and in the UK, up to 2% of these will convert to oesophageal adenocarcinoma per year.4 Therefore, this can mean a lifetime cancer risk of 10–15% for the average UK Barrett's oesophagus patient.5 The UK cancer conversion rate, for some unknown reason, is twofold higher than that in other Western countries.6 While incidences of Barrett's oesophagus and its related adenocarcinoma have been increasing,7,8 there has been little improvement in the abysmal 5‐year survival rates of the cancer patients (currently only 7% in the UK; www.cancerresearchuk.org.uk/aboutcancer/statistics). Low survival rates are compounded by the poor responses of most oesophageal adenocarinomas to neoadjuvant chemotherapy or radiotherapy.9 For these reasons, much interest has centred on the molecular mechanisms leading to cancer formation,10 in the hope that new treatments and earlier diagnoses can be developed to aid patient prognosis.
One molecular pathway that has been increasingly linked to the development of several different types of cancer in recent years, is the NF‐κB signalling pathway.11 As NF‐κB can be an anti‐apoptotic factor, its activation may promote survival in clones of oesophageal cells and hence allow selective outgrowth from surrounding cells. NF‐κB is usually kept inactive in the cytoplasm by its inhibitor I‐κB, but I‐κB is degraded on receipt of activation signals, leading to nuclear translocation of NF‐κB and targeted gene expression. A recent report has presented convincing evidence of a role for NF‐κB in oesophageal tumour development and indeed in survival.12 We have a particular interest in NF‐κB as we have shown it is activated in vitro by bile acid exposure.13 Bile acids are increasingly suspected as being major carcinogens in Barrett's oesophagus patients. We have shown in vitro that after exposure to physiological levels of bile acid, one of the main genes up‐regulated via NF‐κB is interleukin‐8 (IL‐8). IL‐8 is known to be involved in cancer progression and invasion,14 and has recently been suggested to be up‐regulated at either the RNA or protein level, at various stages across the histological series in Barrett's oesophagus.15,16,17
Here, we aimed to confirm the recently reported involvement of NF‐κB and IL‐8 in the histological progression from metaplasia to adenocarcinoma (relative to matched squamous tissues). Crucially, we test here the usefulness of immunohistochemistry (IHC) to monitoring NF‐κB activity in oesophageal adenocarcinoma development. We have used a specific antibody in our IHC, that recognises only the active form of p65. The active p65 antibody recognises the nuclear localisation sequence (NLS) of the p65 subunit and as such its epitope is only vacant when the inhibitor I‐κB has been degraded. Therefore, it was anticipated that this antibody would mainly target nuclear p65 proteins. Previous IHC studies have supported its use in assessing overall NF‐κB activity in tissue sections.18 In addition to NF‐κB, we have assessed both IL‐8 gene and protein expression levels using membrane arrays, real‐time PCR and immunohistochemistry (IHC) to correlate NF‐κB transcriptional activity with the p65 IHC data. Importantly, if IHC can reliably assess both NF‐κB activity and IL‐8 abundance, then given the growing evidence for these proteins playing a role in oesophageal adenocarcinoma development, this may represent a practical way of monitoring cancer risk in Barrett's oesophagus patients.
Materials and methods
Tissue collection
Tissue for this study was obtained from three sources: archival paraffin embedded tissue sections, fresh surgical resections and endoscopic biopsy specimens. Ethical approval was first obtained for this study from the Bridgend Neath Port Talbot and Swansea local research ethics committee. Archival tissue was obtained from the pathology archive, Singleton Hospital. Forty‐one archival samples were identified, 22 with Barrett's oesophagus and 19 with adenocarcinoma. In 23 of these cases, there was also adjacent squamous tissue available. For the RNA study, 11 fresh surgical adenocarcinoma samples were obtained from consenting patients during oesophagectomy at Swansea NHS Trust (adenocarcinoma plus matched squamous tissue). For the pre‐malignant real‐time PCR study of IL‐8 and I‐κB expression, 25 additional consenting patients with pre‐malignant Barrett's oesophagus (dysplasia, n = 8; metaplasia, n = 17) were obtained from upper gastrointestinal endoscopy clinics held at Swansea NHS Trust. Fresh tissue was stored in RNA Later (Ambion, Huntington, UK) at 4°C to preserve its integrity before RNA extraction.
Immunohistochemistry
IHC was performed on tissue sections from 22 metaplastic sections and 19 adenocarcinoma sections (containing areas of squamous tissue in 23 cases). IHC was carried out using a Ventana Benchmark XT automated stainer (Illkirch, France) with prior application of a biotin blocking kit, to prevent any endogenous biotin in the tissue from staining and giving false positive results. The antibodies used included an antibody against the p50 subunit of NF‐κB (AbCam, Cambridge, UK), an antibody against the p65 subunit of NF‐κB (AbCam), a mouse monoclonal against the active form of the p65 subunit of NF‐κB (specifically against the NLS) (Chemicon, Harrow, UK) and an antibody against the IL‐8 protein (AbCam). The p50 antibody, the p65 antibody and the IL‐8 antibody were used at a dilution of 1:100; the p65 NLS antibody was used at a dilution of 1:200. For each test case a negative control was also used which received the same treatment as the test slide except the primary antibody was omitted. Positive controls (breast ductal cancer for p50 and both p65 antibodies and tonsil for IL‐8 antibody) were used to optimise the conditions. Light counterstaining (60 s) of all sections was achieved with in‐house formulated haematoxylin. The stained sections were analysed by two independent observers and a scoring system (relative to the positive control) was used to assess each section. The following scores were applied: score 0, no nuclear or cytoplasmic staining; score 1, weak nuclear or cytoplasmic staining (less than seen in positive control); score 2, moderate nuclear or cytoplasmic staining (equal to that seen in the control); score 3, strong nuclear or cytoplasmic staining (greater than that seen in the control). NF‐κB subunit (p65 and p50) staining was scored in both the nucleus and cytoplasm and IL‐8 staining was only scored in the cytoplasm.
In the case of the sections of Barrett's oesophagus, the staining was assessed in the epithelial areas surrounding the goblet cells, which are characteristic of the condition. All photomicrographs in this article were produced using the Nikon Elipse (E6000) microscope (Nikon, Kingston Upon Thames, UK), a Nikon digital camera (DXM1200) (Nikon) and a Nikon ACT‐1 (version 2.12) image capture system.
RNA extraction
The fresh oesophageal tissue samples were subject to RNA extraction through the use of the TriSpin method as detailed previously.13
Membrane microarrays
The RNA (2 μg) was first reverse transcribed into cDNA using a Retroscript RT kit (Ambion). RNA extracted from oesophageal adenocarcinomas and adjacent squamous mucosa, was subject to gene expression analysis using pathway specific membrane microarrays (Superarray, Frederick, MD, USA) following the manufacturer's recommendations. The arrays used in this study were the Cancer Pathway array, Signal Tranduction array, NF‐κB array and the Cell Cycle array. Each array contained two copies of five housekeeping genes (β‐actin, Gapdh, PPIA and RPL13A), three negative control DNA samples (pUC18) and three blank spots. The housekeeping gene spot intensities allowed standardisation of RNA loading between samples. The negative control spots reduced the chance of false positive hybridisations. The arrays were scanned using a chemiluminescence reader (Chemi‐doc, BioRad, Hemel Hempstead, UK) and each spot intensity was quantified where possible. It was found however, that in many cases, the arrays were not suitable to provide quantitative data due to problems with patchy backgrounds across the arrays that made normalisation difficult. Hence, we relied on real‐time PCR to provide accurate quantitative data. Only obvious expression changes noted on the arrays are listed here, hence more subtle changes may be missed.
Real‐time PCR
IL‐8 and I‐κB expression differences were quantitated across the histological series by real‐time PCR using an iCycler PCR machine (BioRad), Sybrgreen IQ mix (BioRad) and using β‐actin as a housekeeper.
The PCR primers used in real‐time analysis were as follows: β‐actin (forward: GAT GGC CAC GGC TGC TTC; reverse: TGC CTC AGG GCA GCG GAA); I‐κB (forward: ACA CTA GAA AAC TTC AGA TGC; reverse: ACA CAG TCA TCA TAG GGC AG), IL‐8 (forward: CAA TCC TAG TTT GAT ACT CCC; reverse: AAT TAC TAA TAT TGA CTG TGG AG). All PCRs were run in triplicate on the 96‐well plate at 60°C annealing. Relative gene expression between samples was calculated through the inclusion of a cDNA dilution series of a reference sample, amplified with each primer set separately. Real‐time experiments were performed in duplicate on two different occasions (hence six data points), average fold increases were calculated and compared between matched normal and disease tissues. Fold increases in expression were calculated for Barrett's, dysplastic Barrett's and adenocarcinoma tissue, due to the fact that matched adjacent squamous tissue was available for each case.
Statistical analysis
The IHC scoring data and the real‐time fold increases were assessed for significance using a two‐tailed Fisher's exact test. For IHC, relative numbers of strong, moderate and weak scores were compared between the histological subtypes. For the real‐time data fold increases relative to matched squamous tissue were compared between histologies. Significance was only accepted if p<0.05 or p<0.01.
Results
NF‐κB activity in Barrett's oesophagus and adenocarcinoma tissues
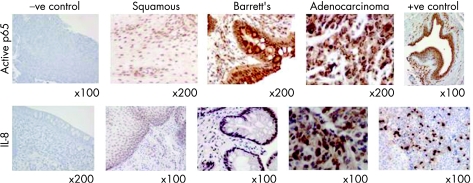
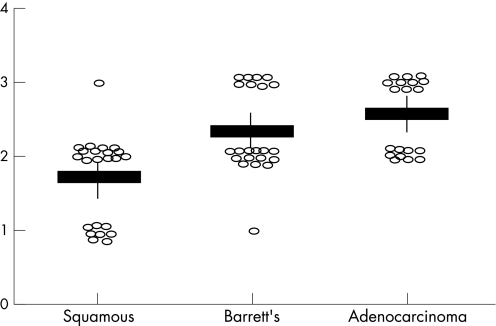
Initial IHC studies were performed to assess total p50 and p65 protein levels across the histological series in Barrett's oesophagus. Overall, as might be expected, there was little visible difference in p50 or p65 total protein levels across the histological series, although there was a suggestion of increasing protein levels in the adenocarcinomas (results not shown). More importantly, to assess the levels of active NF‐κB across the histological series, an antibody against the nuclear localisation sequence (NLS) of p65 was employed. As can be seen in fig 1 (top panel), there was increased antibody binding in the Barrett's tissues and adenocarcinoma tissues compared to the adjacent squamous tissue. This staining was increasingly nuclear in the later histologies and in some cases displayed both punctate staining and distinct perinuclear staining. The antibody scores for each histological grade are shown in table 1 and the nuclear scores are plotted in fig 2. There was a highly significant increase in strong nuclear staining in Barrett's tissues (p = 0.033) and in adenocarcinoma tissues (p = 0.0075), when both were compared to adjacent squamous tissue. There was no difference in nuclear staining in the squamous sections adjacent to adenocarcinoma compared to those squamous sections adjacent to Barrett's oesophagus (weak vs moderate + strong, p = 0.19).
Figure 1 Example immunohistochemistry images with the active nuclear factor‐κB (NF‐κB) and interleukin‐8 (IL‐8) antibodies in squamous, Barrett's and adenocarcinoma sections, showing increasing staining across the histological series. Negative controls (no antibody) are shown for the active p65 antibody (adenocarcinoma material) and the IL‐8 antibody (Barrett's metaplasia material). Positive controls are also shown (breast ductal cancer for active p65 and tonsil for IL‐8).
Table 1 Immunohistochemistry (IHC) scores by histology.
| Histology | p65 Cyto Weak | p65 Cyto Mod | p65 Cyto Strong | p65 Nuc Weak | p65 Nuc Mod | p65 Nuc Strong | IL‐8 Weak | IL‐8 Mod | IL‐8 Strong |
|---|---|---|---|---|---|---|---|---|---|
| Squamous | 17/23 | 6/231 | 0/23 | 8/23 | 14/23 | 1/23 | 20/23 | 3/23 | 0/23 |
| Barrett's | 15/22 | 5/22 | 2/22 | 1/22 | 13/22 | 8/22* | 7/22* | 12/22* | 3/22 |
| Adenocarcinoma | 3/19*† | 12/19 | 4/19 | 0/19* | 9/19 | 10/19* | 8/19 | 6/19 | 5/19* |
Cyto, cytoplasm; Nuc, nucleus.
IHC scores for nuclear factor‐κB (NF‐κB) (p65 active antibody) and interleukin‐8 (IL‐8) across histological series.
Staining scored: weak, moderate (Mod) and strong relative to negative (no antibody) and positive (breast for NF‐κB and tonsil for IL‐8) controls.
*p<0.05 relative to squamous tissue.
†Adenocarcinoma staining is significant relative to both Barrett's tissue and squamous tissue.
Figure 2 Immunohistochemistry (IHC) scores for nuclear factor‐κB (NF‐κB) staining across the histological series. Each patient score is shown as a open circle. Average scores for each histology are shown by horizontal line, standard deviations by vertical line.
IL‐8 protein levels in Barrett's oesophagus and adenocarcinoma tissues
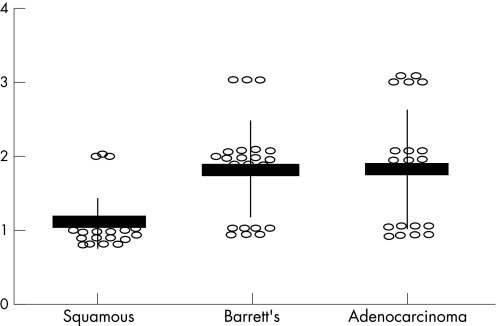
Immunohistochemistry was also used to concurrently assess IL‐8 protein levels with the active p65 levels detailed above, using the same tissue samples. As IL‐8 is transcriptionally controlled by NF‐κB, measuring IL‐8 levels allows us to assess NF‐κB functionality. Figure 1 (lower panel) illustrates the increased IL‐8 protein levels in both Barrett's tissues and adenocarcinoma. Table 1 contains the scores for IL‐8 staining and this data is plotted in fig 3. There was a significant increase in moderate IL‐8 staining in Barrett's tissues (p = 0.04) and an increase in strong staining in adenocarcinoma tissue (p = 0.049) compared to adjacent squamous tissue. There was no difference in IL‐8 staining in the squamous sections adjacent to adenocarcinoma compared to those squamous sections adjacent to Barrett's oesophagus (weak vs moderate + strong, p = 1).
Figure 3 Immunohistochemistry (IHC) scores for interleukin‐8 staining across the histological series. Each patient score is shown as a open circle. Average scores for each histology are shown by horizontal line, standard deviations by vertical line.
Correlation of active p65 staining and IL‐8 staining with clinical data for the cancer patients
Although both NF‐κB and IL‐8 staining showed clear increases during the histological progression, there was no overall correlation between IL‐8 levels and NF‐κB activity. That is, those patients with highest levels of IL‐8 staining did not necessarily show the highest levels of NF‐κB activity (nuclear staining).
There was no association between active p65 nuclear staining or IL‐8 staining and the stage or grade of the adenocarcinoma. Neither was there an association between IL‐8 staining or active NF‐κB staining and age or sex of the Barrett's or adenocarcinoma patients.
Up‐regulation of the IL‐8 gene in adenocarcinoma tissues
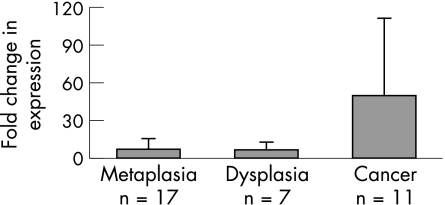
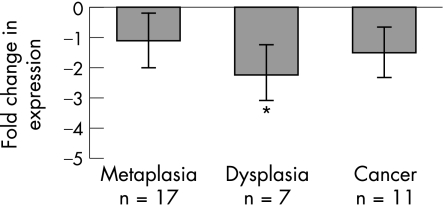
In order to determine if increased IL‐8 protein levels were a result of increases in IL‐8 gene expression and to assess overall NF‐κB transcriptional activity, we assessed IL‐8 gene expression in a number of freshly resected adenocarcinoma tissues (n = 11) and biopsy specimens of Barrett's oesophagus (with and without dysplasia) (n = 25) relative to matched adjacent squamous material. As I‐κB inhibits NF‐κB translocation, and given that the IHC showed striking NF‐κB activity, we decided to assess I‐κB expression levels also. Unfortunately, due to the need for fresh material for RNA extraction, this group of adenocarcinomas and Barrett's tissues was different to those used in the IHC study. The gene expression changes were assessed in two ways. Firstly, we conducted real‐time PCR for IL‐8 and I‐κB on both Barrett's tissue (with and without dysplasia) and adenocarcinoma tissue relative to matched squamous material from the same patient. Secondly, the 11 adenocarcinomas (plus 11 matched squamous tissues) were assessed using four different types of membrane arrays each containing 100 or so cancer‐related genes. Changes in gene expression were noted for each tumour and an overall picture of more global expression changes built up. In terms of the real‐time PCR, fig 4 shows the average IL‐8 gene up‐regulation and fig 5 shows the average I‐κB down‐regulation across the histological series. There was considerable up‐regulation of IL‐8 in metaplastic and dysplastic tissue (averaging 7‐fold), however this rose to an average of 50‐fold in the adenocarcinomas. In contrast, I‐κB was shown to be correspondingly down‐regulated in the same tissues (twofold in the dysplasias).
Figure 4 Real‐time PCR analysis of interleukin‐8 (IL‐8) across histological series. Expression is shown as fold change relative to matched squamous tissue. IL‐8 gene expression shows clear increased expression across the histological series and the fold increase relative to matched squamous tissue is significantly greater in adenocarcinoma tissue relative to Barrett's tissue (p = 0.010).
Figure 5 Real‐time PCR analysis of I‐κB expression across histological series. I‐κB, in contrast to interleukin‐8 (IL‐8), shows down‐regulation in the dysplasia and adenocarcinoma samples. The down‐regulation of I‐κB is significant in the dysplasias relative to the Barrett's tissues (p = 0.035).
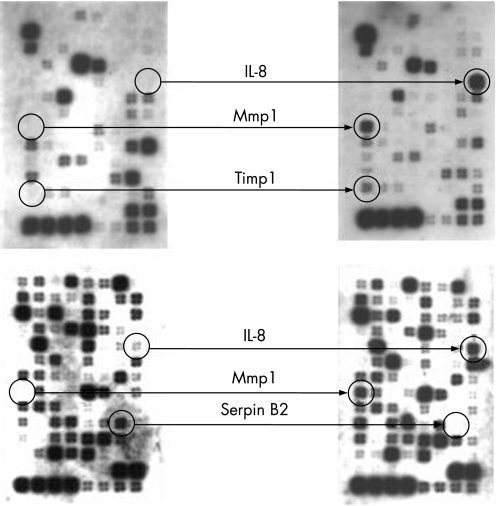
Figure 6 shows some example array images illustrating the prominent up‐regulation of the IL‐8 gene in the adenocarcinomas. Table 2 contains the list of genes commonly up‐regulated in the 11 adenocarcinomas compared to matched squamous tissue and highlights the fact that IL‐8 up‐regulation was the most common gene expression change noted using these arrays (in 72% of adenocarcinomas). Other genes shown to be up‐regulated in the tumours include mmp1, timp1 and β‐catenin. Pai‐2 was shown to be down‐regulated relative to matched squamous tissue.
Figure 6 Example of expression analysis of adenocarcinoma tissue (versus matched squamous material) by membrane array. Left‐hand arrays show squamous tissue, right‐hand arrays show matched tumour tissue. Among other changes noted, interleukin‐8 (IL‐8) shows strong expression in adenocarcinomas, but no expression in squamous tissue. Genes contained on these “cancer pathway” arrays are available at http://www.superarray.com/ArrayList.php?application = CANCER.
Table 2 Gene expression changes in tumour tissues.
| Gene | Function | Array | Up/down |
|---|---|---|---|
| IL‐8 | Inflammation/angiogenesis | Cancer | Up (8) |
| Pai‐2 | Inhibitor of invasion | Cancer | Down (6) |
| Mmp1 | Invasion factor/protease | Cancer | Up (4) |
| Timp 1 | Inhibitor of mmp | Cancer | Up (3) |
| β‐catenin | Adhesion/proliferation | Cancer | Up (3) |
| Properdin | B‐factor | NFκB | Up (2) |
| Cyclin B1 | Mitosis promoting factor | Cell cycle | Up (2) |
| CDK1 | Mitosis promoting factor | Cell cycle | Up (2) |
| Cyclin G2 | Cell cycle protein | Cell cycle | Up (2) |
| Fos | Transcription factor | NFκB | Up (2) |
| ETS2 | Transcription factor | Cancer | Up (2) |
| Serpine B5 | Inhibitor of invasion | Cancer | Down (2) |
| Apaf 1 | Apoptosis | Cancer | Up (2) |
NF‐κB, nuclear factor‐κB.
List of common gene expression changes noted on the membrane arrays in the 11 adenocarcinoma tissues versus the 11 matched squamous tissues. Numbers in parentheses represent the number of tumour samples showing such up/down regulation. IL‐8 is up‐regulated in 8/11 (72%) tumours.
Discussion
This study has highlighted the usefulness of IHC in studying NF‐κB activity and IL‐8 abundance, in the progression from normal squamous oesophagus to metaplasia and adenocarcinoma. Importantly, this study suggests that IHC may be a useful and simple tool for monitoring NF‐κB activity, in a screening programme. The advantages of IHC, over traditional NF‐κB assays such as electrophoretic mobility shift assays include cost, time and the potential for high‐throughput screening. In addition, IHC allows the histological localisation of NF‐κB activity within specific cell‐types. Electrophoretic mobility shift assay methods tend to use homogenised tissues, making cell‐type specific assessment difficult.
The critical involvement of NF‐κB in cancer progression in Barrett's patients has recently been suggested.12 In their paper, Abdel Latif et al show NF‐κB activity (through promoter binding assays) to be present in 40% of metaplastic tissues and 76% of adenocarcinomas. Importantly, the authors were able to show that NF‐κB activity correlated with late stage tumours, enhanced chemo‐resistance and significantly reduced survival. Our data support this association, showing strong nuclear NF‐κB staining in 4% of squamous tissues, 36% of metaplasias and 53% of adenocarcinomas.
Our data also support earlier studies showing that IL‐8 protein and/or mRNA is up‐regulated across the histological series in Barrett's patients.15,16,17 IL‐8 protein levels have previously been shown to be 2–3‐fold up‐regulated17 or 2–4‐fold up‐regulated15 by ELISA methods in Barrett's tissues compared to squamous tissues. The same studies have shown a 10‐fold up‐regulation of IL‐8 protein in adenocarcinomas relative to squamous tissues.17 IL‐8 mRNA has also previously been shown to be up‐regulated by 3‐fold in Barrett's tissues relative to squamous tissue.16 Furthermore, as well as IL‐8 up‐regulation, these previous studies have also shown increased NF‐κB activity (using transcription factor assays and promoter binding) in Barrett's and adenocarcinoma tissues.16,17
The IHC data generated here showed significant increases in active p65 nuclear staining in Barrett's tissues and adenocarcinoma tissues compared to squamous oesophageal tissues. Furthermore, IL‐8 staining was also shown to be stronger in the Barrett's and adenocarcinoma tissues compared to squamous tissue. No significant associations were detected with clinical grade, stage, age or sex. Our study did not show a correlation between IL‐8 IHC staining and NF‐κB IHC staining. The reason for this could be due to small sample size or may relate to the fact that expression of IL‐8 may be induced via other pathways (e.g. MapK).
Both p65 antibodies (total p65 and active p65) showed a curious clumping effect in the cytoplasm of some sections, whereby intense aggregates were seen in some areas of the slide. This was not seen with the other antibodies (p50, IL‐8) on the same sections. This “clumping effect” may either be an artefact of the staining procedure, or may suggest a biological aggregating feature of p65 in these tissues. Furthermore, another unexpected observation was the perinuclear localisation of both the p65 and IL‐8 staining in some sections. This suggests that these proteins may reside transiently at the nuclear membrane. It has already been suggested that p65 (NF‐κB) can interact with the components of the nuclear membrane and hence this may explain its accumulation here in some cases. There are no reports of IL‐8 having a perinuclear location and so this may represent an interesting finding worthy of follow‐up.
Unsurprisingly, we show here that the IL‐8 gene was up‐regulated at the mRNA level in both Barrett's tissue (with and without dysplasia) and in adenocarcinoma, explaining the increased IL‐8 protein levels seen by IHC. Presumably, this IL‐8 up‐regulation is the result of increased NF‐κB activity, as it is a well known transcriptional target of NF‐κB and was shown to be linked to NF‐κB activity in a previous study.17 However, other transcription factors may also play a role in IL‐8 expression. As no microdissection was performed in this study, the fresh tissue samples will contain stromal and inflammatory cells. Hence, it is not possible to say absolutely that the expression changes were arising in the epithelial cells. However, the IHC data in this manuscript clearly supports the epithelial cell origin of active NF‐κB and IL‐8, whose staining was concentrated in the same epithelial cell types.
Membrane arrays were also used with the adenocarcinoma and matched squamous RNA and a number of gene expression differences were identified. Some of these (like IL‐8) were NF‐κB targets and hence further support the role of NF‐κB in this type of cancer. Interestingly, using these arrays (containing 100 cancer related genes per array type), IL‐8 up‐regulation was the most common gene expression change noted, occurring in over 70% of the tumours analysed. In contrast, our real‐time PCR data shows that another NF‐B controlled gene, I‐κB, showed down‐regulation of expression in dysplastic tissue in particular (by a factor of 2‐fold), suggesting that transcriptional repression of I‐κB may contribute to overall NF‐κB activation. In their study, Abdel‐Latif et al also showed loss of I‐κB protein in tumours and suggested enhanced protein degradation, leading to enhanced NF‐κB activation. We now suggest that I‐κB down‐regulation may be important in leading to constitutively active NF‐κB. It has previously been suggested that NF‐κB has two phases of activation. The first phase, responding to new activation signals, causes I‐κB protein degradation via IKK phosphorylation; the second phase, responding to continuous NF‐κB activating signals, leads to I‐κB down‐regulation, leaving NF‐κB in a constitutively active form.19
Take‐home messages
Immunohistochemistry (IHC) for nuclear factor‐κB (NF‐κB) with an antibody against the active form of p65, is a simple and cost effective tool for monitoring NF‐κB activity, which is amenable to high‐throughput screening.
NF‐κB activity, as measured by IHC, increases during the metaplasia to adenocarcinoma sequence in Barrett's oesophagus.
Interleukin‐8 (IL‐8) gene and protein expression increases in parallel to NF‐κB activity; IL‐8 gene up‐regulation is the most common gene expression change noted here using membrane arrays.
I‐κB is down‐regulated during the histological sequence and this may cause constitutive NF‐κB activity.
In conclusion, we show here that immunohistochemistry for NF‐κB activity (using an antibody for the active form of p65) is a potentially useful screening tool for patients with Barrett's oesophagus. Coupled to IL‐8 immunohistochemistry, this approach may be useful in predicting cancer progression in pre‐malignant Barrett's patients.
Abbreviations
IHC - immunohistochemistry
IL - interleukin
NF‐κB - nuclear factor‐κB
NLS - nuclear localisation sequence
Footnotes
Funding: This work was partly funded by the Welsh Office for Research and Development (Grant WS01/1/009).
Competing interests: None.
References
- 1.Cameron A J, Lomboy C T. Barrett's esophagus: age, prevalence and extent of columnar epithelium. Gastroenterology 19921031241–1245. [DOI] [PubMed] [Google Scholar]
- 2.Spechler S J, Zeroogian J M, Antonioli D A.et al Prevalence of metaplasia at the gastro‐esophageal junction. Lancet 19943441533–1536. [DOI] [PubMed] [Google Scholar]
- 3.Cameron A J, Zinsmeister A R, Ballard D J.et al Prevalence of columnar‐lined (Barrett) esophagus—comparison of population‐based clinical and autopsy findings. Gastroenterology 199099918–922. [DOI] [PubMed] [Google Scholar]
- 4.Jankowski J A, Harrison R F, Perry I.et al Barrett's metaplasia. Lancet 20003562079–2085. [DOI] [PubMed] [Google Scholar]
- 5.Wild C P, Hardie L J. Reflux, Barrett's oesophagus and adenocarcinoma: burning questions. Nat Rev Cancer 20033676–684. [DOI] [PubMed] [Google Scholar]
- 6.Jankowski J A Z, Provenzale D, Moayyedi P. Esophageal adenocarcinoma arising from Barrett's metaplasia has regional variations in the west. Gastroenterology 2002122588–590. [DOI] [PubMed] [Google Scholar]
- 7.Pera M, Cameron A J, Trastek V F.et al Increasing incidence of adenocarcinoma of the esophagus and esophogastric junction. Gastroenterology 1993104510–513. [DOI] [PubMed] [Google Scholar]
- 8.Devesa S, Blot W, Fraumeni J., Jr Changing patterns in the incidences of esophageal and gastric carcinoma in the United States. Cancer 1998152049–2053. [PubMed] [Google Scholar]
- 9.Lerut T, Coosemans W, De Leyn P.et al Reflections on three field lymphadenectomy in carcinoma of the esophagus and gastroesophageal junction. Hepatogastroenterology 199946717–725. [PubMed] [Google Scholar]
- 10.Jenkins G J S, Doak S H, Parry J M.et al Barrett's oesophagus: the genetic pathways involved in its progression to adenocarcinoma. Br J Surg 200289824–837. [DOI] [PubMed] [Google Scholar]
- 11.Rayet B, Gelinas C. Aberrant rel/nfκβ genes and activity in human cancer. Oncogene 1999186938–6947. [DOI] [PubMed] [Google Scholar]
- 12.Abdel‐Latif M M, O'Riordan J, Windle H J.et al NF‐kB activation in esophageal adenocarcinoma. Relationship to Barrett's metaplasia, survival and response to neoadjuvant chemotherapy. Ann Surg 2004239491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins G J S, Harries K, Doak S H.et al The bile acid deoxycholic acid at neutral pH activates NFKB and causes upregulation of IL‐8. Carcinogenesis 200425317–323. [DOI] [PubMed] [Google Scholar]
- 14.Bar Eli M. Role of interleukin‐8 in tumor growth and metastasis of human melanoma. Pathobiology 19996712–18. [DOI] [PubMed] [Google Scholar]
- 15.Fitzgerald R C, Abdalla S, Onwuegbusi B A.et al Inflammatory gradient in Barrett's oesophagus: implications for disease complications. Gut 200251316–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Konturek P C, Nikiforuk A, Kania J.et al Activation of NFKB represents the central event in the neoplastic progression associated with Barrett's esophagus: a possible link to the inflammation and over‐expression of cox‐2, ppargamma and growth factors. Dig Dis Sci 2004491075–1083. [DOI] [PubMed] [Google Scholar]
- 17.O'Riordan J M, AbdelLatif M M, Ravi N.et al Proinflammatory cytokine and nuclear factor kappa B expression along the inflammation‐metaplasia‐dysplasia‐adenocarcinoma sequence in the esophagus. Am J Gastroenterol 20051001257–1264. [DOI] [PubMed] [Google Scholar]
- 18.Lessard L, Begin L R, Gleave M E.et al Nuclear localisation of the nuclear factor‐kappaB transcription factors in prostate cancer: an immunohistochemical study. Br J Cancer 2005931019–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ladner K J, Caliguri M A, Guttridge D C. Tumor necrosis factor‐regulated biphasic activation of NF‐kappa B is required for cytokine‐induced loss of skeletal muscle gene products. J Biol Chem 20032782294–2303. [DOI] [PubMed] [Google Scholar]