Abstract
Objectives
Bile reflux contributes to oesophageal injury and neoplasia. COX‐2 is involved in both inflammation and carcinogenesis; however, the precise mechanisms by which bile acids promote COX‐2 expression in the oesophagus are largely unknown. We analysed the molecular mechanisms that govern bile acid‐mediated expression of COX‐2 in Barrett's oesophagus and oesophageal adenocarcinoma (OA).
Design
The effects of bile acids on COX‐2 expression were analysed in immortalised Barrett's oesophagus and OA cells using immunoblotting and transient transfections. Pharmacological inhibitors, phospho‐specific antibodies, dominant‐negative mutants and siRNA techniques were used to identify relevant signalling pathways. Flow cytometry and reactive oxygen species (ROS) scavengers were used to examine ROS involvement. Immunohistochemistry was performed on oesophageal mucosa obtained from an established rat model of bile reflux.
Results
Unconjugated bile acids potently stimulated COX‐2 expression and induced AKT and ERK1/2 phosphorylation in concert with COX‐2 induction. These findings were mimicked in the in vivo rat model. Dominant‐negative (DN) AKT and LY294002 (PI3K inhibitor) or U0126 (MEK‐1/2 inhibitor) blocked chenodeoxycholic acid (CD) and deoxycholic acid (DC) mediated COX‐2 induction. CD and DC also induced CREB phosphorylation and AP‐1 activity. CREB‐specific siRNA and DN AP‐1 blocked CD and DC‐induced COX‐2 induction. Finally, CD and DC increased intracellular ROS, while ROS scavengers blocked COX‐2 induction and the signalling pathways involved.
Conclusions
Unconjugated bile acids induce CREB and AP‐1‐dependent COX‐2 expression in Barrett's oesophagus and OA through ROS‐mediated activation of PI3K/AKT and ERK1/2. This study enhances our understanding of the molecular mechanisms by which bile acids promote the development of oesophageal adenocarcinoma.
Abundant epidemiological evidence links duodenogastrooesophageal reflux with the development of Barrett's oesophagus and oesophageal adenocarcinoma (OA).1,2,3 Chronic exposure to both acid and bile in gastrooesophageal refluxate promotes damage and inflammation in the oesophageal epithelium. A number of studies have examined the cellular mechanisms by which acid promotes neoplastic transformation.4,5,6 Recent evidence suggests that bile acids, major constituents of the duodenogastrooesophageal reflux, can also promote the development of Barrett's oesophagus and OA. Bile reflux is particularly common in individuals with gastrooesophageal reflux disease who subsequently develop Barrett's oesophagus.7,8 Barrett's oesophagus also develops in patients who have undergone partial or total gastrectomy: situations in which bile reflux is common.9 Development of Barrett's oesophagus and subsequently OA occurs in a rat model that uses oesophagojejunostomy to bypass exposure to acid reflux from the stomach.10 In this model, enterooesophageal reflux produces OA in 48% of rats in the absence of exposure to exogenous carcinogens.11 The precise mechanisms by which duodenal reflux cause oesophageal injury and predisposes to OA are uncertain. There is considerable evidence, however, that bile acids contribute to this process. Bile acids can be both potent tumour promoters and carcinogens that mediate activator protein (AP)‐1 activation through extracellular signal‐regulated kinase (ERK)1/2 and protein kinase C (PKC) dependent signalling pathways,12,13,14 and induce genetic instability through DNA damage.15,16,17,18
A large body of knowledge has accumulated regarding the molecular alterations associated with bile reflux in the oesophagus. Experimental evidence suggests that cyclooxygenase‐2 (COX‐2) is involved in the development of Barrett's oesophagus and OA. COX‐2 is frequently overexpressed in OA cells and tissues.19,20 COX‐2 expression also increases progressively in the evolution from Barrett's oesophagus to low‐grade and high‐grade dysplasia and to OA.21 Several studies have demonstrated that bile acids increase COX‐2 expression in human Barrett's oesophagus and OA tissues and in a preclinical model of enterooesophageal reflux.2,22,23,24 Bile acid‐mediated induction of COX‐2 has been reported to be blocked by inhibitors of PKC activity;23 however, the precise mechanisms by which bile acids enhance COX‐2 expression are largely unknown. It is also unclear which bile acids in the refluxate contribute to COX‐2 induction. The present study was designed to investigate the detailed molecular mechanisms by which bile acids regulate COX‐2 expression in the oesophagus.
Bile acids are known to increase intracellular reactive oxygen species (ROS). The cellular effects triggered by bile acids, including cell proliferation, apoptosis and gene regulation, depend on the production of ROS.25,26,27 In rat hepatocytes, bile acids deoxycholic acid (DC) and taurochenodeoxycholic acid (TCDC) increase ROS production, activate receptor tyrosine kinases and stimulate signalling through ERK1/2 and AKT pathways.28 Bile acid‐mediated COX‐2 induction through ROS production in the oesophagus has not been reported.
CREB is a 43 kD basic leucine‐zipper transcription factor that regulates gene expression through the cAMP‐dependent or independent signal transduction pathways.29,30 CREB binds to a cAMP response element (CRE) consensus sequence in the promoters of target genes. Indirect evidence in the literature suggest that, in addition to AP‐1, CREB is an important transcription factor that regulates COX‐2 expression.16,23 The 5′‐flanking promoter region of COX‐2 contains a CRE element31,32 and CREB mediates COX‐2 transcriptional regulation by proteasome inhibitors33 and clostridium difficile toxin A.34
In this study, we demonstrate that unconjugated dihydroxy bile acids, chenodeoxycholic acid (CD) and DC are potent stimulators of COX‐2 induction in Barrett's oesophagus and OA cells, and that these bile acids induce CREB and AP‐1 dependent COX‐2 expression through ROS mediated activation of PI3K/AKT and ERK1/2 pathways.
Materials and methods
Materials
Dulbecco's modified Eagle medium (DMEM) and fetal bovine serum (FBS) (Life Technologies, Inc, New York, USA); LY294002 and U0126 (Calbiochem, San Diego, CA, USA); conjugated and unconjugated bile acids, 2,7‐dichlorodihydrofluorescein diacetate (DCHF‐DA); N‐acetyl‐L‐cysteine (NAC) and sodium formate (Sigma, St Louis, USA); ECL‐chemiluminescence reagents for western blots (Amersham‐Pharmacia, New Jersey, USA); total and phosphorylated ERK1/2 antibodies, total and phosphorylated AKT antibodies, and phosphorylated CREB antibody (Cell Signaling Technology, Beverly, USA); COX‐2 monoclonal antibody (Cayman Chemical, Ann Arbor, USA) and c‐Jun/AP‐1 antibody (Santa Cruz Biotechnology, Inc, Santa Cruz, USA).
Cell culture
Human OA cell lines SEG‐1 and SKGT‐4, derived from well‐differentiated adenocarcinomas arising in Barrett's oesophagus of the distal oesophagus35 were maintained in DMEM supplemented with 10% FBS, 100 units/ml penicillin and 100 μl/ml streptomycin at 37°C in a humidified atmosphere of 95% air and 5% CO2. The h‐TERT‐immortalised BE cell line CP‐A (a gift from Dr Peter Rabinovich, Fred Hutchinson Cancer Center, Seattle, USA) was grown in MCDB‐153 medium supplemented with 5% fetal calf serum, 20 ng/ml epidermal growth factor (Gibco, New York, USA), 140 μg/ml bovine pituitary extract (Sigma, St Louis, USA), 5 μg/ml insulin, 5 μg/ml transferring and 5 ng/ml selenium (Sigma) as described previously.36,37 Treatments with vehicle (0.1% ethanol) or bile acids were performed for 16 hours in reduced serum conditions (0.5% FBS).23,38,39,40 Previous studies have demonstrated maximum stimulation of COX‐2 protein expression by bile acids in cultured SKGT‐4 cells at 12–24 h without affecting cell viability.23 Cellular cytotoxicity was assessed by cell number measurements, trypan blue exclusion analysis and the MTT assay (Promega, Madison, USA). For trypan blue analysis, following treatment with bile acids for 16 h, cells were mixed 1:1 with 0.4% trypan blue and examined for dye exclusion.
Protein isolation and immunoblot analysis
Cells were lysed in buffer containing 30 mM Tris‐HCl (pH 6.8), 150 mM NaCl, 2 mM EDTA, 100 mM NaF, 10 mM sodium pyrophosphate, 2 mM orthovanadate, 1% Triton X‐100, 1% NP‐40, 0.2 mM phenylmethylsulfonyl fluoride and one mini tablet protease inhibitor cocktail (Roche Diagnostics Corp, Indianapolis, USA). Equal amounts of protein were subjected to electrophoresis on 10% Tris‐glycine gels. Western analyses were performed as previously described,36 and immunoreactive bands were visualised by chemiluminescence detection. The relative intensities of non‐saturated signals were quantified by densitometry using a digital imaging system (Alpha Innotech, San Leandro, California, USA).
Plasmids, transient transfection and luciferase reporter assays
The COX‐2 promoter construct (−1432/+59) has been previously described.41 Dominant‐negative AP‐1 (blunted TAM67)42 and the AP‐1 promoter‐luciferase reporter43 were provided by S A Reddy and J Kurie (MD Anderson Cancer Center, Houston, USA). The cDNA plasmid for dominant‐negative mutant of AKT (AKT AAA) was provided by D Koul (UT MD Anderson Cancer Center, Houston, USA).44 Plasmids were prepared using the Genopure Plasmid Midi Kit (Roche, Indianapolis, USA). Beta‐galactosidase expression vector pCH110 (Amersham‐Pharmacia, Arlington Heights, USA) was used to normalise transfection efficiency.
For transient transfection, cells were seeded at 8×105 cells per well in six‐well plates. After overnight culture, cells were transfected with 1 μg of COX‐2 or AP‐1‐luciferase reporter plasmid and 0.2 μg of pCH110 mixed with 3 μl of FuGENE‐6 (Roche Diagnostics Corp, Indianapolis, USA) per manufacturer's protocol. Cells were cultured for an additional 24 h and harvested for luciferase activity (measurement in relative light units normalised to beta‐galactosidase), with a TD‐20/20 luminometer (Turner Designs, Sunnyvale, USA) using the Promega luciferase assay system (Promega Corp, Madison, USA). Values shown represent the mean and standard deviation of at least three independent experiments.
Measurement of ROS generation
A 2,7‐dichlorodihydrofluorescein diacetate (DCHF‐DA) is a non‐polar compound that enters the cell and is cleaved to form DCHF. Trapped DCHF is oxidized by oxygen free radicals to produce fluorescent DCF.34,45 2×105 cells were seeded on six‐well plates 24 h prior to the experiment. A total of 200 μM unconjugated bile acids (CD and DC) or conjugated bile acids (taurocholic (TCA) and glycochenodeoxycholic acid (GCDC)) were added to the cell culture medium for 16 h. Cells were washed three times with cold PBS and incubated with 10 μM DCHF‐DA in serum‐free phenol red‐reduced DMEM at 37°C for 30 min. Cells were then washed with cold PBS three times and scraped from the dishes in 1 ml of cold PBS. The fluorescence intensity of the DCF‐labelled cells was measured by a FACScan flow cytometer (Becton Dickson, San Jose, USA). The mean fluorescent intensity values were calculated using a four‐decade logarithmic scale by the CellQuest (Becton Dickson) software and, for each sample, 1×104 cells were analysed using the green fluorescene emission parameter.
Small interfering RNA (siRNA) synthesis and transfection
Complementary hairpin siRNA oligonucleotides targeting CREB exon 2 and 3 (NM‐004379) and a 19 bp scrambled sequence with 3′‐dT overhangs (negative control for siRNA) were synthesised by Ambion (Austin, Texas, USA). CREB‐siRNA transfections were performed by siPORT™ NeoFX™ (Ambion) per manufacturer's guidelines. After 24 h, cells were transfected with COX‐2 promoter constructs. Twenty four hours later, cells were exposed to 200 μM bile acids. Transfected cells were harvested for measurement of luciferase activity as described above. Values shown represent the mean and standard deviation of at least three independent experiments. To ascertain the specificity of CREB siRNA in silencing CREB gene expression, the cells were also transfected with scrambled siRNA (negative control) and the samples were processed identically as the above.
Rat model of Barrett's oesophagus
Six‐week‐old male Sprague‐Dawley rats (Harlan, Indianapolis, USA) were housed under standard laboratory conditions and allowed to acclimate for 2 weeks before surgical intervention. General anaesthesia was induced by isoflurane (99.9%, 0.5 mL/L volume), followed by maintenance anaesthesia using intramuscular xylazine hydrochloride (12 mg/kg) and ketamine (75 mg/kg). Levrat's oesophagojejunostomy technique was used to induce enterooesophageal reflux as previously described.46 The animal care committee (IACUC) at the Mayo Clinic, Rochester, USA, approved this study.
Rats were euthanised using a combination of CO2 narcosis and intramuscular injection of xylazine hydrochloride (12 mg/kg). The site of anastomosis was identified by locating the polypropylene sutures, freed of adhesions and dissected free of surrounding tissue to the level of the laryngopharynx. The oesophagus was then cut at the level of larynx and 2 mm above the anastomosis site and opened longitudinally. After snap freezing, a representative sample of the oesophagus was stored at −70°C. The remaining oesophagus was fixed in 10% buffered formalin for 24 h and then transferred to 80% ethanol. The oesophagus was longitudinally divided into 6–8 well‐oriented tissue slices, each representing the entire length and full depth of the oesophagus but ⩽1 mm wide. These slices were processed and fixed in paraffin to maintain tissue orientation for histopathological analysis.
Histopathological analysis and immunohistochemistry
Histopathological analysis was carried out on tissue sectioned into 6 μm slices and stained with haematoxylin and eosin. Diagnosis of BE was determined by the presence of columnar metaplasia, including intestinal‐type goblet cells, and confirmed on mucin characteristics, periodic acid‐Schiff and alcian blue (pH 2.5) staining. Immunohistochemistry was performed on formalin‐fixed paraffin‐embedded sections (6 μm) with antibodies against COX‐2 (polyclonal antibody; dilution 1:200; Cayman Chemical, Ann Arbor, USA), phospho‐AKT (rabbit monoclonal antibody; dilution 1:50; Cell Signaling Technology, Boston, USA) and phospho‐CREB (rabbit monoclonal antibody; dilution 1:40; Cell Signaling) after retrieval of antigens using 10 mM sodium citrate buffer as previously described.47
Statistical analysis
Promoter assays are presented in figures as mean ± SD, and represent the results of at least three experiments. Fold reduction in COX‐2 and p‐CREB proteins by specific inhibitors represents the mean values ± SDs of three separate experiments. Significance of differences between groups was judged using a 2‐tailed Student t test. Results were considered statistically significant if the p value was less than 0.05.
Results
Unconjugated dihydroxy bile acids induce COX‐2 expression in EA cells
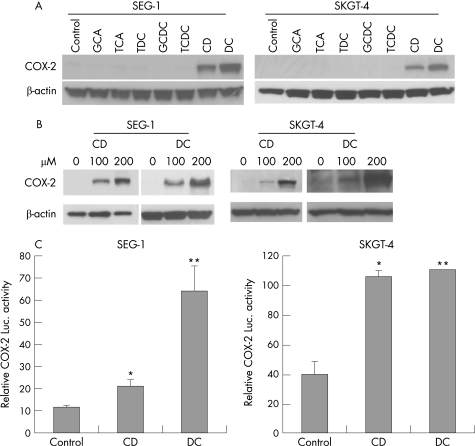
As both conjugated and unconjugated bile acids are present in the gastrooesophageal refluxate, we sought to determine which bile acids affect COX‐2 expression. We exposed SEG‐1 and SKGT‐4 EA cells to various bile acids and their conjugates in vitro. Unconjugated dihydroxy bile acids CD and DC potently induced COX‐2 expression, whereas glyco‐conjugated and tauro‐conjugated bile acids (GCA, TCA, TDC, GCDC and TCDC) had no effect on expression of COX‐2 (fig 1A). The induction of COX‐2 by CD and DC was dose‐dependent (fig 1B).
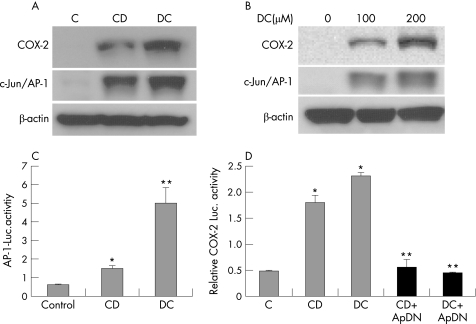
Figure 1 Unconjugated but not conjugated bile acids potently induce cyclooxygenase (COX)‐2 expression in oesophageal adenocarcinoma cells. (A) SEG‐1 and SKGT‐4 cells were treated with 200 μM of conjugated (GCA, TCA, TDC, GCDC and TCDC) and unconjugated (CD and DC) bile acids for 16–24 h. Immunoblots were performed to analyse COX‐2 expression as described in Materials and methods. (B) SEG‐1 and SKGT‐4 cells were stimulated with increasing concentrations of CD and DC and immunoblots were performed as in (A). (C) SEG‐1 and SKGT‐4 cells were cotransfected with 1 μg of human COX‐2 promoter plus 0.2 μg of pSV‐β‐gal and then subsequently treated with 200 μM CD, DC or vehicle (0.1% ethanol). Luciferase reporter activities were measured after 16 h and reported as described in Materials and methods. *p<0.01, ** p<0.001 vs control. CD, chenodeoxycholic acid; DC, deoxycholic acid; GCA, glycocholic acid; GCDC, glycochenodeoxycholic acid; luc, luciferase; TCA, taurodeoxycholic acid; TDC, taurodeoxycholic acid; TCDC, taurochenodeoxycholic acid.
In order to determine whether bile acid‐mediated COX‐2 induction occurs at the transcriptional level, transient transfections were performed in OA cells with human COX‐2 promoter‐luciferase construct. Treatment with CD or DC for 16 h led to 2–6‐fold increase in COX‐2 promoter activity in SEG‐1 cells and 2–3‐fold increase in SKGT‐4 cells (fig 1C). Since COX‐2 induction was highest in SEG‐1 cells, further experiments were performed using this cell system.
PI3K/AKT pathway mediates COX‐2 induction by unconjugated bile acids
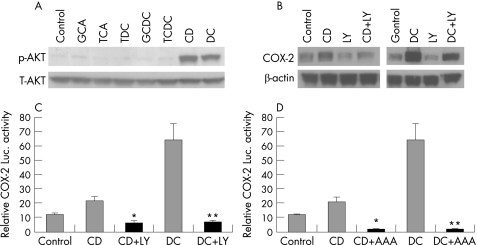
Pharmacological and genetic approaches were next used to evaluate the potential role of the PI3K/AKT signalling pathway in mediating bile acids‐induced COX‐2 expression. In a pattern similar to COX‐2 induction, phosphorylation of AKT was potently induced only by unconjugated bile acids but not by glyco‐conjugated or tauro‐conjugated bile acids, whereas total AKT did not change (fig 2A). LY294002, a PI3K specific inhibitor, blocked CD (3.64±0.89 fold reduction; p = 0.005) or DC (5.09±1.0; p<0.02 fold reduction) mediated COX‐2 expression (fig 2B) and COX‐2 transcription (fig 2C). Co‐expression of dominant‐negative mutant of AKT (AKT AAA) also completely blocked CD or DC mediated COX‐2 transcription (fig 2D).
Figure 2 The PI3K/AKT pathway mediates cyclooxygenase (COX)‐2 induction by CD and DC. (A) SEG‐1 cells were treated with 200 μM of conjugated (GCA, TCA, TDC, GCDC and TCDC) and unconjugated (CD & DC) bile acids for 16–24 h, and immunoblots were performed with phospho‐AKT (S473) and total AKT antibodies as described in Materials and methods. (B) SEG‐1 cells were pre‐incubated with LY294002, a PI3K‐specific inhibitor, for 30 min before stimulating with CD or DC for 16–24 h. COX‐2‐specific immunoblots were performed as described in Materials and methods. LY294002 blocked CD (3.64±0.89 fold reduction; p = 0.005) or DC (5.09±1.0; p<0.02 fold reduction) mediated COX‐2 expression. Results are based on three separate experiments. (C) SEG‐1 cells were cotransfected with 1 μg of human COX‐2 promoter plus 0.2 μg of pSV‐β‐gal, then treated with LY294002 or vehicle, and subsequently challenged with 200 μM CD, DC or vehicle (0.1% ethanol). Luciferase reporter activities were measured after 16 h and reported as described in Materials and methods. *p<0.01 CD+LY vs CD, **p<0.001 DC+LY vs DC. (D) SEG‐1 cells were cotransfected with 1 μg of human COX‐2 promoter, 1 μg of DN AKT (AAA) plus 0.2 μg of pSV‐β‐gal, and then challenged with 200 μM CD, DC or vehicle (0.1% ethanol). Luciferase reporter activities were measured after 16 h and reported as described in Materials and methods. p<0.01 CD+AAA vs CD, **p<0.001 DC+AAA vs DC. CD, chenodeoxycholic acid; DC, deoxycholic acid; GCA, glycocholic acid; GCDC, glycochenodeoxycholic acid; luc, luciferase; TCA, taurodeoxycholic acid; TDC, taurodeoxycholic acid; TCDC, taurochenodeoxycholic acid.
ERK1/2 pathway also mediates COX‐2 induction by unconjugated bile acids
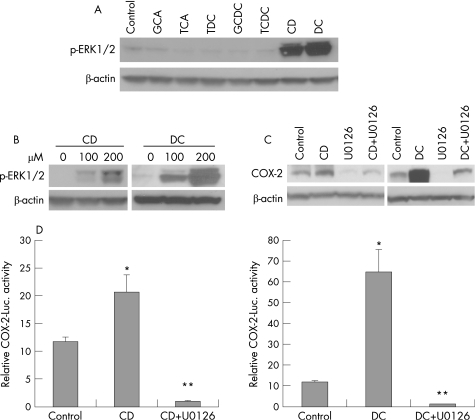
We next evaluated the role of ERK1/2 pathway in mediating bile acids‐induced COX‐2 expression in SEG‐1 cells. In parallel with COX‐2 induction, CD or DC strongly induced dose‐dependent ERK1/2 phosphorylation, while glyco‐conjugated and tauro‐conjugated bile acids had no effect (fig 3A,B). U0126, a MEK‐1/2 inhibitor, almost completely blocked CD (p<0.005) or DC (p<0.001) induced COX‐2 expression (fig 3C) and COX‐2 transcriptional activity (fig 3D).
Figure 3 Cyclooxygenase (COX)‐2 induction is also mediated by ERK1/2. (A) SEG‐1 cells were treated with 200 μM of conjugated (GCA, TCA, TDC, GCDC and TCDC) and unconjugated bile acids (CD and DC) for 16–24 h, and immunoblots were performed with phospho‐ERK1/2 and beta‐actin antibodies as described in Materials and methods. (B) SEG‐1 cells were stimulated with increasing concentrations of CD and DC and immunoblots for phospho‐ERK1/2 were performed as in (A). (C) SEG‐1 cells were pre‐incubated with MEK1/2 specific inhibitor U0126 for 30 min before stimulating with CD or DC for 16–24 h. COX‐2 specific immunoblots were performed as described in Materials and methods. U0126 almost completely blocked CD (p<0.005) or DC (p<0.001) induced COX‐2 expression. p Values are based on data from three separate experiments. (D) SEG‐1 cells were cotransfected with 1 μg of human COX‐2 promoter plus 0.2 μg of pSV‐β‐gal, then treated with U0126 or vehicle, and subsequently challenged with 200 μM CD or vehicle (0.1% ethanol). Luciferase reporter activities were measured after 16 h and reported as described in Materials and methods. (E) SEG‐1 cells were cotransfected with 1 μg of human COX‐2 promoter plus 0.2 μg of pSV‐β‐gal, then treated with U0126 or vehicle, and subsequently challenged with 200 μM DC or vehicle (0.1% ethanol). Luciferase reporter activities were measured after 16 h and reported as described in Materials and methods. *p<0.01 vs control, **p<0.001 vs CD or DC alone. CD, chenodeoxycholic acid; DC, deoxycholic acid; GCA, glycocholic acid; GCDC, glycochenodeoxycholic acid; luc, luciferase; TCA, taurodeoxycholic acid; TDC, taurodeoxycholic acid; TCDC, taurochenodeoxycholic acid.
Role of CREB in the induction of COX‐2 by unconjugated bile acids
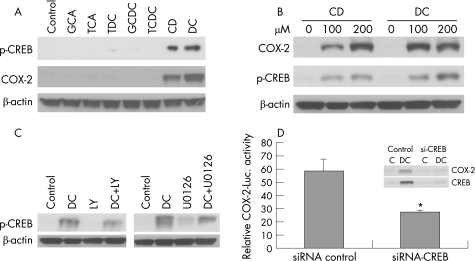
Previous studies have demonstrated that the unconjugated bile acid DC increases CREB DNA binding ability, which controls hepatocyte survival.48 The human COX‐2 promoter contains a cyclic AMP response‐element (CRE; CREB binding site), and CREB is a known downstream target of the ERK1/2 and PI3K/AKT pathways.31,32,49,50 To further determine the potential role of CREB in mediating COX‐2 induction by bile acids, we investigated bile acid‐mediated serine 133 (S133) phosphorylation of CREB, since phosphorylation of this site is essential for CREB‐mediated transcription. Treatment with CD or DC potently increased dose‐dependent CREB S133 phosphorylation, whereas conjugated bile acids had no effects on CREB phosphorylation (fig 4A,B). Next, we determined whether blocking PI3K and ERK1/2 pathways could abrogate bile acid‐induced COX‐2 expression in SEG‐1 cells. Using PI3K and MEK‐1/2 inhibitors, LY294002 and U0126, respectively, we showed that DC‐induced CREB S133 phosphorylation was blocked (2.71±0.40 fold by LY294002, p<0.005; 3.84±0.90 fold by UO126, p<0.001) (fig 4C). Our results suggest that CREB is a transcriptional factor downstream of PI3K/AKT and ERK‐1/2 involved in COX‐2 induction by unconjugated bile acids.
Figure 4 CREB downstream of PI3K/AKT and ERK1/2 mediates cyclooxygebase (COX)‐2 induction by unconjugated bile acids. (A) SEG‐1 cells were treated with 200 μM of conjugated (GCA, TCA, TDC, GCDC and TCDC) and unconjugated bile acids (CD and DC) for 16–24 h, and immunoblots were performed with phospho‐CREB (S133), COX‐2 and beta‐actin antibodies as described in Materials and methods. (B) SEG‐1 cells were stimulated with increasing concentrations of CD and DC, and immunoblots for phospho‐CREB (S133), COX‐2 and beta‐actin were performed as in (A). (C) SEG‐1 cells were pre‐incubated with PI3K specific inhibitor LY294002 for 30 min (left panel) and MEK‐1/2 specific inhibitor U0126 for 30 min (right panel) before stimulating with DC for 16–24 h. Immunoblots for phospho‐CREB (S133) and beta‐actin were performed as described in Materials and methods. DC‐induced CREB S133 phosphorylation was blocked by both inhibitors (2.71±0.40 fold by LY294002, p<0.005; 3.84±0.90 fold by UO126, p<0.001). Data are based on the results of three separate experiments. (D) SEG‐1 cells were initially transfected with either CREB specific siRNA or scrambled siRNA by siPORT™ NeoFX™ and 24 h later cotransfected with 1 μg of human COX‐2 promoter plus 0.2 μg of pSV‐β‐gal. The transfected cells were subsequently challenged with 200 μM DC or vehicle (0.1% ethanol). Luciferase reporter activities were measured after 16 h and reported as described in Materials and methods. (inset) SEG‐1 cells transfected with scrambled siRNA and with CREB‐specific siRNA and treated with DC were lysed and immunoblots for COX‐2 and CREB were performed as described in Materials and methods. Data represent the mean ± SD of three experiments. *p<0.05 vs control. CD, chenodeoxycholic acid; DC, deoxycholic acid; GCA, glycocholic acid; GCDC, glycochenodeoxycholic acid; luc, luciferase; TCA, taurodeoxycholic acid; TDC, taurodeoxycholic acid; TCDC, taurochenodeoxycholic acid.
Further, using CREB‐specific siRNA, we showed that specifically silencing CREB blocks COX‐2 transcription, and abrogates DC‐induced COX‐2 expression in SEG‐1 cells (fig 4D).
Induction of COX‐2 by unconjugated bile acids involves AP‐1 activation
Bile acids are known to activate c‐Jun/AP‐1 and stimulate its DNA binding ability in SKGT4 adenocarcinoma cells.51 We therefore examined whether bile acid‐mediated activation of c‐Jun/AP‐1 induced COX‐2 expression in SEG‐1 cells. In keeping with previous findings, treatment with CD or DC strongly increased c‐Jun/AP‐1 transcriptional activity and c‐Jun/AP‐1 expression in a dose‐dependent manner in parallel with COX‐2 induction (fig 5A,B,C). Transfection of the DN AP‐1 expression vector TAM67 potently reduced CD or DC‐induced COX‐2 transcriptional activity (fig 5D). Thus, our results further suggest that c‐Jun/AP‐1, in addition to CREB, promotes CD or DC‐mediated COX‐2 expression.
Figure 5 AP‐1 mediates cyclooxygenase (COX)‐2 induction by CD and DC. (A) SEG‐1 cells were treated with 200 μM of CD and DC for 16–24 h, and immunoblots were performed with c‐Jun/AP‐1, COX‐2, and beta‐actin antibodies as described in Materials and methods. (B) SEG‐1 cells were stimulated with increasing concentrations of DC and immunoblots were performed with c‐Jun/AP‐1, COX‐2, and beta‐actin antibodies as in (A). (C) SEG‐1 cells were cotransfected with 1 μg of human AP‐1 promoter plus 0.2 μg of pSV‐β‐gal, and then stimulated with 200 μM CD or DC or vehicle (0.1% ethanol). Luciferase reporter activities were measured after 16 h and reported as described in Materials and methods. (D) SEG‐1 cells were cotransfected with 1 μg DN AP‐1 (TAM67) or vector control plus1 μg of human COX‐2 promoter + 0.2 μg of pSV‐β‐gal, and then stimulated with 200 μM CD or DC or vehicle (0.1% ethanol). Luciferase reporter activities were measured after 16 h and reported as described in Materials and methods. p<0.01 vs control, **p<0.005 CD+ApDN and DC+ApDN vs CD and DC alone. CD, chenodeoxycholic acid; DC, deoxycholic acid; luc, luciferase.
Role of ROS in the induction of COX‐2 by bile acids
Previous studies have demonstrated that DC and TDC stimulates ROS generation in hepatocytes, which is essential for receptor tyrosine kinase activation and enhanced signalling through the ERK1/2 and AKT pathways.28 Having established that unconjugated bile acids CD and DC stimulate COX‐2 expression through PI3K and ERK1/2 pathways, we investigated whether ROS are involved upstream of these signalling pathways. Using flow cytometry, we determined the effects of exposure to different bile acids on intracellular ROS levels in SEG‐1 cells. Cells were treated with unconjugated or conjugated bile acids and ROS levels were measured using a non‐fluorescent dye, dichloroflurescent diacetate, which becomes highly fluorescent upon oxidation by intracellular ROS. ROS levels were significantly increased by CD or DC but not by conjugated bile acids TCA and GCDC (fig 6A) in a pattern similar to ERK1/2, AKT, CREB activation and induction of COX‐2.
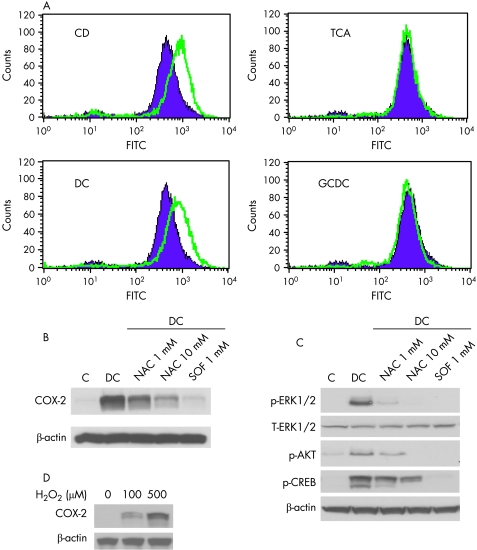
Figure 6 Reactive oxygen species (ROS) mediates unconjugated bile acid induced cyclooxygenase (COX‐2) expression. (A) SEG‐1 cells were treated with 200 μM of unconjugated (CD and DC) and conjugated (TCA and GCDC) bile acids for 16–24 h. A total of 10 μM DCHF‐DA in serum‐free phenol red‐reduced DMEM was then added for 30 min. The mean fluorescence intensity of the DCF‐labelled cells was measured on a FACScan flow cytometer with CellQuest software as described in Materials and methods. The green and purple plots represent mean fluorescence intensity values for treated (green) and untreated (purple) cells, respectively. (B) SEG‐1 cells were preincubated with 1 mM and 10 mM NAC and 1 mM SOF for 30 min. The cells were then stimulated with DC or vehicle (0.1% ethanol), lysates were prepared, and immunoblots were preformed with COX‐2 and beta‐actin antibodies as described in Materials and methods. (C) SEG‐1 cells were treated as in (B), lysates were prepared and immunoblots were preformed with phospho‐ERK1/2, total ERK1/2, phospho‐AKT (S473), phospho‐CREB (S133) and beta‐actin antibodies as described in Materials and methods. (D) SEG‐1 cells were stimulated with increasing concentrations of H2O2 and immunoblots were preformed with COX‐2 and beta‐actin antibodies as described in Materials and methods. CD, chenodeoxycholic acid; DC, deoxycholic acid; GCDC, glycochenodeoxycholic acid; TCA, taurodeoxycholic acid.
Pre‐incubation of SEG‐1 cells with N‐acetyl cysteine (NAC), a thiol reducing agent, and sodium formate (SOF), a hydroxyl radical scavenger, prior to bile acids exposure markedly reduced DC‐mediated COX‐2 expression (fig 6B) and transcriptional activity (data not shown). Both agents also dramatically blocked DC‐induced ERK1/2, AKT and CREB phosphorylation (fig 6C). In addition, exposure of SEG‐1 cells to the strong oxidant H2O2 showed increased expression of COX‐2 (fig 6D), further suggesting that oxidative stress plays a role in up‐regulation of COX‐2 by unconjugated bile acids.
Induction of COX‐2 in immortalised Barrett's epithelial cells
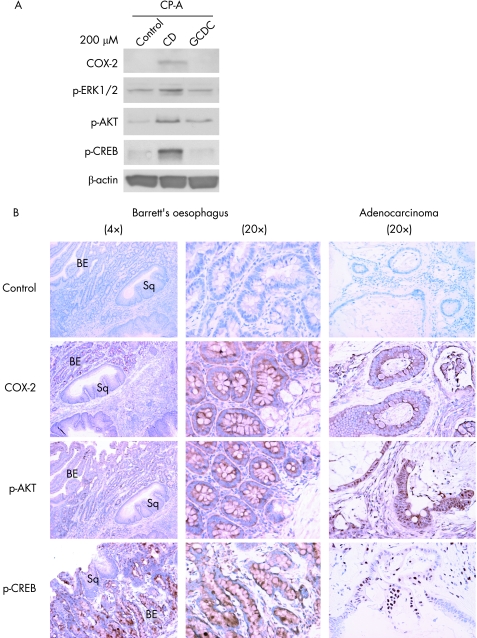
SEG‐1 and SKGT‐4 are useful cell‐culture models for studying tumour progression but results using such models may not be directly applicable to non‐transformed cells. CP‐A is a h‐TERT‐immortalised cell line derived from Barrett's epithelium.36,37 We therefore examined the effect of bile acids on COX‐2 expression in this cell model. In contrast to conjugated bile acid GCDC, the unconjugated bile acid CD increased COX‐2 expression and phosphorylation of ERK1/2, AKT and CREB in CP‐A cells (fig 7A). This suggests that bile acids activate CREB‐dependent COX‐2 expression through ERK1/2 and AKT signalling pathways even in the early stages of neoplastic progression associated with Barrett's oesophagus.
Figure 7 Induction of COX‐2 in immortalised Barrett's oesophagus (Barrett's oesophagus) cells (CP‐A) and expression of COX‐2 in a rat model of Barrett's oesophagus and OA. (A) CP‐A cells were treated with 200 μM of CD or GCDC for 16–24 h, and immunoblots were performed with COX‐2, phospho‐ERK1/2, phospho‐AKT (S473), phospho‐CREB (S133) and beta‐actin antibodies as described in Materials and methods. (B) Immunohistochemical staining for COX‐2, phospho‐AKT (S473) and phospho‐CREB (S133) in tissues derived from a rat model of bile reflux‐induced Barrett's oesophagus (left panel: 4× magnification; middle panel: 20× magnification) and oesophageal adenocarcinoma (right panel: 20× magnification). BE, Barretts's epithelium; Sq, squamous epithelium.
Expression of COX‐2 and activated AKT and CREB in an established rat model of Barrett's oesophagus
We next examined the effects of bile acids on COX‐2 expression in the distal oesophagus using an in vivo model of bile reflux.46 In this model, bile reflux is surgically induced in rats and produces morphological changes in the oesophageal epithelium consistent with Barrett's oesophagus and OA.46 COX‐2, phospho‐AKT and phospho‐CREB were induced in the Barrett's epithelium and oesophageal adenocarcinomas formed in the distal oesophagus during bile‐reflux, but were not present in the adjacent normal squamous epithelium (fig 8B)— results similar to those found in the cell‐culture systems used in this study.
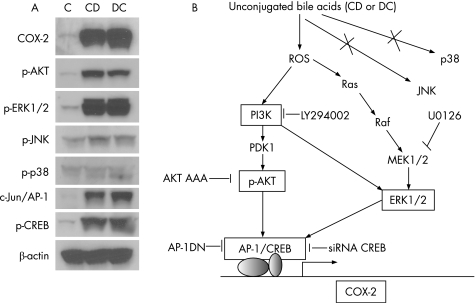
Figure 8 Proposed model of COX‐2 induction by unconjugated bile acids in oesophageal neoplasia. (A) Summary of effects (Western analysis) of unconjugated bile acids on signalling pathways in transformed oesophageal epithelial cells (SEG‐1). (B) Proposed model of pathways by which unconjugated bile acids induce COX‐2 expression based on current data. Unconjugated bile acids generate ROS production, which activate PI3K/AKT and ERK1/2 signalling pathways leading to AP‐1 and CREB dependent COX‐2 induction. The pharmacological inhibitors (LY294002 and U0126) and genetic approaches (DN AKT, DN Ap‐1 and CREB‐siRNA) used to dissect these pathways are depicted here. Previous data23 and data from our laboratory (Supplemental fig 1) indicate that COX‐2 induction may be blocked by inhibitors of PKC. PKC plays a role in the bile‐acid‐induced generation of ROS in colonic epithelial cells (ie acts upstream of ROS), but the precise role of PKC in COX‐2‐induction in oesophageal epithelial cells remains to be elucidated. Crosstalk between PI3K and ERK1/2 is based on supplemental data not presented in the main manuscript but available as Supplemental fig 2.
Discussion
Chronic gastrooesophageal reflux results in Barrett's oesophagus, and the evolution to OA involves a step‐wise progression through dysplasia to invasive carcinoma.52 Population‐based studies show a causal relationship between chronic symptomatic gastrooesophageal reflux and OA in western populations.53,54 Chronic exposure of both acid and bile in the gastrooesophageal refluxate promotes damage and inflammation in the oesophageal epithelium. Bile acids are major constituents of the gastrooesophageal refluxate, and have been associated with the development of Barrett's oesophagus and OA.7,8,9,10,11 We investigated the mechanisms by which bile acids stimulate expression of COX‐2—a pro‐inflammatory marker involved in the neoplastic process. Bile acids strongly induce COX‐2 either by transcriptional or post‐transcriptional mechanisms in multiple gastrointestinal tract cancers, including cancers of the colon, pancreas, stomach, liver, oesophagus and bile ducts.23,39,40,55,56,57,58,59 COX‐2 is significantly upregulated in the oesophageal mucosa of a rat model of duodenooesophageal reflux,60 further suggesting a role of bile acids in the neoplastic progression to OA. In this study, we have demonstrated that the unconjugated bile acids CD and DC strongly induce COX‐2 expression in immortalised Barrett's oesophagus and OA cells. In contrast, glycol‐conjugated and tauro‐conjugated bile acids did not induce COX‐2 expression in these cells. Induction of COX‐2 following exposure to unconjugated bile acids involved ROS‐mediated activation of PI3K/AKT and ERK‐1/2 and their downstream effectors, CREB and AP‐1.
A salient feature of our study is that the unconjugated dihydroxy bile acids CD or DC, but not the conjugated bile acids, dramatically increased intracellular ROS levels, subsequently leading to COX‐2 induction in EA cells. It is known that induction of ROS formation plays a pivotal role in bile acid‐mediated colonic epithelial cell proliferation,25,26,27 where bile acids induce ROS through activation of protein kinase C.25,26 However, little is known about the function of the bile acids in mediating ROS formation in the oesophageal mucosa. The ROS scavengers, NAC and SOF, markedly blocked bile acid‐induced COX‐2 expression in OA cells. Using genetic (DN AKT) and pharmacological (LY294002 and U0126) approaches, we further demonstrated that unconjugated bile acids induce ROS formation thereby stimulating COX‐2 induction in immortalised Barrett's oesophagus and OA cells through the PI3K/AKT and ERK‐1/2 signalling pathways.
The human COX‐2 promoter contains sites for the CRE, AP‐1, AP‐2, NF‐IL6, PEA, Sp‐1, GATA‐1 and NF‐kB, and the AP‐1 site is located in the first intron of COX‐2 gene.31,32,61 AP‐1 is a known transcription factor in the ERK1/2 pathway involved in bile acid‐mediated gene expression in squamous epithelial cells. We have demonstrated that unconjugated bile acids increase c‐Jun/AP‐1 expression and AP‐1‐dependent COX‐2 transcription in OA cells, and that this is completely blocked by DN AP‐1 (TAM67). CD and DC also induced phosphorylation of CREB at serine 133, which is essential for CREB‐mediated transcription. Silencing of CREB by specific siRNA decreased COX‐2 transcription and bile acid‐mediated COX‐2 expression in EA cells. Furthermore, the ROS scavengers NAC and SOF markedly reduced bile acid‐mediated AKT, ERK1/2 and CREB phosphorylation. These results suggest a mechanism by which bile acids induce CREB and AP‐1 dependent COX‐2 expression through ROS mediated activation of PI3K/AKT and ERK1/2 signalling pathways (fig 8).
We further validated our in vitro data using an in vivo rat model of bile reflux, Barrett's oesophagus and oesophageal adenocarcinoma. Using this model, we demonstrated expression of COX‐2, phospho‐AKT and phospho‐CREB in bile‐reflux induced Barrett's mucosa. Prior studies in this rat model demonstrated a strong correlation between COX‐2 expression and development of OA.46
In conclusion, our results indicate that the unconjugated bile acids CD or DC potently upregulate ROS production in the oesophagus, leading to activation of the PI3K and ERK1/2 signalling pathways, and subsequently CREB and AP‐1 dependent COX‐2 expression. A detailed understanding of the mechanisms by which bile acids induce COX‐2 may facilitate the development of chemopreventive strategies to diminish the risk of carcinogenesis within regions of the gastrointestinal tract exposed to bile acids.
Supplementary Material
Acknowledgements
This work was supported by NCI grant R01CA69480 (RSB), American Gastroenterological Association (AGA) Research Scholar Award (SS) and the Texas Gulf Coast Digestive Disease Center (NIDDK).
Abbreviations
AP‐1 - activator protein‐1
CD - chenodeoxycholic acid
CRE - cyclin AMP response element
CREB - CRE binding protein
COX‐2 - cyclooxygenase 2
DC - deoxycholic acid
ERK - extracellular signal‐regulated kinase
DCHF - 2,7‐dichlorohydrofluorescein diacetate
GCA - glycocholic acid
GCDC - glycochenodeoxycholic acid
MEK‐1/2 - mitogen and extracellular signal‐regulated kinase kinase
NAC - N‐acetyl‐L‐cysteine
PI3K - phosphatidylinositol 3‐kinase
OA - oesophageal adenocarcinoma
PKC - protein kinase C
ROS - reactive oxygen species
TCDC - taurochenodeoxycholic acid
TCA - taurocholic acid
TDC - taurodeoxycholic acid
Footnotes
Competing interests: None.
References
- 1.Gillen P, Keeling P, Byrne P J.et al Implication of duodenogastric reflux in the pathogenesis of Barrett's oesophagus. Br J Surg 198875540–543. [DOI] [PubMed] [Google Scholar]
- 2.Kaur B S, Triadafilopoulos G. Acid‐ and bile‐induced PGE(2) release and hyperproliferation in Barrett's esophagus are COX‐2 and PKC‐epsilon dependent. Am J Physiol Gastrointest Liver Physiol 2002283G327–G334. [DOI] [PubMed] [Google Scholar]
- 3.Nehra D, Howell P, Williams C P.et al Toxic bile acids in gastro‐oesophageal reflux disease: influence of gastric acidity. Gut 199944598–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarosi G A, Jr, Jaiswal K, Herndon E.et al Acid increases MAPK‐mediated proliferation in Barrett's esophageal adenocarcinoma cells via intracellular acidification through a Cl‐/HCO3‐ exchanger. Am J Physiol Gastrointest Liver Physiol 2005289G991–G997. [DOI] [PubMed] [Google Scholar]
- 5.Souza R F, Shewmake K, Pearson S.et al Acid increases proliferation via ERK and p38 MAPK‐mediated increases in cyclooxygenase‐2 in Barrett's adenocarcinoma cells. Am J Physiol Gastrointest Liver Physiol 2004287G743–G748. [DOI] [PubMed] [Google Scholar]
- 6.Souza R F, Shewmake K, Terada L S.et al Acid exposure activates the mitogen‐activated protein kinase pathways in Barrett's esophagus. Gastroenterology 2002122299–307. [DOI] [PubMed] [Google Scholar]
- 7.Stein H J, Kauer W K, Feussner H.et al Bile reflux in benign and malignant Barrett's esophagus: effect of medical acid suppression and nissen fundoplication. J Gastrointest Surg 19982333–341. [DOI] [PubMed] [Google Scholar]
- 8.DeMeester T R. Antireflux surgery in the management of Barrett's esophagus. J Gastrointest Surg 20004124–128. [DOI] [PubMed] [Google Scholar]
- 9.Meyer W, Vollmar F. Barrett‐esophagus following total gastrectomy. A contribution to it's pathogenesis. Endoscopy 197911121–126. [DOI] [PubMed] [Google Scholar]
- 10.Nishijima K, Miwa K, Miyashita T.et al Impact of the biliary diversion procedure on carcinogenesis in Barrett's esophagus surgically induced by duodenoesophageal reflux in rats. Ann Surg 200424057–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fein M, Peters J H, Chandrasoma P.et al Duodenoesophageal reflux induces esophageal adenocarcinoma without exogenous carcinogen. J Gastrointest Surg 19982260–268. [DOI] [PubMed] [Google Scholar]
- 12.Jaiswal K, Lopez‐Guzman C, Souza R F.et al Bile salt exposure increases proliferation through p38 and ERK MAPK pathways in a non‐neoplastic Barrett's cell line. Am J Physiol Gastrointest Liver Physiol 2006290G335–G342. [DOI] [PubMed] [Google Scholar]
- 13.Qiao D, Chen W, Stratagoules E D.et al Bile acid‐induced activation of activator protein‐1 requires both extracellular signal‐regulated kinase and protein kinase C signaling. J Biol Chem 200027515090–15098. [DOI] [PubMed] [Google Scholar]
- 14.Romagnolo D F, Chirnomas R B, Ku J.et al Deoxycholate, an endogenous tumor promoter and DNA damaging agent, modulates BRCA‐1 expression in apoptosis‐sensitive epithelial cells: loss of BRCA‐1 expression in colonic adenocarcinomas. Nutr Cancer 20034682–92. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein H, Bernstein C, Payne C M.et al Bile acids as carcinogens in human gastrointestinal cancers. Mutat Res 200558947–65. [DOI] [PubMed] [Google Scholar]
- 16.Debruyne P R, Bruyneel E A, Li X.et al The role of bile acids in carcinogenesis. Mutat Res. 2001;480/481359–369. [DOI] [PubMed] [Google Scholar]
- 17.Hamada K, Umemoto A, Kajikawa A.et al In vitro formation of DNA adducts with bile acids. Carcinogenesis 1994151911–1915. [DOI] [PubMed] [Google Scholar]
- 18.Theisen J, Peters J H, Fein M.et al The mutagenic potential of duodenoesophageal reflux. Ann Surg 200524163–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buskens C J, Sivula A, van Rees B P.et al Comparison of cyclooxygenase 2 expression in adenocarcinomas of the gastric cardia and distal oesophagus. Gut 2003521678–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayakawa T, Fujiwara Y, Hamaguchi M.et al Roles of cyclooxygenase 2 and microsomal prostaglandin E synthase 1 in rat acid reflux oesophagitis. Gut 200655450–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morris C D, Armstrong G R, Bigley G.et al Cyclooxygenase‐2 expression in the Barrett's metaplasia‐dysplasia‐adenocarcinoma sequence. Am J Gastroenterol 200196990–996. [DOI] [PubMed] [Google Scholar]
- 22.Shirvani V N, Ouatu‐Lascar R, Kaur B S.et al Cyclooxygenase 2 expression in Barrett's esophagus and adenocarcinoma: Ex vivo induction by bile salts and acid exposure. Gastroenterology 2000118487–496. [DOI] [PubMed] [Google Scholar]
- 23.Zhang F, Subbaramaiah K, Altorki N.et al Dihydroxy bile acids activate the transcription of cyclooxygenase‐2. J Biol Chem 19982732424–2428. [DOI] [PubMed] [Google Scholar]
- 24.Jang T J, Min S K, Bae J D.et al Expression of cyclooxygenase 2, microsomal prostaglandin E synthase 1, and EP receptors is increased in rat oesophageal squamous cell dysplasia and Barrett's metaplasia induced by duodenal contents reflux. Gut 20045327–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Araki Y, Katoh T, Ogawa A.et al Bile acid modulates transepithelial permeability via the generation of reactive oxygen species in the Caco‐2 cell line. Free Radic Biol Med 200539769–780. [DOI] [PubMed] [Google Scholar]
- 26.Craven P A, Pfanstiel J, DeRubertis F R. Role of reactive oxygen in bile salt stimulation of colonic epithelial proliferation. J Clin Invest 198677850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craven P A, Pfanstiel J, DeRubertis F R. Role of activation of protein kinase C in the stimulation of colonic epithelial proliferation and reactive oxygen formation by bile acids. J Clin Invest 198779532–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang Y, Han S I, Mitchell C.et al Bile acids induce mitochondrial ROS, which promote activation of receptor tyrosine kinases and signaling pathways in rat hepatocytes. Hepatology 200440961–971. [DOI] [PubMed] [Google Scholar]
- 29.Shankar D B, Cheng J C, Kinjo K.et al The role of CREB as a proto‐oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell 20057351–362. [DOI] [PubMed] [Google Scholar]
- 30.Shankar D B, Sakamoto K M. The role of cyclic‐AMP binding protein (CREB) in leukemia cell proliferation and acute leukemias. Leuk Lymphoma 200445265–270. [DOI] [PubMed] [Google Scholar]
- 31.Kong G, Kim H T, Wu K.et al The retinoid X receptor‐selective retinoid, LGD1069, down‐regulates cyclooxygenase‐2 expression in human breast cells through transcription factor crosstalk: implications for molecular‐based chemoprevention. Cancer Res 2005653462–3469. [DOI] [PubMed] [Google Scholar]
- 32.Subbaramaiah K, Norton L, Gerald W.et al Cyclooxygenase‐2 is overexpressed in HER‐2/neu‐positive breast cancer: evidence for involvement of AP‐1 and PEA3. J Biol Chem 200227718649–18657. [DOI] [PubMed] [Google Scholar]
- 33.Chen J J, Huang W C, Chen C C. Transcriptional regulation of cyclooxygenase‐2 in response to proteasome inhibitors involves reactive oxygen species‐mediated signaling pathway and recruitment of CCAAT/enhancer‐binding protein delta and CREB‐binding protein. Mol Biol Cell 2005165579–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H, Rhee S H, Kokkotou E.et al Clostridium difficile toxin A regulates inducible cyclooxygenase‐2 and prostaglandin E2 synthesis in colonocytes via reactive oxygen species and activation of p38 MAPK. J Biol Chem 200528021237–21245. [DOI] [PubMed] [Google Scholar]
- 35.Soldes O S, Kuick R D, Thompson I A., 2ndet al Differential expression of Hsp27 in normal oesophagus, Barrett's metaplasia and oesophageal adenocarcinomas. Br J Cancer 199979595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palanca‐Wessels M C, Barrett M T, Galipeau P C.et al Genetic analysis of long‐term Barrett's esophagus epithelial cultures exhibiting cytogenetic and ploidy abnormalities. Gastroenterology 1998114295–304. [DOI] [PubMed] [Google Scholar]
- 37.Palanca‐Wessels M C, Klingelhutz A, Reid B J.et al Extended lifespan of Barrett's esophagus epithelium transduced with the human telomerase catalytic subunit: a useful in vitro model. Carcinogenesis 2003241183–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pai R, Tarnawski A S, Tran T. Deoxycholic acid activates beta‐catenin signaling pathway and increases colon cell cancer growth and invasiveness. Mol Biol Cell 2004152156–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soma T, Kaganoi J, Kawabe A.et al Chenodeoxycholic acid stimulates the progression of human esophageal cancer cells: A possible mechanism of angiogenesis in patients with esophageal cancer. Int J Cancer 2006119771–782. [DOI] [PubMed] [Google Scholar]
- 40.Tucker O N, Dannenberg A J, Yang E K.et al Bile acids induce cyclooxygenase‐2 expression in human pancreatic cancer cell lines. Carcinogenesis 200425419–423. [DOI] [PubMed] [Google Scholar]
- 41.Song S, Lippman S M, Zou Y.et al Induction of cyclooxygenase‐2 by benzo[a]pyrene diol epoxide through inhibition of retinoic acid receptor‐beta 2 expression. Oncogene 2005248268–8276. [DOI] [PubMed] [Google Scholar]
- 42.Brown P H, Chen T K, Birrer M J. Mechanism of action of a dominant‐negative mutant of c‐Jun. Oncogene 19949791–799. [PubMed] [Google Scholar]
- 43.Lee H Y, Dawson M I, Claret F X.et al Evidence of a retinoid signaling alteration involving the activator protein 1 complex in tumorigenic human bronchial epithelial cells and non‐small cell lung cancer cells. Cell Growth Differ 19978283–291. [PubMed] [Google Scholar]
- 44.Shishodia S, Koul D, Aggarwal B B. Cyclooxygenase (COX)‐2 inhibitor celecoxib abrogates TNF‐induced NF‐kappa B activation through inhibition of activation of I kappa B alpha kinase and Akt in human non‐small cell lung carcinoma: correlation with suppression of COX‐2 synthesis. J Immunol 20041732011–2022. [DOI] [PubMed] [Google Scholar]
- 45.Deng L, Lin‐Lee Y C, Claret F X.et al 2‐acetylaminofluorene up‐regulates rat mdr1b expression through generating reactive oxygen species that activate NF‐kappa B pathway. J Biol Chem 2001276413–420. [DOI] [PubMed] [Google Scholar]
- 46.Buttar N S, Wang K K, Leontovich O.et al Chemoprevention of esophageal adenocarcinoma by COX‐2 inhibitors in an animal model of Barrett's esophagus. Gastroenterology 20021221101–1112. [DOI] [PubMed] [Google Scholar]
- 47.Holt P R, Bresalier R S, Ma C K.et al Calcium plus vitamin D alters preneoplastic features of colorectal adenomas and rectal mucosa. Cancer 2006106287–296. [DOI] [PubMed] [Google Scholar]
- 48.Qiao L, Han S I, Fang Y.et al Bile acid regulation of C/EBPbeta, CREB, and c‐Jun function, via the extracellular signal‐regulated kinase and c‐Jun NH2‐terminal kinase pathways, modulates the apoptotic response of hepatocytes. Mol Cell Biol 2003233052–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martelli A M, Faenza I, Billi A M.et al Intranuclear 3′‐phosphoinositide metabolism and Akt signaling: new mechanisms for tumorigenesis and protection against apoptosis? Cell Signal 2006181101–1107. [DOI] [PubMed] [Google Scholar]
- 50.White P C, Shore A M, Clement M.et al Regulation of cyclin D2 and the cyclin D2 promoter by protein kinase A and CREB in lymphocytes. Oncogene 2006252170–2180. [DOI] [PubMed] [Google Scholar]
- 51.Hirano F, Tanada H, Makino Y.et al Induction of the transcription factor AP‐1 in cultured human colon adenocarcinoma cells following exposure to bile acids. Carcinogenesis 199617427–433. [DOI] [PubMed] [Google Scholar]
- 52.Buttar N, Wang K. Mechanisms of disease: Carcinogenesis in Barrett's esophagus. Nat clin Pract Gastroenterol Hepatol 20041106–112. [DOI] [PubMed] [Google Scholar]
- 53.Lagergren J, Bergstrom R, Lindgren A.et al Symptomatic gastroesophageal reflux as a risk factor for esophageal adenocarcinoma. N Engl J Med 1999340825–831. [DOI] [PubMed] [Google Scholar]
- 54.Ye W, Chow W H, Lagergren J.et al Risk of adenocarcinomas of the esophagus and gastric cardia in patients with gastroesophageal reflux diseases and after antireflux surgery. Gastroenterology 20011211286–1293. [DOI] [PubMed] [Google Scholar]
- 55.de Ledinghen V, Liu H, Zhang F.et al Induction of cyclooxygenase‐2 by tumor promoters in transformed and cytochrome P450 2E1‐expressing hepatocytes. Carcinogenesis 20022373–79. [DOI] [PubMed] [Google Scholar]
- 56.Jurek D, Fleckl E, Marian B. Bile acid induced gene expression in LT97 colonic adenoma cells. Food Chem Toxicol 20054387–93. [DOI] [PubMed] [Google Scholar]
- 57.Sung M W, Roh J L, Park B J.et al Bile acid induces cyclo‐oxygenase‐2 expression in cultured human pharyngeal cells: a possible mechanism of carcinogenesis in the upper aerodigestive tract by laryngopharyngeal reflux. Laryngoscope 20031131059–1063. [DOI] [PubMed] [Google Scholar]
- 58.Yasuda H, Yamada M, Endo Y.et al Elevated cyclooxygenase‐2 expression in patients with early gastric cancer in the gastric pylorus. J Gastroenterol 200540690–697. [DOI] [PubMed] [Google Scholar]
- 59.Yoon J H, Higuchi H, Werneburg N W.et al Bile acids induce cyclooxygenase‐2 expression via the epidermal growth factor receptor in a human cholangiocarcinoma cell line. Gastroenterology 2002122985–993. [DOI] [PubMed] [Google Scholar]
- 60.Zhang F, Altorki N K, Wu Y C.et al Duodenal reflux induces cyclooxygenase‐2 in the esophageal mucosa of rats: evidence for involvement of bile acids. Gastroenterology 20011211391–1399. [DOI] [PubMed] [Google Scholar]
- 61.Kosaka T, Miyata A, Ihara H.et al Characterization of the human gene (PTGS2) encoding prostaglandin‐endoperoxide synthase 2. Eur J Biochem 1994221889–897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.