Abstract
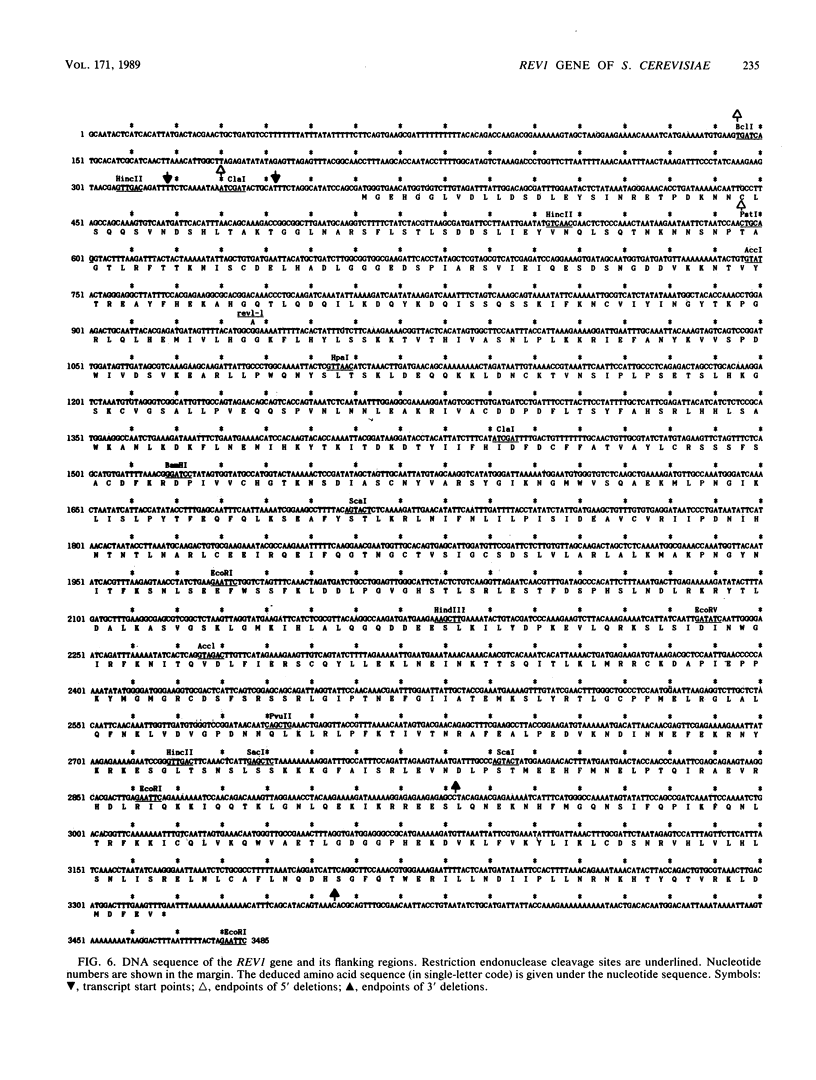
The REV1 gene of Saccharomyces cerevisiae is required for normal induction of mutations by physical and chemical agents. We have determined the sequence of a 3,485-base-pair segment of DNA that complements the rev1-1 mutant. Gene disruption was used to confirm that this DNA contained the REV1 gene. The sequenced segment contains a single long open reading frame, which can encode a polypeptide of 985 amino acid residues. The REV1 transcript is 3.1 kilobase pairs in length. Frameshift mutations introduced into the open reading frame yielded a Rev-phenotype. A base substitution, encoding Gly-193 to Arg-193, was found in this open reading frame in rev1-1. Deletion mutants, lacking segments of the 5' region of REV1, had intermediate mutability relative to REV1 and rev1-1; a complete deletion exhibited lower mutability than rev1-1. REV1 is not an essential gene. An in-frame fusion of the 5' end of the REV1 open reading frame to the lacZ gene produced beta-galactosidase activity constitutively. The predicted REV1 protein is hydrophilic, with a predicted pI of 9.82. No homologies to RAD1, RAD2, RAD3, RAD7, or RAD10 proteins were noted. A 152-residue internal segment displayed 25% identity with UMUC protein.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Angulo J. F., Schwencke J., Moreau P. L., Moustacchi E., Devoret R. A yeast protein analogous to Escherichia coli RecA protein whose cellular level is enhanced after UV irradiation. Mol Gen Genet. 1985;201(1):20–24. doi: 10.1007/BF00397980. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Braus G., Paravicini G., Hütter R. A consensus transcription termination sequence in the promoter region is necessary for efficient gene expression of the TRP1 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1988 Jun;212(3):495–504. doi: 10.1007/BF00330855. [DOI] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Cigan A. M., Donahue T. F. Sequence and structural features associated with translational initiator regions in yeast--a review. Gene. 1987;59(1):1–18. doi: 10.1016/0378-1119(87)90261-7. [DOI] [PubMed] [Google Scholar]
- Cole G. M., Schild D., Lovett S. T., Mortimer R. K. Regulation of RAD54- and RAD52-lacZ gene fusions in Saccharomyces cerevisiae in response to DNA damage. Mol Cell Biol. 1987 Mar;7(3):1078–1084. doi: 10.1128/mcb.7.3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filetici P., Junakovic N., Ballario P. Rapid alkaline preparation for yeast circular covalently closed DNA molecules. Curr Genet. 1985;9(2):123–126. doi: 10.1007/BF00436959. [DOI] [PubMed] [Google Scholar]
- Gannon F., O'Hare K., Perrin F., LePennec J. P., Benoist C., Cochet M., Breathnach R., Royal A., Garapin A., Cami B. Organisation and sequences at the 5' end of a cloned complete ovalbumin gene. Nature. 1979 Mar 29;278(5703):428–434. doi: 10.1038/278428a0. [DOI] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Hu N., Messing J. The making of strand-specific M13 probes. Gene. 1982 Mar;17(3):271–277. doi: 10.1016/0378-1119(82)90143-3. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., McGrath J. P., Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature. 1987 Sep 10;329(6135):131–134. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Kuo C. L., Campbell J. L. Cloning of Saccharomyces cerevisiae DNA replication genes: isolation of the CDC8 gene and two genes that compensate for the cdc8-1 mutation. Mol Cell Biol. 1983 Oct;3(10):1730–1737. doi: 10.1128/mcb.3.10.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C. W., Das G., Christensen R. B. REV7, a new gene concerned with UV mutagenesis in yeast. Mol Gen Genet. 1985;200(1):80–85. doi: 10.1007/BF00383316. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W. Mutagenesis in Saccharomyces cerevisiae. Adv Genet. 1982;21:173–254. doi: 10.1016/s0065-2660(08)60299-0. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W., Nisson P. E., Christensen R. B. UV and chemical mutagenesis in rev7 mutants of yeast. Mol Gen Genet. 1985;200(1):86–91. doi: 10.1007/BF00383317. [DOI] [PubMed] [Google Scholar]
- Lawrence C. W., O'Brien T., Bond J. UV-induced reversion of his4 frameshift mutations in rad6, rev1, and rev3 mutants of yeast. Mol Gen Genet. 1984;195(3):487–490. doi: 10.1007/BF00341451. [DOI] [PubMed] [Google Scholar]
- Lemontt J. F. Genetic and physiological factors affecting repair and mutagenesis in yeast. Basic Life Sci. 1980;15:85–120. doi: 10.1007/978-1-4684-3842-0_7. [DOI] [PubMed] [Google Scholar]
- Lemontt J. F. Mutants of yeast defective in mutation induced by ultraviolet light. Genetics. 1971 May;68(1):21–33. doi: 10.1093/genetics/68.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Madura K., Prakash S. Nucleotide sequence, transcript mapping, and regulation of the RAD2 gene of Saccharomyces cerevisiae. J Bacteriol. 1986 Jun;166(3):914–923. doi: 10.1128/jb.166.3.914-923.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinus M. G. Location of DNA methylation genes on the Escherichia coli K-12 genetic map. Mol Gen Genet. 1973 Dec 14;127(1):47–55. doi: 10.1007/BF00267782. [DOI] [PubMed] [Google Scholar]
- McClanahan T., McEntee K. Specific transcripts are elevated in Saccharomyces cerevisiae in response to DNA damage. Mol Cell Biol. 1984 Nov;4(11):2356–2363. doi: 10.1128/mcb.4.11.2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. E., Brammar W. J., Murray K. Lambdoid phages that simplify the recovery of in vitro recombinants. Mol Gen Genet. 1977 Jan 7;150(1):53–61. doi: 10.1007/BF02425325. [DOI] [PubMed] [Google Scholar]
- Nagpal M. L., Higgins D. R., Prakash S. Expression of the RAD1 and RAD3 genes of Saccharomyces cerevisiae is not affected by DNA damage or during the cell division cycle. Mol Gen Genet. 1985;199(1):59–63. doi: 10.1007/BF00327510. [DOI] [PubMed] [Google Scholar]
- Naumovski L., Chu G., Berg P., Friedberg E. C. RAD3 gene of Saccharomyces cerevisiae: nucleotide sequence of wild-type and mutant alleles, transcript mapping, and aspects of gene regulation. Mol Cell Biol. 1985 Jan;5(1):17–26. doi: 10.1128/mcb.5.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisson P. E., Lawrence C. W. The isolation and characterization of ngm2, a mutation that affects nitrosoguanidine mutagenesis in yeast. Mol Gen Genet. 1986 Jul;204(1):90–97. doi: 10.1007/BF00330193. [DOI] [PubMed] [Google Scholar]
- Njagi G. D., Kilbey B. J. cdc7-1 a temperature sensitive cell-cycle mutant which interferes with induced mutagenesis in Saccharomyces cerevisiae. Mol Gen Genet. 1982;186(4):478–481. doi: 10.1007/BF00337951. [DOI] [PubMed] [Google Scholar]
- Olson M. V., Loughney K., Hall B. D. Identification of the yeast DNA sequences that correspond to specific tyrosine-inserting nonsense suppressor loci. J Mol Biol. 1979 Aug 15;132(3):387–410. doi: 10.1016/0022-2836(79)90267-5. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Genetic applications of yeast transformation with linear and gapped plasmids. Methods Enzymol. 1983;101:228–245. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Patterson M., Sclafani R. A., Fangman W. L., Rosamond J. Molecular characterization of cell cycle gene CDC7 from Saccharomyces cerevisiae. Mol Cell Biol. 1986 May;6(5):1590–1598. doi: 10.1128/mcb.6.5.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozzi G., Prakash S. RAD7 gene of Saccharomyces cerevisiae: transcripts, nucleotide sequence analysis, and functional relationship between the RAD7 and RAD23 gene products. Mol Cell Biol. 1986 May;6(5):1497–1507. doi: 10.1128/mcb.6.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry K. L., Elledge S. J., Mitchell B. B., Marsh L., Walker G. C. umuDC and mucAB operons whose products are required for UV light- and chemical-induced mutagenesis: UmuD, MucA, and LexA proteins share homology. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4331–4335. doi: 10.1073/pnas.82.13.4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson T. A., Prakash L., Prakash S., Osley M. A., Reed S. I. Regulation of CDC9, the Saccharomyces cerevisiae gene that encodes DNA ligase. Mol Cell Biol. 1985 Jan;5(1):226–235. doi: 10.1128/mcb.5.1.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Reynolds P., Higgins D. R., Prakash L., Prakash S. The nucleotide sequence of the RAD3 gene of Saccharomyces cerevisiae: a potential adenine nucleotide binding amino acid sequence and a nonessential acidic carboxyl terminal region. Nucleic Acids Res. 1985 Apr 11;13(7):2357–2372. doi: 10.1093/nar/13.7.2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Prakash L., Dumais D., Perozzi G., Prakash S. Nucleotide sequence of the RAD10 gene of Saccharomyces cerevisiae. EMBO J. 1985 Dec 16;4(13A):3549–3552. doi: 10.1002/j.1460-2075.1985.tb04115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Prakash L., Prakash S. Nucleotide sequence and functional analysis of the RAD1 gene of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):1012–1020. doi: 10.1128/mcb.7.3.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson G. W., Nicolet C. M., Kalainov D., Friedberg E. C. A yeast excision-repair gene is inducible by DNA damaging agents. Proc Natl Acad Sci U S A. 1986 Mar;83(6):1842–1846. doi: 10.1073/pnas.83.6.1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Ruby S. W., Szostak J. W. Specific Saccharomyces cerevisiae genes are expressed in response to DNA-damaging agents. Mol Cell Biol. 1985 Jan;5(1):75–84. doi: 10.1128/mcb.5.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani R. A., Patterson M., Rosamond J., Fangman W. L. Differential regulation of the yeast CDC7 gene during mitosis and meiosis. Mol Cell Biol. 1988 Jan;8(1):293–300. doi: 10.1128/mcb.8.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siede W., Eckardt-Schupp F. DNA repair genes of Saccharomyces cerevisiae: complementing rad4 and rev2 mutations by plasmids which cannot be propagated in Escherichia coli. Curr Genet. 1986;11(3):205–210. doi: 10.1007/BF00420608. [DOI] [PubMed] [Google Scholar]
- Struhl K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8419–8423. doi: 10.1073/pnas.82.24.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]