Abstract
Background
Long term ulcerative colitis (UC) increases the risk of colorectal cancer (CRC). DNA aneuploidy is a common feature of both dysplastic and non‐dysplastic colonic epithelia from patients with longstanding UC, and is regarded as an early sign of possible malignant transformation. The spindle proteins Aurora A, BUB1B and Mad2 have been implicated as contributors to aneuploidy and carcinogenesis.
Aims
To investigate the role of these spindle proteins in relation to DNA aneuploidy and during the progressive morphological changes in ulcerative colitis associated colorectal cancer (UCCRC).
Methods
Tissue microarrays were made from 31 colectomy specimens from patients with longstanding UC. Expression of Aurora A, BUB1B and Mad2 was investigated by immunohistochemistry and their relation to ploidy status, mucosal morphology and Ki67 levels was explored.
Results
Expression of Aurora A and BUB1B was significantly associated with the progressive morphological changes of UCCRC. In the progression from non‐dysplastic to dysplastic mucosa, Aurora A expression decreased while BUB1B expression increased. There was an increasing incidence of aneuploidy with progression towards cancer; expression of all spindle proteins was associated with the level of Ki67 but not with aneuploidy.
Conclusion
Due to the significant differences in Aurora A and BUB1B expression in dysplastic compared non‐dysplastic mucosa, these proteins may serve as putative biological markers for the progressive morphological changes in UC associated carcinogenesis. The close relationship to Ki67 levels reflect that spindle proteins are expressed in tissues with a high proliferative rate; a role for these proteins in the development of aneuploidy was not found.
Ulcerative colitis (UC) is a chronic disease which is associated with an increased risk of developing colorectal cancer (CRC). The overall prevalence of CRC in patients with UC is 3.7%,1 and the risk of cancer increases with duration of the disease.2 UC associated colorectal cancer (UCCRC) is the cause of death in every sixth patient with UC.3 Surveillance colonoscopy with biopsies is the current screening method to detect dysplastic lesions in patients with UC. However, mucosal dysplasia as a marker for cancer has several implications,4 and there exist conflicting reports concerning the significance of dysplasia as a precancerous lesion.5,6 Furthermore, surveillance colonoscopy has a poor success rate in preventing cancer in patients with UC,7 and current guidelines on CRC screening state that there is no evidence that endoscopic surveillance reduces cancer mortality in patients with UC.8 Obviously there is a need for improved screening methods and there is an ongoing search for biological markers to detect patients with UC at greatest risk of developing cancer. One such marker is DNA aneuploidy that is present in more than half of colorectal adenocarcinomas developing in UC,9 and aneuploidy has been shown to precede mucosal dysplasia.10,11,12,13 A compromised spindle checkpoint, the primary mechanism to ensure that two daughter cells receive the same amount of DNA after mitosis, has been suggested to contribute to aneuploidy and carcinogenesis.14 Mad2 (mitosis arrest‐deficient 2) and BUB1B (budding uninhibited by benzimidazole) are vital parts of this checkpoint, and together they control the separation of sister chromatids. The Aurora family of threonine kinases, and especially Aurora A (STK6/15, ARK1, Aurora 2), has received attention for its role in the development of aneuploidy,15 and has been shown to induce centrosome amplification and aneuploidy in malignant human cell lines.16 Hence, in this study we wanted to investigate the role of Aurora A, BUB1B and Mad2 proteins in the development of aneuploidy and during the progressive morphological stages of UC associated carcinogenesis.
Methods
Colectomy specimens, histopathology
We examined the colectomy specimens resected from 31 patients with longstanding UC in the period 1985–94 at the Department of Surgery, Rikshospitalet, University of Oslo. Tissue specimens from each bowel were harvested from eight consecutive locations from the ascending colon (I–III), transverse colon (IV–V) and descending colon (VI–VIII). Specimens were fixed in 70% ethanol and embedded in paraffin. Tissue sections were cut at 4 μm thickness, stained with H&E and evaluated independently by two experienced pathologists (OPFC, SNA) according to Riddell et al.17 The best areas with retained mucosal architecture and representative morphological changes were marked for tissue microarray core sampling. Ten normal mucosal samples, taken from the resection margins of sporadic colon cancer colectomy specimens, were used as controls.
Immunohistochemistry
Tissue microarrays were made using a Beecher tissue microarrayer. Core size was 0.6 mm, and at least two tissue cores from each mucosal region of interest were sampled. After being exposed to 0.5% H2O2 solution followed by antigen retrieval (Tris‐EDTA buffer at pH 9.0), sections were incubated for one hour at room temperature with the primary antibodies (Aurora A, 1:50 dilution, monoclonal antibody, Novocastra NCL‐L‐AK2, Newcastle, UK; BUB1B, 1:50 dilution, polyclonal antibody, BD Transduction 612503, Franklin Lakes, NY, USA; Mad2, 1:200 dilution, polyclonal antibody; Immuquest IQ239, Cleveland, UK; Ki67, 1:100 dilution, monoclonal antibody, gift from Dr J Gerdes, Germany). After incubation with horseradish peroxidase‐labelled polymer conjugated secondary antibody (EnVision, DakoCytomation, Via Real Carpintera), DAB+ was applied onto the slides for seven minutes, rinsed in distilled water and finally counterstained with haematoxylin and mounted. Fresh fixed tonsillary tissue was used as positive control; the primary antibody was substituted by IgG‐matched immune serum as a negative control. Details concerning antibody specificity and immunohistochemical analysis of spindle proteins has been described by us previously.17a
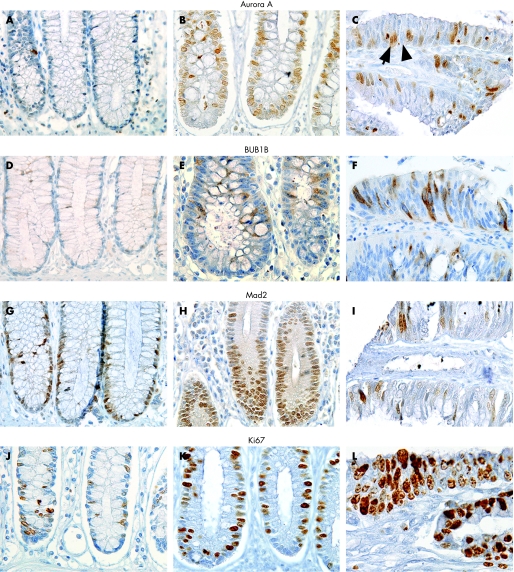
One possible limitation using tissue microarrays compared to conventional tissue sections is the representativity of the tissue obtained by the needle cores. This is especially important when studying features that are unevenly distributed along architectural structures as colonic crypts. To avoid any misinterpretation, each tissue sample was represented by cores including whole crypts, or at least one core from the lower and one core from the upper part. Cores from mucosal samples that did not display these criteria were excluded. Two tissue cores from each sample represent the antigen expression of the whole tissue with greater than 95% accuracy.18 An average of 300 randomly selected, mucosal epithelial cells were counted for each tissue sample. Staining intensity for each protein was homogeneous and cells were evaluated as either positive (arrow) or negative (arrowhead, fig 1c). The number of positive cells among the 300 cells was registered and the resulting percentage calculated; this level indicates protein expression in this article.
Figure 1 Immunohistochemical analyses with antibodies against Aurora A (A–C), BUB1B (D–F), Mad2 (G–I) and Ki67 (J–L) in normal mucosa (left column), non‐dysplastic mucosa of patients with longstanding ulcerative colitis (middle column) and cancer (right column). Original magnification ×480.
DNA ploidy analysis
Mucosa adjacent to that used for histopathological examination was subjected to mechanical disaggregation in phosphate buffered saline (PBS) by a scalpel. The resulting cell suspensions were filtered through 70 μm nylon mesh and centrifuged at 1500 rpm. The pellets were resuspended in 1 ml of PBS; the cells were disaggregated by pipetting and fixed in 70% ice‐cold ethanol and stored at 4°C. Cells were prepared for DNA content analysis using the procedure of Vindeløv et al.19 A FACStar Plus flow cytometer equipped with 488 nm argon ion laser (BDIS, Spectra Physics, Mountain View, CA, USA) was used to analyse cell samples for DNA content based on propidium iodide fluorescence emission. A case report including results of the ploidy analysis from one of the patients in this study has been published.20
Statistical analysis
The relationship between protein expression and histopathological features was evaluated by non‐parametric methods (Mann–Whitney (MW) test, median test and Spearman's rho correlation) and the general linear mixed model (GLMM). The GLMM can be viewed as an extension of a univariate logistic regression where the fact that individuals contribute with several observations (ie, biopsy samples) is adjusted for. A p‐value of ⩽0.05 was considered statistically significant. Analyses were performed using SPSS V.12.0. (Chicago, IL, USA); GLMM was done using R software (http://www.r‐project.org).
Investigations were in accordance with the Declaration of Helsinki and were approved by the regional ethical committee for scientific studies on human tissues (S‐060629).
Results
Eight of the 31 patients had cancer (25%) in one or more locations of the bowel, with the following pTNM status: three patients with T3N2M0, two patients with T1N0M0, one with T3N1Mx and one with T2N0Mx. One patient underwent a palliative resection (T4NxMx). Furthermore, 14 patients had dysplastic areas (45%), 12 patients had areas indefinite for dysplasia (39%) and 30 patients (97%) had colonic areas free of dysplasia (table 1). A total of 258 paraffin embedded tissue blocks were available for immunohistochemical analysis. Nine blocks were excluded due to insufficient tissue material, thus 249 tissue blocks were available for core sampling. On the basis of the histopathological evaluation of the tissue core samples, they were grouped according to mucosal morphology (table 1).
Table 1 Overview of morphological classification, ploidy and immunohistochemical analyses of tissue samples gathered from 31 patients with longstanding UC.
| Diagnosis | Patients | Samples | Samples for ploidy analysis | Aneuploid samples | *Protein expression (%) | |||
|---|---|---|---|---|---|---|---|---|
| Aurora A | BUB1B | Mad2 | Ki67 | |||||
| Normal | 10 | 10 | 0 | – | 13 | 4 | 19 | 14 |
| No dysplasia | 30 | 165 | 159 | 30 (19%) | 40 | 7 | 43 | 27 |
| Ind. dysplasia | 12 | 30 | 29 | 8 (28%) | 31 | 10 | 46 | 27 |
| Dysplasia | 14 | 33 | 32 | 9 (28%) | 24 | 10 | 41 | 28 |
| Cancer | 8 | 11 | 11 | 6 (55%) | 45 | 12 | 32 | 25 |
| Total | ‡ | 249 | 231 | 53 (23%) | ||||
| p‐Value† | <0.001 | <0.001 | 0.8 | 0.18 | ||||
*Reflects the mean % of positive epithelial cells for each sample.
†p‐Value describing the difference in protein expression in dysplastic versus non‐dyplastic mucosa.
‡Total 31 patients, 19 patients had ⩾2 morphological diagnoses in their colons.
Of the 249 core samples, 165 were negative for dysplasia, 30 were indefinite for dysplasia, 27 showed low and 6 samples showed high grade dysplasia, 11 were cancers and 10 were normal mucosal controls. Due to the low number of samples in the high grade dysplasia group (n = 6), the groups with low and high grade dysplasia were considered as one entity in the statistical analysis (table 1).
Eighteen of the 31 analysed bowels harboured one or more locations with DNA aneuploidy. In the normal controls and eight tissue samples from UC patients there was not enough tissue available for ploidy analysis. However, DNA ploidy analysis by flow cytometry has been performed on 10 other normal mucosal samples taken from the resection margins of colon cancer colectomy specimens; none of these samples were aneuploid (unpublished data). Of 231 samples analysed, 53 (23%) showed DNA aneuploidy, with an increasing incidence of aneuploidy with progression towards cancer (p = 0.04, median test).
Antibodies against Aurora A and Mad2 were localised primarily to the nucleus, and such staining pattern was considered specific (fig 1). Additional staining of the cytoplasm was seen in mitotic cells for both proteins. The antibody against BUB1B showed this protein to be localised to the cytoplasm which was considered specific (fig 1). Additional nuclear staining was seen in mitotic cells; a detailed description of the subcellular localisation of spindle proteins assessed by immunohistochemistry has been described by us previously.17a
Protein expression: relationship to mucosal morphology and DNA ploidy
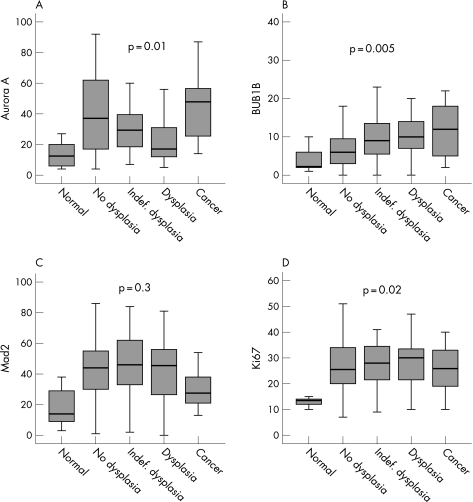
The expression levels of all proteins were considerably higher in the non‐dysplastic mucosal samples from UC patients compared to normal mucosa from patients without UC (Aurora A, Mad2 and Ki67, p<0.001; BUB1B, p = 0.08, MW). Furthermore, higher levels of BUB1B were found in non‐dysplastic areas of patients with cancer compared with non‐dysplastic areas of patients without cancer (p = 0.02, MW). There was also a strong tendency that Mad2 expression in non‐dysplastic mucosa was higher in cancer patients (p = 0.06, MW), while Aurora A expression showed no difference (p = 0.8, MW). During the progressive morphological steps towards cancer, Aurora A levels showed a biphasic expression curve (p = 0.01, fig 2A, median test), while BUB1B and Ki67 levels increased stepwise towards cancer (BUB1B, p = 0.005, fig 2B; Ki67, p = 0.02, fig 2D, median test). Although Mad2 did not show a similar significant expression pattern towards cancer (p = 0.3, median test), there was an obvious drop in cancerous tissue (fig 2C).
Figure 2 Protein expression of Aurora A (A), BUB1B (B), Mad2 (C) and Ki67 (D) in mucosal samples from patients with longstanding ulcerative colitis. Numbers on the y‐axis represent protein expression (percentage of positive cells).
When comparing protein levels in non‐dysplastic versus dysplasia mucosa, Aurora A expression dropped significantly towards dysplasia (p<0.001, MW), BUB1B expression increased (p<0.001, MW) while the levels of Mad2 and Ki67 did not change (p = 0.8 and p = 0.18 respectively, MW). The expression of all proteins were correlated with the level of Ki67 (p<0.001 for all, correlation coefficients (CC) 0.23–0.35). Aurora A expression was correlated with Mad2 levels (p = 0.001, CC = 0.25), as was BUB1B expression (p<0.001, CC = 0.33). However, no correlation was found between Aurora A and BUB1B expression (p = 0.8). There was an increasing incidence of aneuploidy with progression towards cancer (p = 0.04, median test), but no association between protein expression and DNA ploidy was revealed (Aurora A, p = 0.3; BUB1B, p = 0.4; Mad2, p = 0.2; MW). Bowel localisation (I–VIII) did not influence the degree of protein expression (Aurora A, p = 0.7; BUB1B, p = 0.6; Mad2, p = 0.09; median test).
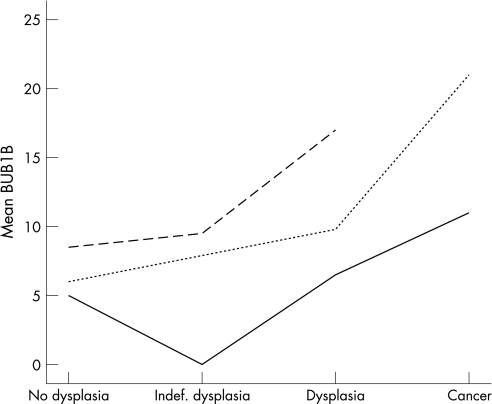
Although the level of protein expression varied between patients, resulting in the wide box‐plots seen in fig 2, the tendency, for example, for BUB1B expression to increase in dysplastic and cancerous mucosa was consistent when the expression levels of individual patients were compared (fig 3). However, these interpatient differences in the level of protein expression, that possibly could mask their general biological appearance, can be adjusted for by using a GLMM. GLMM accounts for the fact that each patient contributes with several samples in the collection of data, and was applied to reanalyse the difference in protein expression in dysplastic versus non‐dysplastic mucosa. Contrary to the Mann–Whitney test, a significant increase in Ki67 levels was revealed in dysplastic compared to non‐dysplastic mucosa (p = 0.01 with GLMM, p = 0.18 with MW). For the other proteins no significant changes were revealed, and GLMM did not alter the conclusions of the ploidy analysis.
Figure 3 The different mean levels of BUB1B protein expression in three patients with longstanding ulcerative colitis. The spread between patients is obvious, but the tendency of increased BUB1B expression in the dysplasia–carcinoma sequence is clearly shown.
Discussion
The primary goal of our investigations was to study the expression of Aurora A and spindle checkpoint proteins BUB1B and Mad2 and their associations with the development of aneuploidy and dysplasia in UCCRC. Chromosomal alterations are found in 85% of UCCRC,21 the remaining 15% showing near diploid DNA content with mainly errors in the DNA mismatch repair mechanisms. Chromosomal instability precedes dysplasia and cancer in UC,10,22 and based on these results DNA aneuploidy could serve as a potential marker for cancer progression. It is reasonable to speculate that the ongoing inflammation with repair of damaged and regenerating epithelial cells in UC increases the workload of the spindle checkpoint machinery of the epithelium. In our study, Aurora A, BUB1B and Mad2 were highly expressed even in non‐dysplastic mucosa from UC patients compared to normal controls, reflecting this up‐regulated checkpoint function. Furthermore, BUB1B and Mad2 expression were higher in non‐dysplastic mucosa of colons from cancer patients compared to colons without cancer development, reflecting a broader “field change” of increased expression in the entire colon of the cancer patients.23 Aberrant Aurora A expression is reported to cause chromosome missegregation and aneuploidy by several mechanisms. In vitro studies by Anand et al24 show that raised levels of Aurora A override an activated spindle checkpoint, allowing cells to enter anaphase despite a defective spindle formation. This leads to division failure, multinucleation and polyploidisation. Furthermore, its overexpression induces multiple centrosomes forming multipolar mitotic spindles,25 and is also reported to activate telomerase, resulting in “sticky‐end chromosomes”26 and aneuploidy. Several studies indicate a role for BUB1B in the development of aneuploidy,14,27,28,29 and in mammary epithelial cells increased Mad2 expression resulted in chromosome missegregation and aneuploidy.30 Based on these data, we anticipated finding a relationship between DNA ploidy and spindle protein expression, but such an association was not found. However, other reports are in agreement with our findings. Studies of Barrett's oesophagus, in which aneuploidy is a common feature,31 failed to show any relationship between aneuploidy and Mad2 or BUB1B gene expression. In testicular germ cell tumours, centrosome amplification and aneuploidy were unrelated to Aurora A expression.32 However, one should exercise some caution in the interpretation of the relationship between protein expression and ploidy data, since the immunohistochemical analyses was not performed on the exact same cells utilised for ploidy analysis.
In view of the ongoing search for biological markers to improve the methods of UCCRC screening, our findings of significant differences in the expression levels of Aurora A and BUB1B in dysplastic versus non‐dysplastic mucosal lesions, respectively, deserve some attention. BUB1B expression increased from non‐dysplastic to dysplastic lesions, while Aurora A decreased. This inverse relationship could serve as an additional marker in the evaluation of mucosal biopsy specimens from UC patients with extensive inflammation and ulceration, which is known to complicate the interpretation of morphological changes.33 A previous study of Aurora A expression in the adenoma–carcinoma sequence of sporadic colorectal cancer,34 showed an increased expression of Aurora A in adenomas compared to normal mucosa. Furthermore, the highest expression level of Aurora A was reported in cancerous lesions, consistent with our findings. In contrast, reduced Aurora A levels were associated with progression from in situ to invasive cancer in breast cancer,35 and a similar pattern of decreased Aurora A expression was seen in the progression from non‐invasive to invasive ovarian cancer.36 Together these studies show some inconsistency of Aurora A expression in the progressive steps towards cancer. Even though there are biological similarities in the stages of malignant transformation irrespective of cancer tissue,37 the expression of Aurora A and its impact on tumour progression appear to be different among various cancer types.
BUB1B and Mad2 proteins are closely related in the spindle checkpoint.38,39,40 The close relationship between BUB1B and Mad2 expression in our study confirms this. However, the levels of protein expression vary considerably between Mad2 and BUB1B in the same morphological group (fig 2). This may reflect the alternative functions of these proteins that are unrelated to the spindle checkpoint. Mad2 is reported to interact with oestrogen receptors41 and nuclear pore complexes possibly involved in cell cycle regulation and nuclear transport,42 whereas BUB1B is involved in the apoptosis of polyploid cells43 and in the cross‐talk between the spindle and DNA damage checkpoints.44
The processing of biomedical data, in which several samples were gathered from one individual, requires a sophisticated statistical tool. In this context, we want to highlight GLMM, being an advanced, regressional analysis that adjusts for the fact that multiple samples were taken from one patient. In our material there were obvious interpatient differences in the level of protein expression (fig 3) that could possibly mask associations between protein expression and morphology. When reanalysing the data with GLMM, a significant increase in the Ki67 level was revealed in dysplastic compared to non‐dysplastic mucosa (p = 0.01 with GLMM compared to p = 0.18 with the MW test). However, GLMM has some limitations because it requires a defined relationship between the dependent variable (in this case diagnosis) and covariates (in this case protein expression). Furthermore, GLMM cannot analyse dependent variables with more than two categories making GLMM less suitable for analyses including all the progressive, morphological changes characteristic for UCCRC. We conclude that a combination of the standardised, non‐parametric methods together with GLMM is the most comprehensive way to analyse these types of data.
To our knowledge, this is the first study describing the expression of Aurora A and the spindle checkpoint proteins BUB1B and Mad2 in UCCRC. All spindle proteins were associated with Ki67 levels, and expression of levels of Aurora A and BUB1B could serve as biological markers for the progressive morphological changes of UC associated carcinogenesis due to their aberrant expression in dysplastic mucosa. Furthermore, there was an increase in aneuploidy during the progression towards cancer, but a relationship between protein expression and DNA ploidy was not found. More extensive studies are necessary to fully understand the function of these proteins and their role in the process of chromosome missegregation and aneuploidy.
Take‐home messages
Expression of spindle proteins Aurora A and BUB1B appears to be associated with the progressive morphological changes of the inflammation–dysplasia–carcinoma sequence of ulcerative colitis (UC) associated carcinogenesis.
Increased expression of BUB1B and decreased expression of Aurora A is associated with dysplastic mucosal changes of patients with longstanding UC.
The expression of all spindle proteins is associated with the level of Ki67.
The incidence of aneuploidy increased towards cancer, but a correlation between protein levels and aneuploidy was not found.
Acknowledgements
The authors thank laboratory scientist Sara Moberg, Institute of Forensic Medicine, Rikshospitalet‐Radiumhospitalet Medical Center, for excellent technical support.
Abbreviations
CC - correlation coefficient
CRC - colorectal cancer
GLMM - general linear mixed model
MW - Mann–Whitney
UC - ulcerative colitis
UCCRC - ulcerative colitis associated colorectal cancer
Footnotes
Funding: The sole sponsor for this work was the Norwegian Cancer Society, which had no role in the collection, analyses or interpretation of the data or in writing the report.
Competing interests: None.
References
- 1.MacDougall I P. The cancer risk in ulcerative colitis. Lancet 196419655–658. [DOI] [PubMed] [Google Scholar]
- 2.Eaden J A, Abrams K R, Mayberry J F. The risk of colorectal cancer in ulcerative colitis: a meta‐analysis. Gut 200148526–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gyde S, Prior P, Dew M J.et al Mortality in ulcerative colitis. Gastroenterology 19828336–43. [PubMed] [Google Scholar]
- 4.Chambers W M, Warren B F, Jewell D P.et al Cancer surveillance in ulcerative colitis. Br J Surg 200592928–936. [DOI] [PubMed] [Google Scholar]
- 5.Ullman T, Croog V, Harpaz N.et al Progression of flat low‐grade dysplasia to advanced neoplasia in patients with ulcerative colitis. Gastroenterology 20031251311–1319. [DOI] [PubMed] [Google Scholar]
- 6.Befrits R, Ljung T, Jaramillo E.et al Low‐grade dysplasia in extensive, long‐standing inflammatory bowel disease: a follow‐up study. Dis Colon Rectum 2002145615–620. [DOI] [PubMed] [Google Scholar]
- 7.Jonsson B, Ahsgren L, Andersson L O.et al Colorectal cancer surveillance in patients with ulcerative colitis. Br J Surg 199481689–691. [DOI] [PubMed] [Google Scholar]
- 8.Winawer S J, Fletcher R H, Miller L.et al Colorectal cancer screening: clinical guidelines and rationale. Gastroenterology 1997112594–642. [DOI] [PubMed] [Google Scholar]
- 9.Meling G I, Clausen O P, Bergan A.et al Flow cytometric DNA ploidy pattern in dysplastic mucosa, and in primary and metastatic carcinomas in patients with longstanding ulcerative colitis. Br J Cancer 199164339–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rubin C E, Haggitt R C, Burmer G C.et al DNA aneuploidy in colonic biopsies predicts future development of dysplasia in ulcerative colitis. Gastroenterology 19921031611–1620. [DOI] [PubMed] [Google Scholar]
- 11.Itzkowitz S. Colon carcinogenesis in inflammatory bowel disease: applying molecular genetics to clinical practice. J Clin Gastroenterol 200336S70–S74. [DOI] [PubMed] [Google Scholar]
- 12.Lindberg J O, Stenling R B, Rutegard J N. DNA aneuploidy as a marker of premalignancy in surveillance of patients with ulcerative colitis. Br J Surg 199986947–950. [DOI] [PubMed] [Google Scholar]
- 13.Lofberg R, Lindquist K, Veress B.et al Highly malignant carcinoma in chronic ulcerative colitis without preceding dysplasia or DNA aneuploidy. Report of a case. Dis Colon Rectum 19923582–86. [DOI] [PubMed] [Google Scholar]
- 14.Kops G J, Foltz D R, Cleveland D W. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci USA 20041018699–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol 200515241–250. [DOI] [PubMed] [Google Scholar]
- 16.Zhou H, Kuang J, Zhong L.et al Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet 199820189–193. [DOI] [PubMed] [Google Scholar]
- 17.Riddell R H, Goldman H, Ransohoff D F.et al Dysplasia in inflammatory bowel disease: standardized classification with provisional clinical applications. Hum Pathol 198314931–968. [DOI] [PubMed] [Google Scholar]
- 17a.Burum‐Auensen E, De Angelis P M, Schjolberg A R.et al Subcellular localization of the spindle proteins Aurora A. Mad2, and BUBRI assessed by immunohistochemistry. J Histochem Cytochem 200755477–486. [DOI] [PubMed] [Google Scholar]
- 18.Camp R L, Charette L A, Rimm D L. Validation of tissue microarray technology in breast carcinoma. Lab Invest 2000801943–1949. [DOI] [PubMed] [Google Scholar]
- 19.Vindeløv L L, Christensen I J, Nissen N I. A detergent‐trypsin method for the preparation of nuclei for flow cytometric DNA analysis. Cytometry 19833323–327. [DOI] [PubMed] [Google Scholar]
- 20.Clausen O P, Andersen S N, Stromkjaer H.et al A strategy combining flow sorting and comparative genomic hybridization for studying genetic aberrations at different stages of colorectal tumorigenesis in ulcerative colitis. Cytometry 20014346–54. [DOI] [PubMed] [Google Scholar]
- 21.Willenbucher R F, Aust D E, Chang C G.et al Genomic instability is an early event during the progression pathway of ulcerative‐colitis‐related neoplasia. Am J Pathol 19991541825–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabinovitch P S, Dziadon S, Brentnall T A.et al Pancolonic chromosomal instability precedes dysplasia and cancer in ulcerative colitis. Cancer Res 1999595148–5153. [PubMed] [Google Scholar]
- 23.Itzkowitz S H, Yio X. Inflammation and cancer IV. Colorectal cancer in inflammatory bowel disease: the role of inflammation. Am J Physiol Gastrointest Liver Physiol 2004287G7–17. [DOI] [PubMed] [Google Scholar]
- 24.Anand S, Penrhyn‐Lowe S, Venkitaraman A R. AURORA‐A amplification overrides the mitotic spindle assembly checkpoint, inducing resistance to Taxol. Cancer Cell 2003351–62. [DOI] [PubMed] [Google Scholar]
- 25.Warner S L, Bearss D J, Han H.et al Targeting Aurora‐2 kinase in cancer. Mol Cancer Ther 20032589–595. [PubMed] [Google Scholar]
- 26.Yang H, Ou C C, Feldman R I.et al Aurora‐A kinase regulates telomerase activity through c‐Myc in human ovarian and breast epithelial cells. Cancer Res 200464463–467. [DOI] [PubMed] [Google Scholar]
- 27.Cahill D P, Lengauer C, Yu J.et al Mutations of mitotic checkpoint genes in human cancers. Nature 1998392300–303. [DOI] [PubMed] [Google Scholar]
- 28.Musio A, Montagna C, Zambroni D.et al Inhibition of BUB1 results in genomic instability and anchorage‐independent growth of normal human fibroblasts. Cancer Res 2003632855–2863. [PubMed] [Google Scholar]
- 29.Rao C V, Yang Y M, Swamy M V.et al Colonic tumorigenesis in BubR1+/−ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci USA 20051024365–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pati D, Haddad B R, Haegele A.et al Hormone‐induced chromosomal instability in p53‐null mammary epithelium. Cancer Res 2004645608–5616. [DOI] [PubMed] [Google Scholar]
- 31.Doak S H, Jenkins G J, Parry E M.et al Differential expression of the MAD2, BUB1 and HSP27 genes in Barrett's oesophagus—their association with aneuploidy and neoplastic progression. Mutat Res 2004547133–144. [DOI] [PubMed] [Google Scholar]
- 32.Mayer F, Stoop H, Sen S.et al Aneuploidy of human testicular germ cell tumors is associated with amplification of centrosomes. Oncogene 2003223859–3866. [DOI] [PubMed] [Google Scholar]
- 33.Collins R H, Jr, Feldman M, Fordtran J S. Colon cancer, dysplasia, and surveillance in patients with ulcerative colitis. A critical review. N Engl J Med 19873161654–1658. [DOI] [PubMed] [Google Scholar]
- 34.Takahashi T, Futamura M, Yoshimi N.et al Centrosomal kinases, HsAIRK1 and HsAIRK3, are overexpressed in primary colorectal cancers. Jpn J Cancer Res 2000911007–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoque A, Carter J, Xia W.et al Loss of aurora A/STK15/BTAK overexpression correlates with transition of in situ to invasive ductal carcinoma of the breast. Cancer Epidemiol Biomarkers Prev 2003121518–1522. [PubMed] [Google Scholar]
- 36.Gritsko T M, Coppola D, Paciga J E.et al Activation and overexpression of centrosome kinase BTAK/Aurora‐A in human ovarian cancer. Clin Cancer Res 200391420–1426. [PubMed] [Google Scholar]
- 37.Vogelstein B, Kinzler K W. Cancer genes and the pathways they control. Nat Med 200410789–799. [DOI] [PubMed] [Google Scholar]
- 38.Jallepalli P V, Lengauer C. Chromosome segregation and cancer: cutting through the mystery. Nat Rev Cancer 20011109–117. [DOI] [PubMed] [Google Scholar]
- 39.Amon A. The spindle checkpoint. Curr Opin Genet Dev 1999969–75. [DOI] [PubMed] [Google Scholar]
- 40.Sudakin V, Chan G K, Yen T J. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol 2001154925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poelzl G, Kasai Y, Mochizuki N.et al Specific association of estrogen receptor beta with the cell cycle spindle assembly checkpoint protein, MAD2. Proc Natl Acad Sci USA 2000972836–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell M S, Chan G K, Yen T J. Mitotic checkpoint proteins HsMAD1 and HsMAD2 are associated with nuclear pore complexes in interphase. J Cell Sci 2001114953–963. [DOI] [PubMed] [Google Scholar]
- 43.Shin H J, Baek K H, Jeon A H.et al Dual roles of human BubR1, a mitotic checkpoint kinase, in the monitoring of chromosomal instability. Cancer Cell 20034483–497. [DOI] [PubMed] [Google Scholar]
- 44.Fang Y, Liu T, Wang X.et al BubR1 is involved in regulation of DNA damage responses. Oncogene 2006253598–3605. [DOI] [PubMed] [Google Scholar]