Abstract
Background
The CpG island methylator phenotype (CIMP or CIMP‐high) with widespread promoter methylation is a distinct epigenetic phenotype in colorectal cancer. In contrast, a phenotype with less widespread promoter methylation (CIMP‐low) has not been well characterised. O‐6‐methylguanine‐DNA methyltransferase (MGMT) promoter methylation and silencing have been associated with G>A mutations and microsatellite instability‐low (MSI‐low).
Aim
To examine molecular correlates with MGMT methylation/silencing in colorectal cancer.
Methods
Utilising MethyLight technology, we quantified DNA methylation in MGMT and eight other markers (a CIMP‐diagnostic panel; CACNA1G, CDKN2A (p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1) in 920 population‐based colorectal cancers.
Results
Tumours with both MGMT methylation and loss were correlated positively with MSI‐low (p = 0.02), CIMP‐high (⩾6/8 methylated CIMP markers, p = 0.005), CIMP‐low (1/8–5/8 methylated CIMP markers, p = 0.002, compared to CIMP‐0 with 0/8 methylated markers), KRAS G>A mutation (p = 0.02), and inversely with 18q loss of heterozygosity (p = 0.0002). Tumours were classified into nine MSI/CIMP subtypes. Among the CIMP‐low group, tumours with both MGMT methylation and loss were far more frequent in MSI‐low tumours (67%, 12/18) than MSI‐high tumours (5.6%, 1/18; p = 0.0003) and microsatellite stable (MSS) tumours (33%, 52/160; p = 0.008). However, no such relationship was observed among the CIMP‐high or CIMP‐0 groups.
Conclusion
The relationship between MGMT methylation/silencing and MSI‐low is limited to only CIMP‐low tumours, supporting the suggestion that CIMP‐low in colorectal cancer may be a different molecular phenotype from CIMP‐high and CIMP‐0. Our data support a molecular difference between MSI‐low and MSS in colorectal cancer, and a possible link between CIMP‐low, MSI‐low, MGMT methylation/loss and KRAS mutation.
Keywords: colon cancer, MGMT, CpG island methylator phenotype, CIMP, microsatellite instability
Transcriptional inactivation by cytosine methylation at the promoter CpG islands of tumour suppressor genes is an important carcinogenic mechanism.1 A number of tumour suppressor genes have been shown to be silenced by promoter methylation in colorectal cancers.1,2,3 In fact, a subset of colorectal cancers exhibit promoter methylation in multiple genes, which is referred to as the CpG island methylator phenotype (CIMP).2,4 CIMP‐positive colorectal tumours have a distinct clinical, pathologic and molecular profile, such as associations with proximal tumour location, female sex, poor differentiation, microsatellite instability (MSI), and high BRAF and low TP53 mutation rates.5,6,7,8,9,10,11,12 Promoter CpG island methylation has been shown to occur early in colorectal carcinogenesis.13,14
Although CIMP (which we designate as “CIMP‐high”, to be distinguished from “CIMP‐low”) appears to be a distinct biological phenotype in colorectal cancer, the existence of CIMP‐low (with less extensive CIMP‐specific promoter methylation) is still controversial. We have previously shown that CIMP‐low is associated with male sex and KRAS mutations compared to CIMP‐high and CIMP‐0 (absence of methylation in five CIMP‐specific promoters).15 However, differences between CIMP‐low and CIMP‐0 are not as clear‐cut as those between CIMP‐low and CIMP‐high,15 and additional evidence is necessary to establish CIMP‐low as a different phenotype from CIMP‐high and CIMP‐0.
O‐6‐methylguanine‐DNA methyltransferase (MGMT) acts to repair inappropriately methylated guanine residues in DNA. Chronic exposure to alkylating/methylating agents can lead to increased MGMT activity,16 and MGMT expression protects from spontaneous G:C to A:T transition mutations.17 Promoter methylation and silencing of MGMT are commonly present in colorectal cancer, and associated with G>A mutations in the KRAS and TP53 genes.18,19MGMT promoter methylation in normal‐appearing colonic mucosa (perhaps as a field effect) may be a predisposing factor for the development of colorectal neoplasia.20,21MGMT methylation in colorectal cancer has been suggested to predict non‐recurrence after chemotherapy, while it does not predict non‐recurrence among colorectal cancer patients who have not received chemotherapy.22 In agreement with that report, another study has shown that MGMT loss of expression does not affect survival among stage C colon cancer patients who have not received chemotherapy.23 Compared to the high performance characteristics of CIMP‐specific promoters (CACNA1G, CDKN2A(p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1), the sensitivity and specificity of MGMT methylation for CIMP‐high are low (62% and 66%, respectively),10,24 raising the possibility that MGMT methylation may be a marker for less extensive promoter methylation (ie, CIMP‐low). In addition, MGMT methylation and loss have been associated with MSI‐low in colorectal cancer.23,25 However, no study to date has examined MGMT methylation and silencing in relation to combined MSI and CIMP status. Previous studies on molecular correlates with MGMT methylation in colorectal cancer have been based on small numbers of cases and/or non‐quantitative methylation‐specific PCR.
In this study, using MethyLight technology and a large number of population‐based colorectal cancer samples, we examined MGMT methylation and silencing in relation to various molecular features, particularly combined MSI and CIMP status. MethyLight assays have been shown to be robust and reproducible in quantifying methylation in DNA from paraffin‐embedded tumour tissue.26 Discovering molecular correlates is important in cancer research because this may: (1) provide clues to carcinogenic mechanisms; (2) propose or support a new molecular subtype; (3) alert investigators to be aware of potential confounding in association studies; and (4) suggest surrogate markers in clinical or research settings.15,27
Methods
Study group
We utilised the databases of two large prospective cohort studies: the Nurses' Health Study (n = 121 700 women followed since 1976),28 and the Health Professional Follow‐up Study (n = 51 500 men followed since 1986).29 Informed consent was obtained from all participants prior to inclusion in the cohorts. A subset of the cohort participants developed colorectal cancers during prospective follow‐up. Thus, these colorectal cancers represented population‐based, relatively unbiased samples (compared to retrospective or single‐hospital‐based samples). Previous studies on the Nurses' Health Study and the Health Professionals Follow‐up Study have described baseline characteristics of cohort participants and incident colorectal cancer cases, and confirmed that our colorectal cancer cases were well representative as a population‐based sample.28,29 We collected information from cohort participants through questionnaire although our questionnaire was not designed for the identification of hereditary non‐polyposis colorectal cancer (HNPCC). We collected paraffin‐embedded tissue blocks from hospitals where cohort participants with colorectal cancers had undergone resections of primary tumours. We excluded cases if adequate paraffin‐embedded tumour tissue was not available or if tumours were previously treated by chemotherapy or radiation. As a result, a total of 920 colorectal cancer cases (410 from the men's cohort and 510 from the women's cohort) were included. Besides the 920 tumours analysed in this study, we excluded 25 cancers in which there was no tissue available other than tumour tissue at metastatic sites, and also excluded 19 patients who had received chemotherapy prior to tumour resection. Among our cohort studies, there was no significant difference in demographic features between cases with tissue analysed and those without tissue data.30 Many of the cases have been previously characterised for status of CIMP, MSI, KRAS and BRAF.15,24 However, we have not examined MGMT methylation and silencing in relation to MSI/CIMP status or various other molecular variables. Follow‐up of these cohorts was still ongoing and analysis on patient outcomes was not possible at the time of the study. Tissue collection and analyses were approved by the Dana‐Farber Cancer Institute and Brigham and Women's Hospital Institutional Review Boards.
Histopathologic evaluations
Haematoxylin and eosin (H&E) stained tissue sections were examined under a light microscope by a pathologist (SO) in a blinded fashion without knowledge of clinical and other laboratory data. Tumours were classified into well/moderately differentiated (<50% solid areas) and poorly differentiated tumours (⩾50% solid areas).12 In addition, the extent and type of mucinous component in each tumour were evaluated, and tumours were classified into five categories: (1) tumours with no mucinous or signet ring cell component (non‐mucinous tumours); (2) tumours with 1–49% mucinous component but no signet ring cells; (3) tumours with ⩾50% mucinous component but no signet ring cells; (4) tumours with 1–49% signet ring cell component; and (5) tumours with ⩾50% signet ring cell component.
Genomic DNA extraction and whole genome amplification
Genomic DNA was extracted from dissected tumour tissue sections using the QIAmp DNA Mini Kit (Qiagen, Valencia, CA, USA) as previously described.31 Normal DNA was obtained from colonic tissue at resection margins. Whole genome amplification (WGA) of genomic DNA was performed by PCR using random 15‐mer primers31 for subsequent MSI analysis and KRAS and BRAF sequencing. Previous studies by us and others showed that WGA did not significantly affect KRAS mutation detection or microsatellite analysis.31,32
Microsatellite instability and 18q loss of heterozygosity analyses
Methods for analysing for MSI status have been previously described.33 In addition to the recommended NCI (National Cancer Institute) panel consisting of D2S123, D5S346, D17S250, BAT25 and BAT26,34 we also used BAT40, D18S55, D18S56, D18S67 and D18S487 (ie, a 10‐marker panel).33 A high degree of MSI (MSI‐H) was defined as the presence of instability in ⩾30% of the markers. A low degree of MSI (MSI‐L) was defined as the presence of instability in <30% of the markers, and microsatellite stable (MSS) tumours were defined as tumours without an unstable marker. We defined MSI‐L cases very strictly. We repeated PCR every time there was an altered peak inconclusive for instability, to confirm instability and exclude sporadic PCR artifact. There were a total of 131 (15%) MSI‐H among 889 tumours with MSI status determined. Among the MSI‐H tumours, the frequencies of MLH1 methylation, TGFBR2 mutation and BRAF mutation were 73% (96/131), 73% (96/131) and 46% (59/128), respectively. Methods for analysis of TGFBR2 mutation have been previously described,35 and the presence of a peak at an altered size in tumour DNA relative to normal DNA was interpreted as positivity for a mutation in the TGFBR2 mononucleotide repeat.
Methods for 18q loss of heterozygosity (LOH) analysis (on microsatellite markers D18S55, D18S56, D18S67 and D18S487) have been previously described.33 The presence of LOH at each locus was defined as 40% or greater reduction of one of two allele peaks in tumour DNA relative to normal DNA, in two duplicated runs to exclude allele dropout and sporadic PCR bias. Overall 18q LOH positivity was strictly defined as the presence of at least two informative markers with LOH, and 18q LOH negativity as the absence of LOH in all (at least two) informative markers. These stringent criteria enabled us to select biologically homogeneous groups of tumours. When we used less stringent criteria in which 18q LOH positivity was defined as ⩾1 informative markers with LOH and 18q LOH negativity as ⩾1 informative markers without evidence of LOH, we could include a larger number of cases; however, the relationship between 18q LOH and MGMT became weaker (data not shown).
Sequencing of KRAS and BRAF
Methods of PCR and sequencing targeted for KRAS codons 12 and 13, and BRAF codon 600 have been previously described.15,31 Pyrosequencing was performed using the PSQ96 HS System (Biotage AB and Biosystems, Uppsala, Sweden) according to the manufacturer's instructions.
Real‐time PCR (MethyLight) for quantitative DNA methylation analysis
Sodium bisulfite treatment on genomic DNA was performed as previously described.26 Real‐time PCR to measure DNA methylation (MethyLight) was performed as previously described.36,37 Utilising ABI 7300 (Applied Biosystems, Foster City, CA, USA) for quantitative real‐time PCR, we amplified MGMT and eight other promoters (CACNA1G, CDKN2A(p16), CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1); methylation in the latter eight promoters has been shown to be sensitive and specific for CIMP,24 and thus we used these eight markers as a CIMP diagnostic panel. COL2A1 (the collagen 2A1 gene) was used to normalise for the amount of input bisulfite‐converted DNA.26,37 Primers and probes were previously described as follows: CACNA1G, CRABP1 and NEUROG110,11; MGMT, CDKN2A and COL2A137; MLH126; and IGF2, RUNX3 and SOCS1.11 The percentage of methylated reference (PMR; ie, degree of methylation) at a specific locus was calculated by dividing the GENE:COL2A1 ratio of template amounts in a sample by the GENE:COL2A1 ratio of template amounts in SssI‐treated human genomic DNA (presumably fully methylated) and multiplying this value by 100.36 A PMR cut‐off value of 4 was based on previously validated data.10,26,36,37 We set a PMR cut‐off value of 6 for CRABP1 and IGF2, based on PMR distribution. Precision and performance characteristics of bisulfite conversion and subsequent MethyLight assays have been previously evaluated and the assays have been validated.26
In particular, we validated the MGMT PMR cutoff of 4 by examining PMR values in relation to loss of expression. Among 567 tumours with valid MGMT methylation and expression data, the frequencies of MGMT loss were as follows: 14% (46/331) in tumours with PMR = 0; 18% (5/28) in tumours with PMR of 0–1; 33% (3/9) in tumours with PMR of 1–4; 60% (6/10) in tumours with PMR of 4–10; and 79% (150/189) in tumours with PMR>10.
CIMP‐high was defined as the presence of ⩾6/8 methylated CIMP markers (excluding MGMT), CIMP‐low as the presence of 1/8 to 5/8 methylated markers, and CIMP‐0 as the absence (0/8) of methylated markers, based on the data that CIMP‐high and CIMP‐low are associated with BRAF mutations and KRAS mutations, respectively.15,24
Immunohistochemistry for MGMT
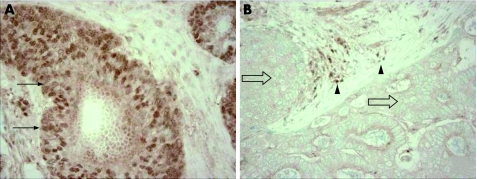
For MGMT immunohistochemistry, antigen retrieval was performed; deparaffinised tissue sections were treated in 1 mM EDTA buffer (pH 8) in a pressure cooker by a microwave for 20 min. Tissue sections were incubated with 3% H2O2 (20 min) to block endogenous peroxidase for 20 min, and then incubated with 2% horse serum (Vector Laboratories, Burlingame, California, USA) in phosphate‐buffered saline (20 min). Primary antibody against MGMT (clone MT3.1, Lab Vision, Fremont, CA; dilution 1:25) was applied for 1 h at room temperature. Secondary antibody (Vector Laboratories) (30 min), and then avidin biotin complex conjugate (Vector Laboratories) (30 min) were applied. Sections were visualised by diaminobenzidine (DAB) (2 min) and methyl‐green counterstain. Normal colonic epithelial cells and inflammatory cells served as internal positive controls when a tumour lost MGMT expression (fig 1). Appropriate positive and negative controls were included in each run of immunohistochemistry. All immunohistochemically‐stained slides were interpreted by a pathologist (SO) blinded from clinical and other molecular data.
Figure 1 Evaluation of MGMT expression by immunohistochemistry. (A) MGMT expression is present in colorectal cancer cells (arrows). (B) Colorectal cancer cells show loss of MGMT expression (empty arrows). Inflammatory cells serve as an internal positive control (solid arrowheads).
Statistical analysis
In statistical analysis, χ2 test (or Fisher's exact test when the number in any category was less than 10) was performed for categorical data, using the SAS program (Version 9.1, SAS Institute, Cary, NC). All p values were two‐sided, and statistical significance was set at p⩽0.05.
Results
MGMT promoter methylation and loss of expression
Utilising MethyLight technology, we quantified DNA methylation in MGMT and a panel of eight promoters (CACNA1G, CDKN2A, CRABP1, IGF2, MLH1, NEUROG1, RUNX3 and SOCS1); the latter eight markers have been shown to be sensitive and specific for CIMP10,11,24 and were thus used as a CIMP diagnostic panel. Among the 920 tumours, 354 (38%) showed MGMT promoter methylation. We also assessed MGMT expression by immunohistochemistry (fig 1). Among the 599 tumours with valid expression data, 224 (37%) showed loss of expression, most consistent with gene silencing or deletion. Table 1 summarises the relationship between clinicopathologic variables and MGMT methylation (or loss of expression).
Table 1 Frequency of MGMT promoter methylation and loss of expression in colorectal cancer.
| MGMT promoter methylation | MGMT expression | ||||
|---|---|---|---|---|---|
| Total examined, n | Methylated (%) | Total examined, n | Loss (%) | p Value | |
| All cases | 920 | 354 (38%) | 599 | 224 (37%) | |
| Sex | |||||
| Men | 410 | 150 (37%) | 250 | 91 (36%) | |
| Women | 510 | 204 (40%) | 349 | 133 (38%) | |
| Tumour differentiation | |||||
| Well/moderate | 815 | 311 (38%) | 519 | 190 (37%) | |
| Poor | 84 | 34 (40%) | 54 | 22 (41%) | |
| Non‐mucinous tumours | 593 | 211 (36%) | 393 | 133 (34%) | Referent |
| Mucinous 1–49% (no signet ring cell) | 185 | 85 (46%) | 123 | 59 (48%) | 0.005 |
| Mucinous ⩾50% (no signet ring cell) | 81 | 41 (51%) | 44 | 16 (36%) | |
| Signet ring cells 1–49% | 46 | 13 (28%) | 35 | 16 (46%) | |
| Signet ring cells ⩾50% | 15 | 4 (27%) | 4 | 0 | |
| Location | |||||
| Right | 255 | 103 (40%) | 176 | 70 (40%) | |
| Left (excluding rectum) | 175 | 62 (35%) | 121 | 50 (41%) | |
| Rectum | 107 | 37 (35%) | 65 | 18 (28%) | |
Only a statistically significant p value is described.
Molecular correlates with MGMT methylation or loss of expression
Table 2 shows the relationship between various molecular features and MGMT promoter methylation (or loss of expression) in colorectal cancer. MGMT promoter methylation was highly correlated with MGMT loss of expression (p<0.0001). MGMT methylation was positively correlated with MLH1 methylation (p = 0.02), unlike in a previous study.38 MSI‐L showed slightly higher frequencies of MGMT methylation (45%) and loss (49%) than MSS tumours (37% showing MGMT methylation and 36% showing MGMT loss), although differences were not statistically significant. Both MGMT methylation and loss were significantly more common in CIMP‐high and CIMP‐low tumours than in CIMP‐0 tumours. 18q LOH‐positive tumours showed a significantly lower frequency of MGMT methylation (30%, p = 0.0008) and loss (27%, p = 0.004) than 18q LOH‐negative tumours (45% showing MGMT methylation and 42% showing MGMT loss).
Table 2 Frequency of MGMT methylation and loss of expression in colorectal cancer.
| MGMT methylation | MGMT expression | |||||
|---|---|---|---|---|---|---|
| Total examined, n | Methylated (%) | p Value | Total examined, n | Loss (%) | p Value | |
| MGMT expression | ||||||
| Loss | 210 | 156 (74%) | <0.0001 | |||
| Intact | 357 | 43 (12%) | Referent | |||
| MLH1 methylation | ||||||
| (+) | 115 | 56 (49%) | 0.02 | |||
| (−) | 805 | 298 (37%) | Referent | |||
| MSI status | ||||||
| MSI‐high | 131 | 57 (44%) | 94 | 37 (39%) | ||
| MSI‐low | 73 | 33 (45%) | 47 | 23 (49%) | ||
| MSS | 685 | 255 (37%) | 444 | 158 (36%) | ||
| TGFBR2 (only MSI‐H tumours) | ||||||
| Mutated | 93 | 41 (44%) | 70 | 31 (44%) | ||
| Wild‐type | 38 | 16 (42%) | 21 | 6 (29%) | ||
| CIMP status | ||||||
| CIMP‐high | 136 | 68 (50%) | <0.0001 | 101 | 42 (42%) | 0.04 |
| CIMP‐low | 353 | 154 (44%) | 0.0002 | 199 | 93 (44%) | 0.003 |
| CIMP‐0 | 431 | 132 (31%) | Referent | 267 | 161 (30%) | Referent |
| KRAS | ||||||
| Any mutation | 321 | 135 (42%) | 206 | 92 (45%) | 0.003 | |
| G>A mutation | 195 | 84 (43%) | 132 | 62 (47%) | 0.003 | |
| Non‐G>A mutation | 126 | 51 (40%) | 74 | 30 (41%) | ||
| Wild‐type | 553 | 205 (37%) | 368 | 119 (32%) | Referent | |
| BRAF | ||||||
| Mutated | 116 | 46 (39%) | 82 | 23 (28%) | ||
| Wild‐type | 758 | 294 (38%) | 492 | 188 (38%) | ||
| KRAS/BRAF | ||||||
| K(+)B(+) | 6 | 3 (50%) | 3 | 2 (67%) | ||
| K(+)B(−) | 315 | 132 (42%) | 203 | 90 (44%) | 0.02 | |
| K(−)B(+) | 110 | 43 (39%) | 79 | 21 (27%) | ||
| K(−)B(−) | 443 | 162 (37%) | 289 | 98 (34%) | Referent | |
| 18q LOH | ||||||
| (+) | 237 | 71 (30%) | 0.0008 | 169 | 45 (27%) | 0.004 |
| (−) | 217 | 98 (45%) | Referent | 145 | 61 (42%) | Referent |
Only statistically significant p values are described. CIMP, CpG island methylator phenotype; LOH, loss of heterozygosity; MSI, microsatellite instability; MSS, microsatellite stable.
Tumours with KRAS G>A mutation showed a higher frequency of MGMT loss (47%, p = 0.003) than tumours with wild‐type KRAS (32% showing MGMT loss). We also examined the frequencies of KRAS G>A mutation in tumours with or without MGMT methylation (or expression). The frequencies of KRAS G>A mutation were not significantly different between MGMT‐methylated tumours (25%, 84/340) and MGMT‐unmethylated tumours (21%, 111/534). However, the frequency of KRAS G>A mutation was significantly higher in MGMT‐lost tumours (29%, 62/211; p = 0.006) than MGMT‐expressing tumours (19%, 70/363).
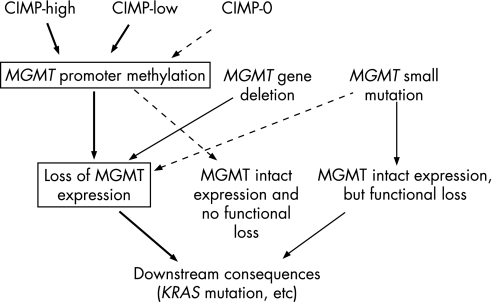
Tumours with both MGMT methylation and loss of expression
As shown in table 2, the correlation between MGMT methylation and loss of expression was tight but not perfect. Figure 2 illustrates various molecular aberrations surrounding MGMT promoter methylation and loss of expression. About 26% of MGMT‐lost tumours exhibited no significant MGMT methylation (maybe due to gene deletion or mutation that impaired gene expression), and about 12% of MGMT‐expressing tumours did show MGMT methylation (maybe partial or monoallelic methylation that did not considerably affect expression). We calculated the frequency of tumours with both MGMT methylation and loss of expression among colorectal cancers with various molecular features. Tumours with both MGMT methylation and loss likely represented a more homogeneous group of tumours than tumours with MGMT methylation alone, or tumours with MGMT loss alone. Table 3 summarises the molecular correlates with simultaneous MGMT methylation and loss of expression. MSI‐L tumours showed a significantly higher frequency of MGMT methylation/loss (41%, p = 0.02) than MSS tumours (25%).
Figure 2 Various molecular aberrations surrounding MGMT promoter methylation and loss of expression in colorectal cancer. Promoter methylation and gene deletion (or small mutation) can occur in two different MGMT alleles in the same tumour. Thus, this figure is a simplified illustration. Tumours with both MGMT methylation and loss of expression represent a more homogeneous group of tumours than tumours with MGMT methylation alone or tumours with MGMT loss alone.
Table 3 Frequency of tumours with both MGMT methylation and loss in colorectal cancer.
| Totalexamined,n | MGMT methylatedand lost (%) | p Value | |
|---|---|---|---|
| MSI status | |||
| MSI‐high | 91 | 29 (32%) | |
| MSI‐low | 46 | 19 (41%) | 0.02 |
| MSS | 421 | 106 (25%) | Referent |
| CIMP status | |||
| CIMP‐high | 101 | 35 (35%) | 0.005 |
| CIMP‐low | 199 | 66 (33%) | 0.002 |
| CIMP‐0 | 267 | 55 (21%) | Referent |
| KRAS | |||
| Any mutation | 197 | 65 (33%) | 0.03 |
| G>A mutation | 127 | 45 (35%) | 0.02 |
| Non‐G>A mutation | 70 | 20 (29%) | |
| Wild‐type | 349 | 85 (24%) | Referent |
| KRAS/BRAF | |||
| K(+)B(+) | 3 | 2 (67%) | |
| K(+)B(−) | 194 | 63 (32%) | 0.04 |
| K(−)B(+) | 76 | 15 (20%) | Referent |
| K(−)B(−) | 273 | 70 (26%) | |
| 18q LOH | |||
| (+) | 163 | 28 (17%) | 0.0002 |
| (−) | 142 | 51 (36%) | Referent |
Only statistically significant p values are described. CIMP, CpG island methylator phenotype; LOH, loss of heterozygosity; MSI, microsatellite instability; MSS, microsatellite stable.
MGMT methylation and loss in nine MSI/CIMP subtypes of colorectal cancer
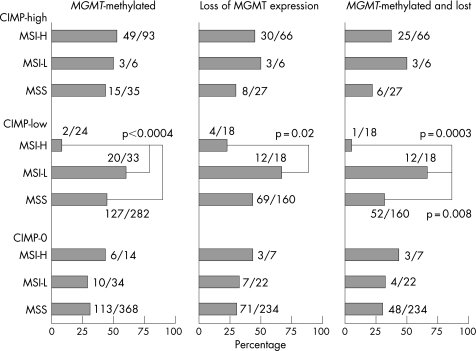
Molecular classification of colorectal cancer based on MSI and CIMP status is increasingly important because MSI status and CIMP status reflect global genomic and epigenomic aberrations in tumour cells. Thus, we classified tumours into nine subtypes according to both MSI and CIMP status as follows: MSI‐H CIMP‐high (n = 93), MSI‐L CIMP‐high (n = 6), MSS CIMP‐high (n = 35), MSI‐H CIMP‐low (n = 24), MSI‐L CIMP‐low (n = 33), MSS CIMP‐low (n = 282), MSI‐H CIMP‐0 (n = 14), MSI‐L CIMP‐0 (n = 34) and MSS CIMP‐0 (n = 368). By virtue of the large sample size, we were able to evaluate the frequencies of MGMT methylation and/or loss in all of the nine subtypes, even in rare subtypes such as MSI‐L CIMP‐high and MSI‐H CIMP‐0 (fig 3).
Figure 3 Frequencies of MGMT methylation and/or loss of expression in nine MSI/CIMP subtypes of colorectal cancer. A significant relationship between MGMT and MSI status is present only among the CIMP‐low tumour group.
In contrast to the CIMP‐high and CIMP‐0 groups, the CIMP‐low groups exhibited a striking difference in the frequencies of MGMT methylation and/or loss between MSI‐H and MSI‐L tumours (fig 3). In the CIMP‐low groups, MSI‐H tumours showed much lower frequencies of MGMT methylation (8.3%), loss of expression (22%) and methylation/loss (5.6%) than MSI‐L tumours (61% showing methylation, p<0.0001; 67% showing loss of expression, p = 0.002; and 67% showing methylation/loss, p = 0.0003). These data indicate that MSI‐L is not a mixture of misdiagnosed MSI‐H and MSS tumors. These relationships between MGMT and MSI status were present only among CIMP‐low tumours, suggesting that CIMP‐low tumours constituted a different subset of colorectal cancers from CIMP‐high and CIMP‐0 tumours. Our data also suggest that MSI‐L tumours were different from MSS tumours at least among the CIMP‐low tumour group.
Within the CIMP‐high group, there was no significant difference in the frequencies of MGMT methylation or loss between MSI‐H, MSI‐L and MSS tumours. Similarly, within the CIMP‐0 group, there was no significant difference in the frequencies of MGMT methylation or loss between MSI‐H, MSI‐L and MSS tumours.
Discussion
We conducted this study to examine the molecular correlates with MGMT promoter methylation and gene silencing in colorectal cancer, using a large number of samples and robust methylation detection methods. Discovering molecular correlates is important, because it may: (1) provide clues to pathogenesis; (2) propose or support a new molecular subtype; (3) alert investigators to be aware of potential confounding in association studies; and (4) suggest surrogate markers in clinical or research settings.27,35 We used quantitative PCR assays (MethyLight)36 to determine the degree of DNA methylation, which is robust enough to reproducibly differentiate low‐level methylation from high‐level methylation.26 Our resource of a large number of samples from two large prospective cohorts (relatively unbiased samples compared to retrospective or single‐hospital‐based samples) has enabled us to precisely estimate the frequency of colorectal cancers with specific molecular features (eg, MGMT methylation, CIMP‐high, MSI‐H, etc).
In particular, we sought to decipher the relationship between MGMT and MSI/CIMP status. Molecular classification based on MSI and CIMP status is increasingly important39 because MSI and CIMP status reflect global genomic and epigenomic aberrations in cancer cells. We have found that, among CIMP‐low tumours, MGMT methylation and silencing are correlated positively with MSI‐L and inversely with MSI‐H. However, no such relationship was present among CIMP‐high and CIMP‐0 tumours. A previous study demonstrated an insignificant trend towards an inverse relationship between MGMT methylation and MSI‐H MINT++ (ie, CIMP)40 (this MINT++ could represent both CIMP‐high and CIMP‐low, because MINT markers are not specific for CIMP‐high but are also frequently methylated in CIMP‐low tumours11). Our data indicate unique molecular features of CIMP‐low, which differ from CIMP‐high and CIMP‐0. Our data also support a possible link between CIMP‐low, MSI‐L, MGMT methylation/loss and KRAS mutation.
We have previously shown that CIMP‐low in colorectal cancer is associated with male sex and KRAS mutations, while CIMP‐high is associated with female sex and BRAF mutations, and CIMP‐0 is associated with wild‐type KRAS/BRAF genes and shows no sex predilection.15 However, the difference between CIMP‐low and CIMP‐0 was not as clear‐cut as the difference between CIMP‐high and CIMP‐low.15 Our data from the current study provide additional supporting evidence for a molecular difference between CIMP‐low and CIMP‐0. Further studies are necessary to identify the best set of markers for the diagnosis of CIMP‐low, because the eight marker panel we used is sensitive and specific for CIMP‐high10,24 but perhaps not the best markers to separate CIMP‐low from CIMP‐0.
We have shown an association between MGMT methylation/loss and MSI‐L, which is in agreement with the previous studies.23,25 In addition, we have shown that the association between MGMT methylation/loss and MSI‐L is only present in CIMP‐low tumours but not in CIMP‐high or CIMP‐0 tumours. As shown in fig 3, MSI‐L is not a mixture of under‐diagnosed MSI‐H and MSS. MSI‐L has been associated with shorter survival in stage C colon cancer compared to MSS tumours.23 A cDNA microarray expression study has also supported MSI‐L as a distinct phenotype from MSS and MSI‐H.41 These data collectively support molecular and biological differences between MSI‐L and MSS in colorectal cancer. Additional studies are necessary to find underlining molecular defects for MSI‐L and better biomarkers for MSI‐L (other than microsatellites), since there has been a controversy whether MSI‐low exists as a distinct molecular phenotype from MSS.42,43 It also remains to be elucidated why the significant relationship between MGMT and MSI‐L is limited to CIMP‐low tumours.
We have shown the positive correlation between MGMT methylation and MLH1 methylation, in contrast to a previous study.38 We have shown that MLH1 methylation is very specific for CIMP‐high,10 and that MGMT methylation is also correlated with CIMP‐high (in this study). Thus, in our large scale study, the observed correlation between MGMT methylation and MLH1 methylation is logical, and likely mediated by CIMP‐high.
We have detected other interesting molecular correlates between MGMT methylation/loss and various molecular features, including KRAS mutation, KRAS G>A mutation and 18q LOH (inverse correlation). Esteller et al18 have previously described a relationship between KRAS G>A mutation and MGMT methylation, and the frequency of MGMT methylation was more common in colorectal cancers with KRAS G>A mutation (71%, 36/51), compared to tumours with KRAS non‐G>A mutation (32%, 12/37 showing MGMT methylation) and tumours with wild‐type KRAS (35%, 55/156 showing MGMT methylation). In the current study, we could show the significant relationship between MGMT loss of expression and KRAS G>A mutation (p = 0.003) (or any KRAS mutation, p = 0.003). Halford et al44 demonstrated that MGMT loss of expression in colorectal cancer was significantly more common in tumours with G>A mutation in APC, CTNNB1 (the β‐catenin gene) or KRAS.
MGMT methylation and/or CIMP have been related to the serrated pathway of colorectal carcinogenesis.25,39,45,46MGMT methylation has been detected in 22% of hyperplastic polyps, in 25% of sessile serrated adenomas,47 in 16–22% of serrated adenomas with a variable degree of dysplasia, and in 50% of serrated adenocarcinomas.48 However, since MGMT methylation and CIMP are positively correlated, it is also possible that serrated neoplasias may be correlated with CIMP and only indirectly with MGMT methylation through CIMP. Nonetheless, MSI‐L occurs more commonly (∼30%) in serrated adenocarcinomas while MSI‐L is less common (∼14%) in non‐serrated adenocarcinomas,49 favouring the suggestion that factors inducing MSI‐L are important for the development of serrated neoplasias.46
In conclusion, MGMT methylation and loss of expression are correlated with MSI‐L, CIMP‐high and CIMP‐low in colorectal cancer. In particular, among CIMP‐low tumours, MGMT methylation and loss are correlated positively with MSI‐L and inversely with MSI‐H. No such relationship is present among CIMP‐high or CIMP‐0 tumours, supporting the suggestion that CIMP‐low may be a different phenotype from CIMP‐high and CIMP‐0. Our data also support a molecular difference between MSI‐L and MSS in colorectal cancer, as well as a possible pathogenetic link between CIMP‐low, MSI‐L, MGMT methylation/loss and KRAS mutation.
Note added in proofs: As additional supporting evidence for molecular differences between CIMP‐low and CIMP‐0, we have recently shown that 18q LOH in non‐MSI‐high tumour is correlated positively with CIMP‐0 and inversely with CIMP‐low and CIMP‐high.50 With regard to the frequency of 18q LOH, CIMP‐low is similar to CIMP‐high, but different from CIMP‐0.
Acknowledgements
We deeply thank the Nurses' Health Study and Health Professionals Follow‐up Study cohort participants who have generously agreed to provide us with biological specimens and information through responses to questionnaires. We thank Graham Colditz, Walter Willett and many other staff members who implemented and have maintained the cohort studies. We deeply thank Peter Laird, Daniel Weisenberger and Mihaela Campan for assisting in the development of the MethyLight assay.
Abbreviations
CACNA1G - calcium channel, voltage‐dependent, T type alpha‐1G subunit
CDKN2A - cyclin‐dependent kinase inhibitor 2A (p16/INK4A)
CIMP - CpG island methylator phenotype
CRABP1 - cellular retinoic acid binding protein 1
DAB - diaminobenzidine
HNPCC - hereditary non‐polyposis colorectal cancer
IGF2 - insulin‐like growth factor 2
LOH - loss of heterozygosity
MGMT - O‐6‐methylguanine‐DNA methyltransferase
MSI - microsatellite instability
MSI‐H - microsatellite instability‐high
MSI‐L - microsatellite instability‐low
MSS - microsatellite stable
NCI - National Cancer Institute
NEUROG1 - neurogenin 1
PMR - percentage of methylated reference (degree of methylation)
RUNX3 - runt‐related transcription factor 3
SOCS1 - suppressor of cytokine signaling 1
TGFBR2 - transforming growth factor‐beta receptor type 2
WGA - whole genome amplification
Footnotes
Funding: This work was supported by the US National Institute of Health (NIH) grants P01 CA87969 and P01 CA55075.
Competing interests: None.
References
- 1.Laird P W. Cancer epigenetics. Hum Mol Genet 200514(Spec No 1)R65–R76. [DOI] [PubMed] [Google Scholar]
- 2.Issa J P. CpG island methylator phenotype in cancer. Nat Rev Cancer 20044988–993. [DOI] [PubMed] [Google Scholar]
- 3.Wong J J, Hawkins N J, Ward R L. Colorectal cancer: a model for epigenetic tumorigenesis. Gut 200756140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toyota M, Ahuja N, Ohe‐Toyota M.et al CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A 1999968681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Rijnsoever M, Grieu F, Elsaleh H.et al Characterisation of colorectal cancers showing hypermethylation at multiple CpG islands. Gut 200251797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toyota M, Ohe‐Toyota M, Ahuja N.et al Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A 200097710–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hawkins N, Norrie M, Cheong K.et al CpG island methylation in sporadic colorectal cancers and its relationship to microsatellite instability. Gastroenterology 20021221376–1387. [DOI] [PubMed] [Google Scholar]
- 8.Samowitz W, Albertsen H, Herrick J.et al Evaluation of a large, population‐based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology 2005129837–845. [DOI] [PubMed] [Google Scholar]
- 9.Kambara T, Simms L A, Whitehall V L J.et al BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut 2004531137–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogino S, Cantor M, Kawasaki T.et al CpG island methylator phenotype (CIMP) of colorectal cancer is best characterised by quantitative DNA methylation analysis and prospective cohort studies. Gut 2006551000–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weisenberger D J, Siegmund K D, Campan M.et al CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 200638787–793. [DOI] [PubMed] [Google Scholar]
- 12.Ogino S, Odze R D, Kawasaki T.et al Correlation of pathologic features with CpG island methylator phenotype (CIMP) by quantitative DNA methylation analysis in colorectal carcinoma. Am J Surg Pathol 2006301175–1183. [DOI] [PubMed] [Google Scholar]
- 13.Minoo P, Jass J. Senescence and serration: a new twist to an old tale. J Pathol 2006210137–140. [DOI] [PubMed] [Google Scholar]
- 14.Jass J R. Serrated adenoma of the colorectum and the DNA‐methylator phenotype. Nat Clin Pract Oncol 20052398–405. [DOI] [PubMed] [Google Scholar]
- 15.Ogino S, Kawasaki T, Kirkner G J.et al CpG island methylator phenotype‐low (CIMP‐low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn 20068582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lees N P, Harrison K L, Hall C N.et al Human colorectal mucosal O6‐alkylguanine DNA‐alkyltransferase activity and DNA‐N7‐methylguanine levels in colorectal adenoma cases and matched referents. Gut. Published Online First 4 Aug 2006. doi: 10, 1136/gut. 2006. 097899 [DOI] [PMC free article] [PubMed]
- 17.Aquilina G, Biondo R, Dogliotti E.et al Expression of the endogenous O6‐methylguanine‐DNA‐methyltransferase protects Chinese hamster ovary cells from spontaneous G:C to A:T transitions. Cancer Res 1992526471–6475. [PubMed] [Google Scholar]
- 18.Esteller M, Toyota M, Sanchez‐Cespedes M.et al Inactivation of the DNA repair gene O6‐methylguanine‐DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K‐ras in colorectal tumorigenesis. Cancer Res 2000602368–2371. [PubMed] [Google Scholar]
- 19.Esteller M, Risques R A, Toyota M.et al Promoter hypermethylation of the DNA repair gene O(6)‐methylguanine‐DNA methyltransferase is associated with the presence of G:C to A:T transition mutations in p53 in human colorectal tumorigenesis. Cancer Res 2001614689–4692. [PubMed] [Google Scholar]
- 20.Shen L, Kondo Y, Rosner G L.et al MGMT promoter methylation and the field defect in sporadic colorectal cancer. J Natl Cancer Inst 2005971330–1338. [DOI] [PubMed] [Google Scholar]
- 21.Giovannucci E, Ogino S. DNA methylation, field effects, and colorectal cancer. J Natl Cancer Inst 2005971317–1319. [DOI] [PubMed] [Google Scholar]
- 22.Nagasaka T, Sharp G B, Notohara K.et al Hypermethylation of O6‐methylguanine‐DNA methyltransferase promoter may predict nonrecurrence after chemotherapy in colorectal cancer cases. Clin Cancer Res 200395306–5312. [PubMed] [Google Scholar]
- 23.Kohonen‐Corish M R, Daniel J J, Chan C.et al Low microsatellite instability is associated with poor prognosis in stage C colon cancer. J Clin Oncol 2005232318–2324. [DOI] [PubMed] [Google Scholar]
- 24.Ogino S, Kawasaki T, Kirkner G J.et al Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer using a large population‐based sample. J Mol Diagn 20079305–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitehall V L, Walsh M D, Young J.et al Methylation of O‐6‐methylguanine DNA methyltransferase characterizes a subset of colorectal cancer with low‐level DNA microsatellite instability. Cancer Res 200161827–830. [PubMed] [Google Scholar]
- 26.Ogino S, Kawasaki T, Brahmandam M.et al Precision and performance characteristics of bisulfite conversion and real‐time PCR (MethyLight) for quantitative DNA methylation analysis. J Mol Diagn 20068209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogino S, Kawasaki T, Kirkner G J.et al Down‐regulation of p21 (CDKN1A/CIP1) is inversely associated with microsatellite instability and CpG island methylator phenotype (CIMP) in colorectal cancer. J Pathol 2006210147–154. [DOI] [PubMed] [Google Scholar]
- 28.Colditz G A, Hankinson S E. The Nurses' Health Study: lifestyle and health among women. Nat Rev Cancer 20055388–396. [DOI] [PubMed] [Google Scholar]
- 29.Giovannucci E, Liu Y, Rimm E B.et al Prospective study of predictors of vitamin D status and cancer incidence and mortality in men. J Natl Cancer Inst 200698451–459. [DOI] [PubMed] [Google Scholar]
- 30.Chan A T, Ogino S, Fuchs C S. Aspirin use and risk of colorectal cancer according to cyclooxygenase‐2 expression. New Engl J Med 20073562131–2142. [DOI] [PubMed] [Google Scholar]
- 31.Ogino S, Kawasaki T, Brahmandam M.et al Sensitive sequencing method for KRAS mutation detection by pyrosequencing. J Mol Diagn 20057413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dietmaier W, Hartmann A, Wallinger S.et al Multiple mutation analyses in single tumor cells with improved whole genome amplification. Am J Pathol 199915483–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogino S, Brahmandam M, Cantor M.et al Distinct molecular features of colorectal carcinoma with signet ring cell component and colorectal carcinoma with mucinous component. Mod Pathol 20061959–68. [DOI] [PubMed] [Google Scholar]
- 34.Boland C R, Thibodeau S N, Hamilton S R.et al A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998585248–5257. [PubMed] [Google Scholar]
- 35.Ogino S, Kawasaki T, Ogawa A.et al TGFBR2 mutation is correlated with CpG island methylator phenotype (CIMP) in microsatellite instability‐high colorectal cancer. Hum Pathol 200738(4)614–620. [DOI] [PubMed] [Google Scholar]
- 36.Eads C A, Danenberg K D, Kawakami K.et al MethyLight: a high‐throughput assay to measure DNA methylation. Nucleic Acids Res 200028E32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Widschwendter M, Siegmund K D, Muller H M.et al Association of breast cancer DNA methylation profiles with hormone receptor status and response to tamoxifen. Cancer Res 2004643807–3813. [DOI] [PubMed] [Google Scholar]
- 38.Fox E J, Leahy D T, Geraghty R.et al Mutually exclusive promoter hypermethylation patterns of hMLH1 and O6‐methylguanine DNA methyltransferase in colorectal cancer. J Mol Diagn 2006868–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jass J R. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology 200750113–130. [DOI] [PubMed] [Google Scholar]
- 40.Whitehall V L, Wynter C V, Walsh M D.et al Morphological and molecular heterogeneity within nonmicrosatellite instability‐high colorectal cancer. Cancer Res 2002626011–6014. [PubMed] [Google Scholar]
- 41.Mori Y, Selaru F M, Sato F.et al The impact of microsatellite instability on the molecular phenotype of colorectal tumors. Cancer Res 2003634577–4582. [PubMed] [Google Scholar]
- 42.Tomlinson I, Halford S, Aaltonen L.et al Does MSI‐low exist? J Pathol 20021976–13. [DOI] [PubMed] [Google Scholar]
- 43.Jass J R. Re: Tomlinson, et al. Does MSI‐low exist. J Pathol 2002197 pp 6-13 J Pathol 2003199267–9 author reply 970. [DOI] [PubMed] [Google Scholar]
- 44.Halford S, Rowan A, Sawyer E.et al O6‐methylguanine methyltransferase in colorectal cancers: detection of mutations, loss of expression, and weak association with G:C>A:T transitions. Gut 200554797–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jass J R, Biden K G, Cummings M C.et al Characterisation of a subtype of colorectal cancer combining features of the suppressor and mild mutator pathways. J Clin Pathol 199952455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makinen M J. Colorectal serrated adenocarcinoma. Histopathology 200750131–150. [DOI] [PubMed] [Google Scholar]
- 47.O'Brien M J, Yang S, Clebanoff J L.et al Hyperplastic (serrated) polyps of the colorectum: relationship of CpG island methylator phenotype and K‐ras mutation to location and histologic subtype. Am J Surg Pathol 200428423–434. [DOI] [PubMed] [Google Scholar]
- 48.Dong S M, Lee E J, Jeon E S.et al Progressive methylation during the serrated neoplasia pathway of the colorectum. Mod Pathol 200518170–178. [DOI] [PubMed] [Google Scholar]
- 49.Tuppurainen K, Makinen J M, Junttila O.et al Morphology and microsatellite instability in sporadic serrated and non‐serrated colorectal cancer. J Pathol 2005207285–294. [DOI] [PubMed] [Google Scholar]
- 50.Ogino S, Kawasaki T, Kirkner G J.et al 18q loss of heterozygosity in microsatellite stable colorectal cancer is correlated positively with CpG island methylator phenotype‐negative (CIMP‐negative) and inversely with CIMP‐low and CIMP‐high. BMC Cancer 2007772. [DOI] [PMC free article] [PubMed] [Google Scholar]