Abstract
Retinal ganglion cell genesis requires the proneural bHLH transcription factor Math5 (Atoh7), but little is known about the regulatory elements that control its expression. Here, we investigate Math5 gene regulation using transgenic mice. These mice express GFP in the prenatal retina, live-labeling RGC axon migration and innervation of the brain. Unexpectedly, these Math5-GFP transgenes are also found in Math1 expression domains throughout the nervous system, intriguing since Math5 and Math1 normally exhibit nonoverlapping expression. Furthermore, Math5-GFP and Math1 are regulated similarly, by both Pax6 and Math1 itself, in the lower rhombic lip and dorsal spinal cord. We also show that Pax6 binds to particular Math5 and Math1 regulatory sequences in vitro. Together these data suggest that these atonal semi-orthologues may share conserved regulatory elements that are normally silent in the Math5 gene.
Keywords: Math5, Math1, visual system, optic nerve, cochlear nucleus, Pax6
Introduction
The mammalian neural retina is composed of seven cell types, six neuronal and one glial, that differentiate from a common progenitor pool within defined temporal windows (Cepko et al., 1996; Livesey and Cepko, 2001). Proper spatial and temporal development of retinal neurons is attributed, in part, to the proper expression of proneural basic helix-loop-helix (bHLH) transcription factors in progenitors (Cepko, 1999). The bHLH factor Math5 (mouse atonal homologue 5) is expressed in retinal progenitors prenatally, beginning at E11 and continuing through P0 (Brown et al., 1998). Math5 is required for the development of RGCs, which transmit visual information to the brain via the optic nerve. In the absence of Math5 RGCs fail to form, and although the eyes appear normal externally, these mice completely lack optic nerves (Brown et al., 2001; Wang et al., 2001). Math5 expression is also critical for the timing of RGC differentiation; in its absence this temporal window is shifted such that these cells adopt late fates, predominantly Müller glia (Brzezinski, 2005; Le et al., 2006).
An understanding of the regulatory networks that control Math5 expression is crucial to elucidating its role in neurogenesis. Math5 requires the paired-domain transcription factor Pax6 for initial activation (Brown et al., 1998; Marquardt et al., 2001) and is repressed by the bHLH factor Hes1 (Lee et al., 2005). However, the cis-regulatory elements regulating Math5 expression are not yet well defined. In Xenopus, Ath5 expression (Xath5) is regulated by upstream bHLH-dependent and independent elements (Hutcheson et al., 2005). Both proximal bHLH-specific binding sites and more distal cis-regulatory Xath5 sequences each independently drive transgenic expression of a GFP reporter in the Xenopus retina. Importantly, a phylogenetically conserved 5′ distal element is required for retinal expression in the mouse retina and sufficient to drive retinal expression in the frog eye (Hutcheson et al., 2005; Riesenberg et al., 2007).
Here, we explore the in vivo expression patterns of Math5-GFP transgenes containing different combinations of 5′ and 3′ Math5 non-coding DNA sequences. While endogenous Math5 expression is confined to retinal progenitors, Math5-GFP persists in mature RGCs and along the length of their axons in the developing brain. We demonstrate that these transgenic mice are useful for observing RGC axon outgrowth and the establishment of the optic nerves, chiasm, and tracts within the brain. Math5-GFP is also ectopically expressed in multiple expression domains of Math1, another orthologue of Drosophila atonal (Jarman et al., 1993; Ben-Arie et al., 1996). Math5 and Math1 are critical regulators of sensory neuron circuit formation in the visual, auditory, and proprioceptive systems (Bermingham et al., 1999; Ben-Arie et al., 2000; Hassan and Bellen, 2000; Brown et al., 2001; Wang et al., 2001; Saul et al., 2007). However, these genes have mutually exclusive expression patterns; thus, the observation of Math5-GFP and Math1 coexpression was further investigated. We find that Math5-GFP is regulated similarly to Math1, by Pax6 in the lower rhombic lip and by Math1 itself in the lower rhombic lip and dorsal spinal cord. These findings provide insight into the divergence of this gene family during vertebrate evolution.
Results
Math5-GFP expression in the developing visual system
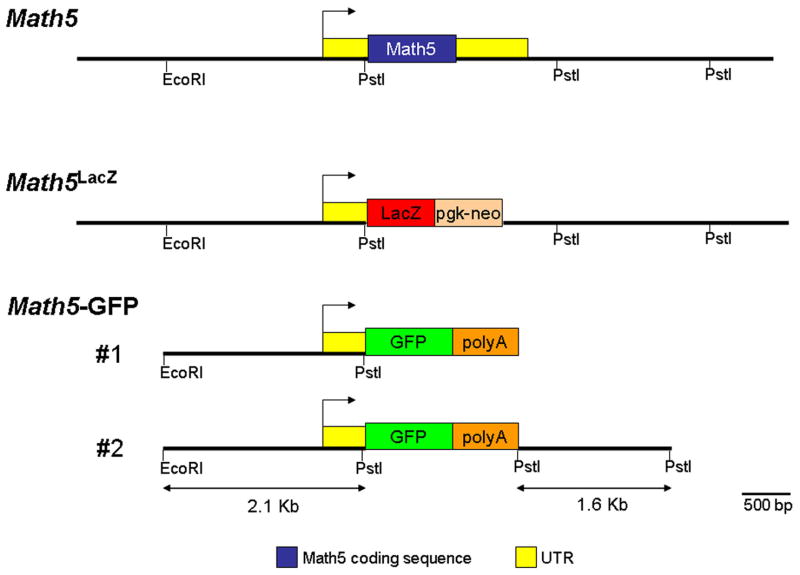
Several transgenic mouse lines expressing green fluorescent protein (GFP) under the control of non-coding 5′ (Math5-GFP1) or 5′ and 3′ (Math5-GFP2) regulatory elements of the Math5 gene have been generated (Riesenberg et al., 2007). Both Math5-GFP transgenes contain 2.1 Kb of 5′ Math5 non-coding sequence that ends 14 base pairs upstream of the ATG start codon (Fig. 1). In addition, Math5-GFP2 contains 1.6 Kb of Math5 3′ DNA inserted downstream of the GFP coding region (Fig. 1). The 5′ 2.1 Kb sequence is sufficient to drive GFP expression in the mouse retina, beginning at E11.5 (Hutcheson et al., 2005; Riesenberg et al., 2007 and Fig. 3A,4B). However, other aspects of Math5 cis-regulation have not been investigated.
Fig. 1. Math5-GFP transgenic reporters.
Diagram of the Math5 gene, Math5LacZ targeted deletion and the Math5-GFP1 and Math5-GFP2 transgenes. The 5′ DNA common to both transgenes was generated from an EcoRI to PstI 2.1 Kb fragment. The 3′ DNA contained in the Math5-GFP2 transgene was generated from a PstI to PstI 1.6 Kb fragment.
Fig. 3. Comparison of Math5-GFP and Math5LacZ expression in the retina.
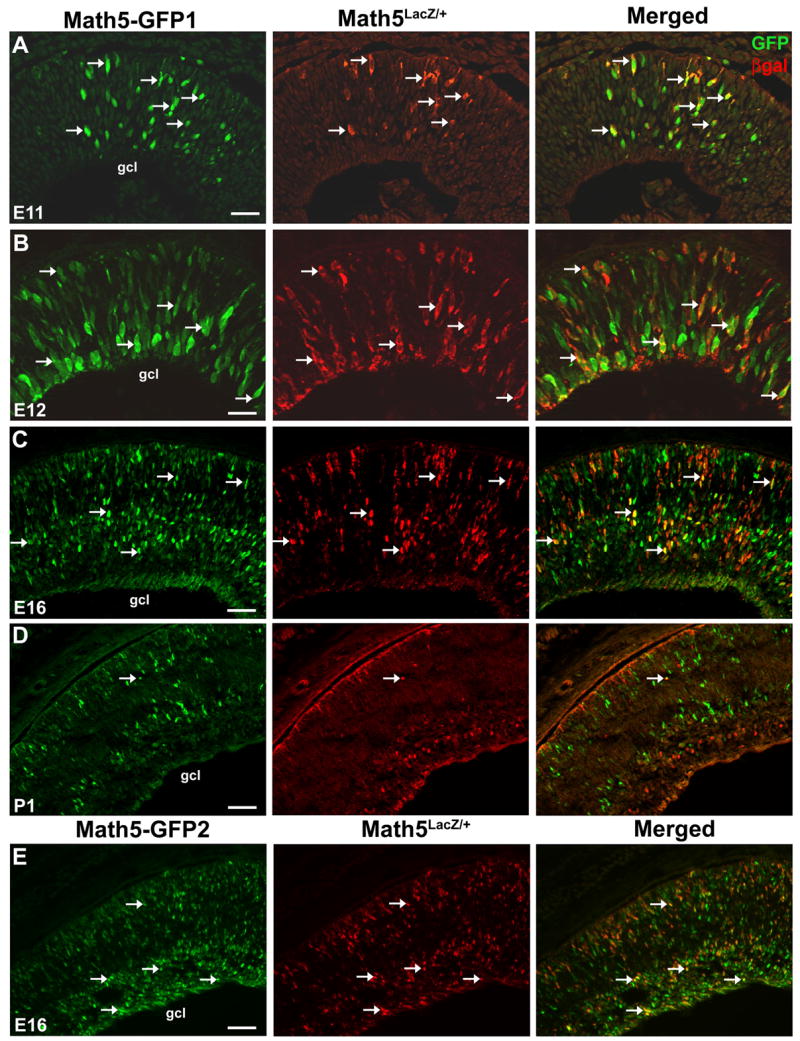
(A–D) Math5-GFP1; Math5LacZ/+ retinal cryosections co-labeled for GFP and βgal expression. At E11.5 and E12.5, βgal+ cells co-localize with Math5-GFP (A,B arrows), and some GFP+ only cells are evident. At E16.5, Math5-GFP1 and Math5LacZ are coexpressed in a subset of retinal cells (C, arrows), while others express GFP or βgal alone. At P1, few co-labeled cells are observed (D, arrows). Also, more GFP+ cells reside in the neuroblast and developing photoreceptor layer at P1, while more βgal+ cells are in the ganglion cell layer. (E) Math5-GFP2 is expressed in the retina in a similar pattern to Math5-GFP1 at E16. Sclera is at the top of all panels. Magnification bars in A,B (25 μm), C,E (50 μm) and D (100 μm). gcl- ganglion cell layer.
Fig. 4. Ectopic Math5-GFP expression in the developing nervous system.
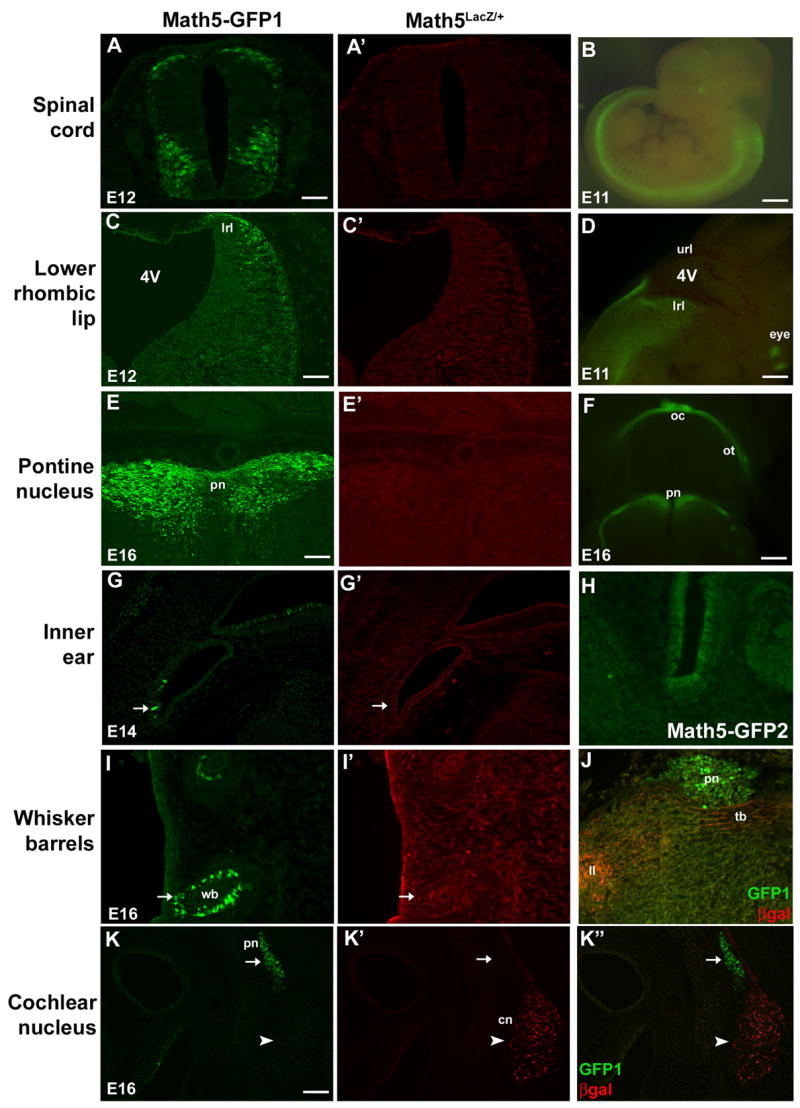
Math5-GFP1; Math5LacZ/+ embryos co-labeled for GFP and βgal. GFP expression (arrows) was observed in discrete domains devoid of Math5LacZ expression, including the spinal cord (A,A′) and lower rhombic lip (C,C′) at E12.5, inner ear hair cells at E14.5 (G,G′), pontine nuclei (E,E′) and whisker barrel cells (I,I′) at E16.5. The spinal cord, lower rhombic lip, and pontine expression domains are also observable using live fluorescence (B,D,F). Math5LacZ is expressed in the cochlear nucleus (arrowhead in K′,K″) and axons of cochlear nucleus neurons, which project to the lateral lemniscus and trapezoid body (J). Math5-GFP1 is not found in the cochlear nucleus neurons or axons (arrow in K,K″; J). The Math5-GFP2 pattern is identical to Math5-GFP1, except for the absence of GFP expression in inner ear hair cells (H). Magnification bars in A (50 μm), B (1 mm), C (100 μm), D (400 μm), E (50 μm), F (800 μm), and I (100 μm). 4V- fourth ventricle; cn- cochlear nucleus; hc- inner ear hair cell; ll- lateral lemniscus; lrl- lower rhombic lip; pn- pontine nucleus; tb- trapezoid body; wb- whisker barrels.
Thus, we examined Math5-GFP1 expression in the embryonic retinae of transgenic animals. Although Math5 mRNA is only expressed by retinal progenitors that are becoming terminally mitotic (Le et al., 2006), Math5-GFP purdures longer, like Math5LacZ (Brown et al., 2001). As a consequence, differentiated RGCs express GFP throughout their nucleus and cytoplasm, and along the length of their axons. This allows visualization of RGC axons as they progress towards their targets in the brain. RGCs, whose cell bodies are located within the ganglion cell layer of the retina, send axons out through the optic disk into the optic nerve, which connects to the optic chiasm and optic tract (Rodieck, 1998). At E16.5, Math5-GFP1 can be observed in the retina using live GFP fluorescence (Fig. 2A). Examination of the ventral surface of the brain reveals GFP in the optic nerve, chiasm, and tract (Fig. 2B). In the lateral diencephalon, we observe the entire length of the optic tract (Fig. 2C). RGC axons in the optic tract make specific synaptic connections to the lateral geniculate nucleus (lg), superior colliculus (sc), and other processing centers to faithfully transmit visual information from the external environment to the brain (Rodieck, 1998). At E16.5, we observe GFP expression in the developing lg (Fig. 2C,E) and sc (Fig. 2D) by live fluorescence or anti-GFP immunolabeling. Figure 2F displays the relative positions of the optic tract, lg, and sc in a coronal section of the E16.5 brain (Schambra et al., 1992). Overall, we observe GFP along the entire length of RGC axons throughout the period of optic nerve formation.
Fig. 2. Math5-GFP1 expression in the developing visual system.
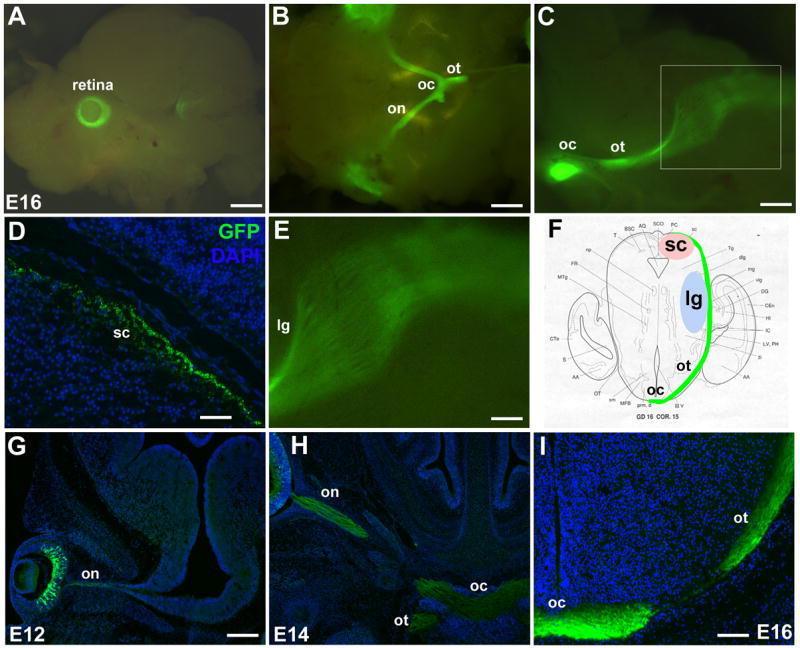
Math5-GFP1 mice express GFP in developing RGCs and their axons. At E16.5, live embryo fluorescence and immunolabeling of GFP in the retina (A), optic nerve, chiasm and tract (B), and the major RGC targets, the superior colliculus (D) and lateral geniculate nucleus (C,E), reveals the entire length of RGC axons. The boxed area in C is shown at higher magnification in E. F is a diagram of the optic tract and its major innervation targets at E16. The progression of RGC axon migration and optic nerve formation can be followed from E12–E16 (G–I). Magnification bars in A (1 mm), B (800 μm), C (400 μm), D (50 μm), E (200 μm), G (200 μm), and I (100 μm). gcl- ganglion cell layer; lg- lateral geniculate nucleus; on- optic nerve; oc- optic chiasm; ot- optic tract; sc- superior colliculus.
The progression of RGC axon outgrowth is readily observed in Math5-GFP1 embryos, beginning with activation of Math5-GFP1 in the optic cup at E11.5 (Fig. 3A,4B). We characterized the progression of GFP-positive axons through the developing brain by immunolabeling for GFP. At E12.5, GFP-positive axons can be seen as they migrate within the optic nerve (Fig. 2G), prior to their extension to the optic chiasm. By E14.5, GFP expression within the optic chiasm and optic tracts is apparent (Fig. 2H). By E16.5, many GFP+ axons are present in the optic tracts (Fig. 2I) and some have reached the sc and lg (Fig. 2D–F). By P1, GFP is still expressed in the retina (Fig. 3C) and RGC axons, but expression is clearly diminished, and by P5 retinal GFP expression is no longer observable (not shown). Math5-GFP2 transgenic mice also express GFP in the developing optic nerve, chiasm, and tracts of the developing brain (not shown). We conclude that the Math5-GFP transgenes allow for visualization of migrating RGC axons from cell bodies in the retina to axon terminations in the developing brain.
Next, we compared Math5-GFP1 expression to that of Math5LacZ in the retina. The Math5LacZ allele was created by inserting the bacterial LacZ gene into the Math5 locus by homologous recombination. Thus, βgal protein reports endogenous Math5 expression, although it purdures longer than Math5 mRNA in differentiated RGCs (Brown et al., 2001). In Math5-GFP1; Math5LacZ/+ E11.5 and E12.5 eyes, essentially all βgal+ cells coexpress GFP (arrows in 3A,B), although a few GFP only cells can be seen. At E16.5, we still observe GFP+/βgal+ colabeled cells (arrows in 3C), but also more GFP+/βgal− and GFP−/βgal+ cells. By P1, an expression difference between the reporters is very evident. At this age, most βgal+ cells reside in the ganglion cell layer, while most GFP+ cells are located either in the neuroblast layer or the forming photoreceptor layer, with only a minor subset of cells coexpressing GFP and βgal (arrow in Fig. 3D). The Math5-GFP2 transgene exhibits a similar difference in expression with Math5LacZ at E16.5 (Fig. 3E). This expression difference suggests that both Math5-GFP transgenes may not contain all spatiotemporal information necessary to recapitulate endogenous Math5 expression. Alternatively, because two gene reporters (GFP and βgal) were compared, expression differences might also be attributed to unequal purdurance of each reporter protein.
Non-retinal expression of Math5-GFP
Math5 mRNA and Math5LacZ are expressed in the developing retina (Brown et al., 1998; Brown et al., 2001; Wang et al., 2001), but Math5-GFP1 is also expressed in several regions of the developing peripheral and central nervous systems. From E10.5 to E12.5, we observe GFP expression in the forebrain (not shown), spinal cord and lower rhombic lip (Fig. 4A–D). In the spinal cord, Math5-GFP1 is expressed in both the dorsal-lateral rim and in a thick ventral band of cells (Fig. 4A,B). In the rhombic lip, Math5-GFP1 expression is restricted to the lower lip (Fig. 4C,D), which gives rise to the precerebellar nuclei, including the pontine and cochlear nuclei (Engelkamp et al., 1999; Landsberg et al., 2005; Farago et al., 2006). Math5-GFP1 is not found in the upper rhombic lip, which develops into the external granular layer (EGL) of the cerebellum (Alder et al., 1996). Math5-GFP1 is also expressed in the pontine nucleus at E16.5 (4E,F), inner ear hair cells from E14.5 to E16.5 (Fig. 4G), whisker barrels from E16.5 through P1 (Fig. 4I), and the molars and the pineal gland at E16.5 (not shown). To determine if these regions normally express Math5, we compared Math5-GFP1 to Math5LacZ in these domains. Math5LacZ is not expressed in any of these non-retinal domains (Fig. 4, second column). This further suggests that Math5-GFP1 is deregulated, allowing expression outside the normal Math5 expression domains. Math5 lineage tracing has described retinal, auditory system, cerebellar cortex, cerebral cortex, and hippocampal labeled adult cells (Yang et al., 2003; Brzezinski, 2005; Saul et al., 2007), but none of the ectopic Math5-GFP expression domains were found. Although Math5-GFP1 expression was seen in a few cells of the developing neocortex, we did not find GFP in the E12.5-P1 cerebellar cortex or hippocampus (not shown). However, the ages for endogenous Math5 expression in these nonretinal domains have not been determined.
To elucidate the role of putative 3′ regulatory elements in the activation or restriction of Math5 expression, we compared both the retinal and non-retinal expression of the Math5-GFP1 and Math5-GFP2 transgenes. The GFP2 construct contains an additional 1.6 Kb of 3′ Math5 non-coding DNA inserted 3′ to the GFP coding sequence (Fig. 1). Since Drosophila atonal and mouse Math1 contain 3′ regulatory modules (Sun et al., 1998; Helms et al., 2000), we hypothesized that Math5 3′ DNA might be required to prohibit the ectopic expression domains observed in Math5-GFP1 transgenic embryos. However, the non-retinal expression domains of both Math5-GFP constructs are nearly identical. The GFP2 transgene is also expressed in the lower rhombic lip, pontine nucleus, spinal cord, and whisker barrels, at the same ages as Math5-GFP1 (data not shown). Unlike GFP1, Math5-GFP2 is not expressed in inner ear hair cells (Fig. 4H). This suggests the presence of an inner ear-specific repressor in the 3′ DNA.
In the course of these experiments, we observed a previously uncharacterized Math5LacZ expression domain in the cochlear nucleus of the developing hindbrain (Fig. 4I′)(Saul et al., 2007). The cochlear nucleus receives input from the spiral (cochlear) ganglion, which integrates auditory signals received by the cochlear hair cells in the Organ of Corti. Axons from the cochlear nucleus neurons travel through the trapezoid body and lateral lemniscus to auditory processing centers including the superior olivary nucleus, the nucleus of the trapezoid body, and the inferior colliculus (Cant and Benson, 2003). Therefore, RGCs and cochlear nucleus neurons are functionally similar in that they are projection neurons for the visual and auditory sense organs, respectively. We observe Math5LacZ expression in cochlear nucleus neurons (Fig. 4K′) and their axonal projections for the trapezoid body and lateral lemniscus (Fig. 4J). Interestingly, Math5-GFP1 is not coexpressed with Math5LacZ in cochlear nucleus neurons or axons from E14.5–E16.5 (Fig. 4K, K′, K″, J, not shown). Math5-GFP2 is also not expressed in the cochlear nucleus (not shown), indicating that the cochlear nucleus enhancer lies outside of the 5′ and 3′ Math5 noncoding DNA examined here. Since the GFP1 and GFP2 transgenes are expressed in identical patterns, except that GFP1 is also present in inner ear hair cells, all remaining analyses were done with Math5-GFP1.
Math5-GFP1 is expressed in a subset of Math1 expression domains
Both Math5-GFP transgenes are expressed in discrete locations outside the developing visual system. The most closely related bHLH factor to Math5 is Math1, which is expressed in several progenitor populations that give rise to components of the proprioceptive system throughout the nervous system. These include inner ear hair cells, whisker barrels, the rhombic lip, and the dorsal spinal cord (Akazawa et al., 1995; Ben-Arie et al., 2000; Bermingham et al., 2001; Machold and Fishell, 2005; Wang et al., 2005). Sensory inputs, including Merkel cells in whisker barrels and hair cells in the inner ear, transmit positional information to the brain, which is then processed in a complex circuit including the pontine nuclei, cerebellum, and cerebral cortex (Bermingham et al., 2001). Since we observed Math5-GFP expression in several regions known to express Math1, we hypothesized that Math5-GFP1 and Math1 are expressed in the same cell lineages. However, Math5-GFP1 is not expressed in every Math1 domain, since GFP expression was not found in the upper rhombic lip, which differentiates into the EGL of the cerebellum (Ben-Arie et al., 1996).
To demonstrate coexpression of Math5-GFP1 and Math1 in these regions, we performed anti-GFP, anti-Math1 double-antibody labeling experiments. Math5-GFP1 and Math1 are coexpressed in cells of the spinal cord (Fig. 5A–B″) and lower rhombic lip from E10.5–E12.5 (Fig. 5D–E″), inner ear hair cells at E14.5 (Fig. 5G–G″), and whisker barrel Merkel cells at E16.5 (Fig. 5H–H″). In hair cells and Merkel cells, Math5-GFP1 and Math1 highly overlap. However, Math5-GFP1 is expressed in only a subset of Math1+ cells in the dorsal spinal cord and lower rhombic lip (Fig. 5A″, inset 5B″,5D″, inset 5E″). At E10.5 and E12.5, Math1-expressing progenitors in the dorsal spinal cord and lower rhombic lip are located closer to the ventricular zone, while Math5-GFP1 cells lie more laterally and ventrally (Fig. 5A–B″, 5D–E″). We hypothesized that these GFP-positive cells represent migrating cells of the Math1 lineage. In the spinal cord of E12.5 Math1LacZ embryos, migrating cells that no longer express Math1 protein do express the more stable βgal reporter (Ben-Arie et al., 2000). To determine whether these GFP+/Math1− cells in the dorsal spinal cord and lower rhombic lip had previously expressed Math1, we compared Math5-GFP1 and Math1LacZ expression in E12.5 Math5-GFP1;Math1LacZ/+ embryos. Indeed, GFP and βgal are largely coexpressed in the dorsal spinal cord and lower rhombic lip (Fig. 5C–C″, 5F–F″). Together, these findings indicate Math5-GFP expression in cells of the Math1-lineage in multiple expression domains.
Fig. 5. Math5-GFP1 is expressed in cell lineages of Math1.
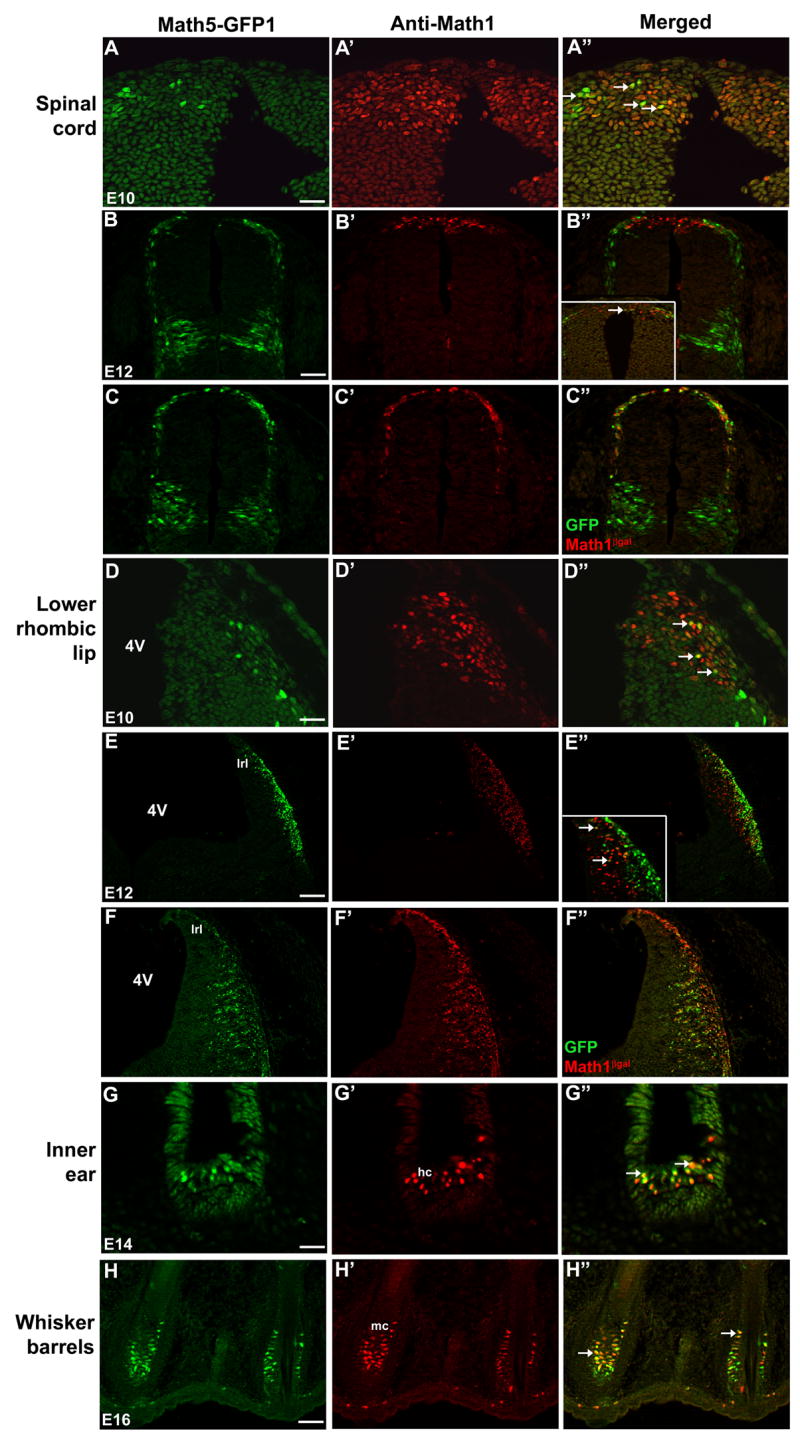
(A–B″,D–E″) Colabeling of cryosections from Math5-GFP1 embryos. GFP and Math1 proteins are coexpressed in cells of the E10.5 and E12.5 spinal cord (A–B″, arrows in A″ and B″ inset) and lower rhombic lip (D–E″, arrows in D″ and E″ inset). In E12.5 Math5-GFP1;Math1LacZ/+ embryos, the high degree of βgal-GFP co-labeling in the spinal cord (C–C″) and lower rhombic lip (F–F″) demonstrates Math5-GFP1 expression within the Math1 cell lineage. (G–H″) Math5-GFP1 and Math1LacZ are also coexpressed in hair cells of the inner ear and Merkel cells of whisker barrels (arrows in G″ and H″). Magnification bars in A,D,G (25 μm), B,C,E,F (50 μm), H (100 μm). 4V- fourth ventricle; hc- hair cells; lrl- lower rhombic lip; mc- Merkel cells.
Regulation of Math5-GFP1
Math5 and Math1 are semi-orthologues of Drosophila atonal that are expressed in non-overlapping domains in mice (Hassan and Bellen, 2000). Our observation that Math5-GFP is expressed in a subset of Math1 expression domains suggests that during vertebrate evolution one or more regulatory enhancers remain conserved between Math1 and Math5 regulatory DNA. Therefore, we investigated whether Math5-GFP1 might be regulated analogously to Math1 in the lower rhombic lip and dorsal spinal cord.
Math1 expression is controlled by two enhancers (A and B) located ~3 Kb 3′ to the Math1 translation stop site (Helms et al., 2000). These enhancers are required for the proper expression of Math1 in its normal pattern, and Math1 autoregulates its own expression in the dorsal neural tube by binding to a bHLH-specific E-box in Enhancer B (Helms et al., 2000). First, we tested whether Math1 might cross-regulate Math5-GFP1 in the lower rhombic lip. Extensive overlap of Math5-GFP and Math1LacZ was observed at E12.5 (Figs. 5F, 6A). In Math5-GFP1;Math1LacZ/LacZ mutants, GFP expression was obviously decreased in the lower rhombic lip (compare 6C,C′ to 6A). While this indicates partial cross-regulation of Math1 upon Math5-GFP1, it also implies that other factors might regulate some aspects of the Math5-GFP hindbrain domain.
Fig. 6. Math5-GFP1 expression is regulated by Pax6 and Math1 in the lower rhombic lip and spinal cord.
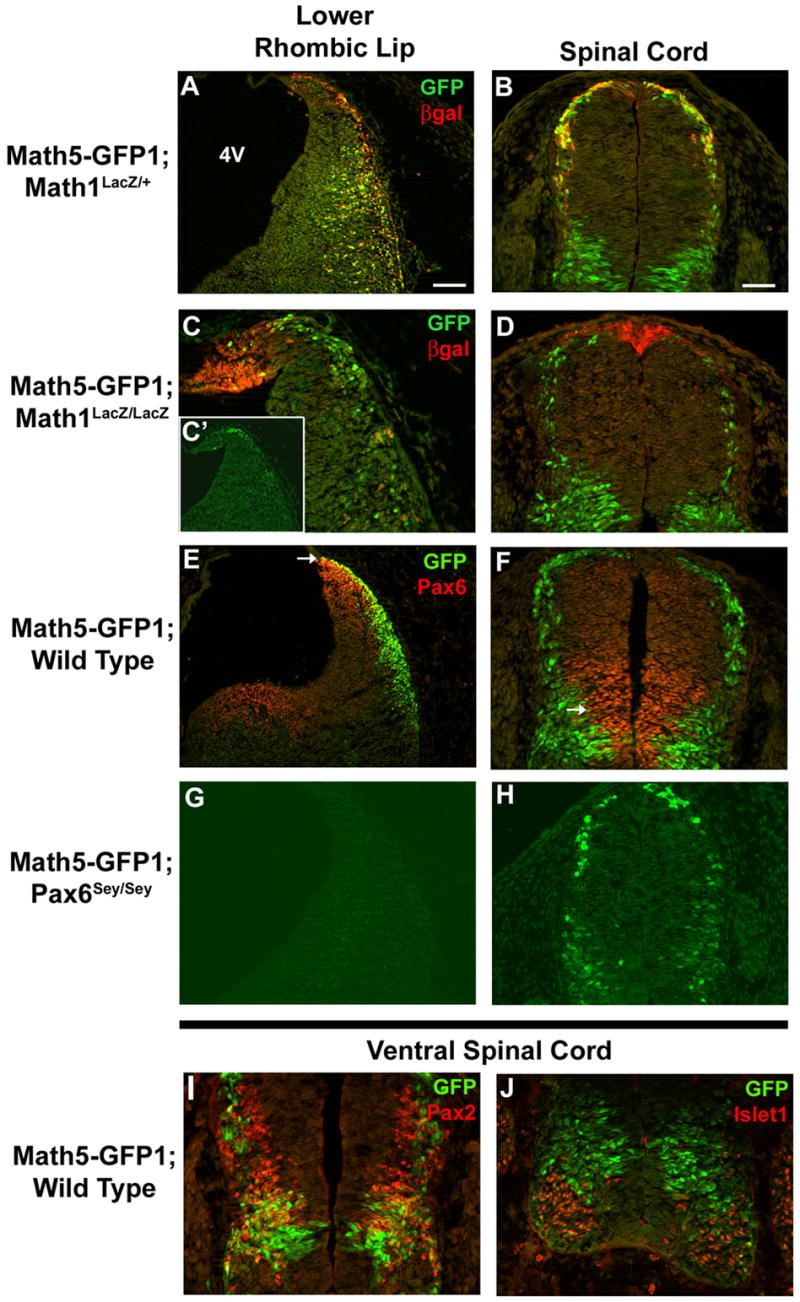
(A–D) E12.5 Math5-GFP1; Math1LacZ/+ and Math5-GFP1; Math1LacZ/LacZ embryos were assessed for GFP expression in the lower rhombic lip and dorsal spinal cord. A and B demonstrate nearly complete co-localization of GFP and βgal proteins in Math1 heterozygotes. Math5-GFP1 expression, particularly coexpression with βgal+ cells is decreased in the lower rhombic lip (C,C′) and dorsal spinal cord (D) of Math1-null embryos. C is the same magnification as A, while C′ is a higher magnification. (E–H) E12.5 Math5-GFP1;Pax6+/+ and Math5-GFP1;Pax6sey/sey mice were analyzed for GFP expression in the lower rhombic lip and dorsal spinal cord. GFP and Pax6 are coexpressed in particular neurons of the lower rhombic lip (E) and spinal cord (F) at E12.5 (arrows in E and F). In Pax6-null embryos, Math5-GFP1 expression is absent in the lower rhombic lip (G), but is expressed in the spinal cord (H). (I–J) In the ventral spinal cord, GFP-positive cells fall mainly within the Pax2 domain (I) and outside the Islet1 domain (J). Magnification bars in A (100 μm) and B (50 μm). 4V-fourth ventricle.
Math1 expression is also Pax6-dependent in the lower rhombic lip (Walther and Gruss, 1991; Landsberg et al., 2005), making Pax6 an attractive candidate to also regulate Math5-GFP. In the E12.5 wild type rhombic lip, Math5-GFP1- and Pax6-expressing cells exhibit some overlap (Fig. 6E). Therefore, we scrutinized Math5-GFP1 expression in E12.5 Math5-GFP1;Pax6Sey/Sey embryos, to determine if those Math5-GFP cells that do not require Math1 might instead require Pax6. Indeed, Math5-GFP1 expression is absent in the lower rhombic lip of Pax6-null mice (Fig. 6G). Together these data suggest that Math5-GFP transgenes have overlapping, but genetically separable, regulation by Math1 and Pax6 in the lower rhombic lip.
Then we examined Math5-GFP1 transgene regulation in the spinal cord. Again we tested for cross-regulation by Math1, since here we also found Math5-GFP coexpression with Math1LacZ (Figs. 5D, 6B). In the dorsal spinal cord of Math5-GFP1;Math1LacZ/LacZ mutant embryos, some Math5-GFP cells were still observed, although greatly reduced in number in the dorsal-most region (Fig. 6D). Furthermore, co-localization of GFP and βgal was nearly absent (compare Fig. 6D to 6B), indicating the loss of Math5-GFP1 expression specifically in the Math1-lineage. Although only a few spinal cord cells coexpress Pax6 and Math5-GFP1 (Fig. 6F), we compared Pax6 spinal cord regulation to compare this outcome with that for the hindbrain. In Pax6Sey/Sey embryos, Math5-GFP1 was still expressed throughout the spinal cord, although the number of GFP+ cells appeared somewhat reduced ventrally (Fig. 6H). To define the Math5-GFP1 ventral spinal cord domain better, we compared GFP expression to that of the transcription factors Pax2 and Islet1, which mark clusters of ventral interneurons and motor neurons, respectively. In the ventral spinal cord, Math5-GFP1 largely colocalized with Pax2 (Fig. 6I), but exhibited no coexpression with Islet1 (6J). Pax6 is required for the generation of a population of Pax2+/En1+ ventral spinal cord interneurons (Burrill et al., 1997), meaning the loss of Pax6 may indirectly reduce the number of Math5-GFP1 cells in this domain. Overall, we conclude that Math5-GFP is regulated by both Math1 and Pax6 in the lower rhombic lip of the hindbrain, but only Math1-dependent in the dorsal spinal cord.
Finally, we asked whether Math5 and Math1 retain conserved nucleotide sequences by comparing Math5 5′ and Math1 3′ noncoding DNA. Figure 7A diagrams both Math5 5′ 2.1 Kb and Math1 3′ 4.5 Kb regulatory DNA. In the upstream region of Math5, two highly conserved regions among the Xenopus, mouse and human Ath5 genes are shown (yellow boxes in Fig. 7A)(Brown et al., 2002; Hutcheson et al., 2005; Riesenberg et al., 2007). Within these two evolutionarily conserved regions are four E-boxes, bHLH consensus binding sites (blue boxes) (Murre et al., 1989; Hutcheson et al., 2005). For Math1, Helms et al. (2000) demonstrated that two enhancers (A and B, located 3 Kb downstream) activate Math1 expression (green boxes in Figs. 7A,B). As mentioned, Math1 positively autoregulates its expression in the lower rhombic lip and spinal cord through one E-box binding site in Enhancer B (blue box). In addition, putative Pax6 paired-domain binding sites in Math5 and Math1 regulatory DNA were identified using the Transfac MATCH program (black and grey boxes in Figs. 7A,B). Overall, 20 putative Pax6 sites are predicted in 3 Kb upstream of the Math5 start codon (Riesenberg et al., 2007), but only those sites relevant for comparison with Math1 regulatory DNA are shown here (Figs. 7A–C). Math5 site 5-J, shown in black, is highly conserved among at least four vertebrate species and specifically binds Pax6 protein in vitro (Riesenberg et al., 2007). Additional predicted Pax6 binding sites depicted in grey for Math5 and Math1 DNA are listed in Figure 7C, along with their Transfac core score and the prediction matrix used.
Fig. 7. Comparative analyses of Math5 and Math1 regulatory DNA.
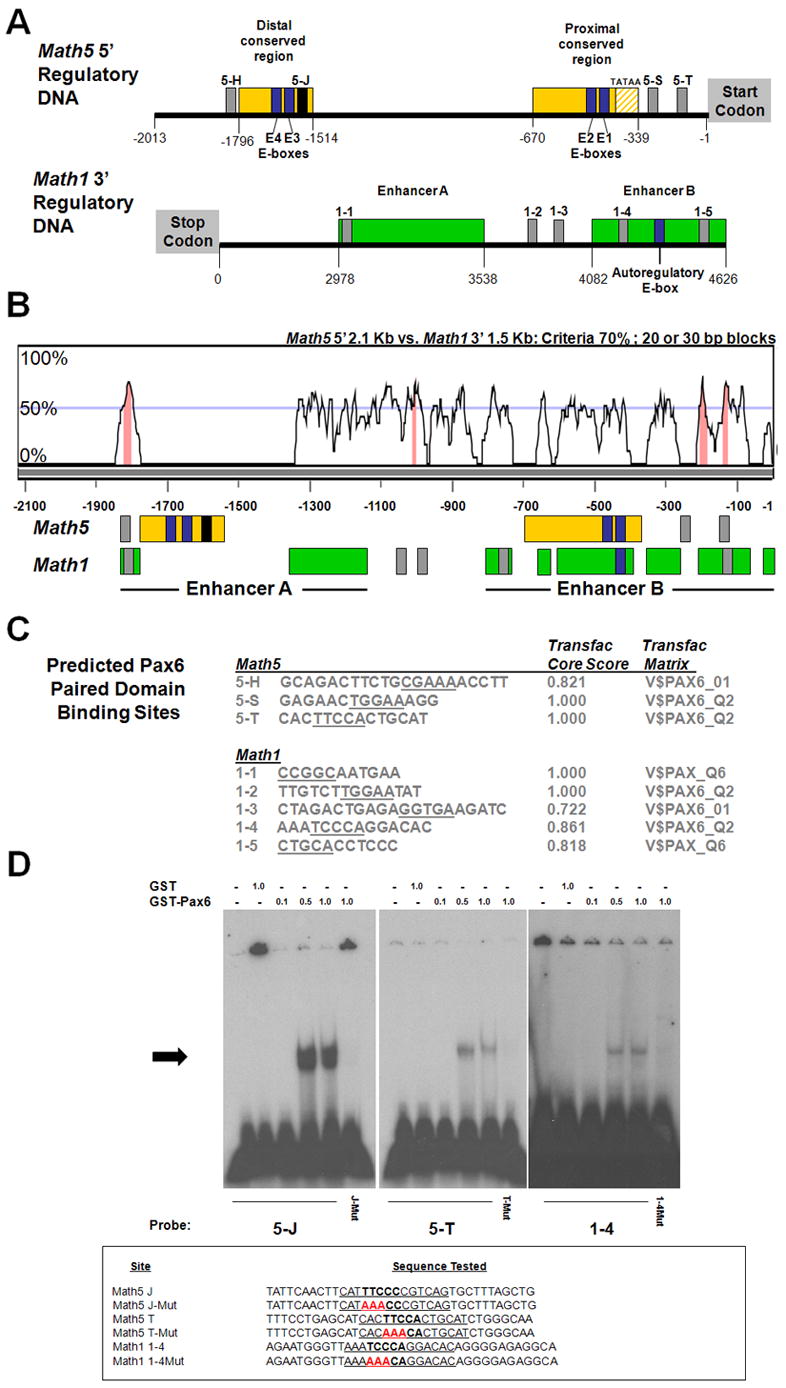
(A) Two phylogenetically conserved regions (yellow boxes) in 2.1 Kb of Math5 DNA 5′ to the start codon. Blue boxes E1–E4 represent four conserved E-box binding sites reported in Hutcheson et al. (2005) and Riesenberg et al. (2007). The black box 5-J denotes a functional Pax6 binding site (Riesenberg et al., 2007 and Panel D). Grey boxes 5-H, 5-S, and 5-T are putative Pax6 binding sites in Math5 5′ regulatory DNA. The two 3′ Math1 enhancers are located ~3 Kb downstream of the Math1 stop codon. The Math1 auto-regulatory E-box resides in Enhancer B (Helms et al., 2000). Grey boxes 1–1 to 1–5 are predicted Pax6 binding sites in Math1 3′ DNA. (B) VISTA analysis comparing 2.1 Kb of Math5 5′ DNA (X-axis) with the 1.6 Kb Math1 3′ enhancers (Y-axis), utilizing a 20 bp calculation window. Several regions contain ≥70% nucleotide identity (pink shading). Immediately below, the position of Math1 and Math5 enhancers and putative Pax6 or bHLH binding sites is indicated, including gaps of nonalignment. Notably, the Math5 E1 and Math1 auto-regulatory E-boxes align, while the Math5 distal conserved region (containing a retinal enhancer) lies within a stretch of nonalignment between Math5 and Math1. (C) The nucleotide sequences of predicted Pax6 binding sites, with core nucleotides underlined, along with the Transfac Core score and the prediction matrix used. (D) EMSA of GST-Pax6 paired domain fusion protein with binding sites 5-J, 5-T, and 1–4, with 5-J serving as a positive control. For each binding site, the left lane contains free annealed ds probe, the second lane probe and 1 μg GST protein, and the next three lanes probe and 0.1, 0.5 or 1.0 μg of GST-Pax6. Pax6 binding to site 1–4 is the weakest, since this gel shift was exposed to x-ray film 5 times longer than the others. However, specific binding is lost at all three sites when 3/5 core nucleotides are mutated (red bases in J-Mut, T-Mut, 1-4Mut).
To compare these Math5 and Math1 noncoding sequences directly, we performed a VISTA alignment of the Math5 5′ 2.1 Kb and Math1 3′ 1.5 Kb sequences (Fig. 7B). Many regions exhibited >50% identity, using 20 bp calculation windows. The relevant regulatory regions and putative binding sites from panel A are shown under the graphical depiction of this nucleotide alignment. Across this alignment four stretches of DNA contain ≥70% nucleotide identity. Interestingly, the Math5 distal conserved region, containing a retinal enhancer (Hutcheson et al., 2005; Riesenberg et al., 2007), lies within a long stretch of nonalignment between Math5 and Math1. Conversely, the most proximal Math5 E-box (E1) aligns with the Math1 autoregulatory E-box (Fig. 7B). This shared feature could account for conserved expression between Math1 and Math5-GFP1, via cross-regulation by Math1. Intriguingly, two pairs of predicted Pax6 binding sites (5-H to 1–1 and 5-T to 1–5) appear to be conserved between Math1 and Math5 regulatory DNA (Fig. 7B).
To test whether Pax6 can bind any of the predicted sites (grey boxes) in vitro, electrophoretic mobility shift assays (EMSAs), using a GST-Pax6 paired domain fusion protein, were performed on predicted Math5 sites 5-H, 5-S, 5-T and all five Math sites, using Math5 site 5-J as a positive control (Fig. 7D). Only sites 5-T and 1–4 were specifically bound by 0.5–1 μg Pax6 protein (Fig. 7D). When three of five core nucleotides were mutated within each site, Pax6 paired domain binding was completely lost (Fig. 7D). While the two pairs of putative Pax6 binding sites that aligned between Math5 and Math1 do not appear functional, both semi-orthologues are directly regulated by Pax6. In conclusion, we demonstrate correlation between in vivo regulation of Math5-GFP1 and Math1 (by Pax6 and Math1) with bioinformatic and in vitro protein-DNA binding data.
Discussion
A new tool for the study of RGC axon outgrowth and visual system patterning
In this paper, we examined Math5 gene regulation in vivo using transgenic GFP reporter mice. We observed that Math5-GFP transgenes delineate migrating RGC axons from the developing retina into the brain. At E12.5, one day after RGC differentiation begins, we observed RGCs axons extended outside the eye. By 16.5 GFP-labeled RGC axons had arrived at their two major targets, the lateral geniculate nucleus and superior colliculus. Our transgenic mouse model offers multiple advantages for observing RGC axon outgrowth, including high specificity, in vivo labeling, and live fluorescence. In Math5-GFP embryos, RGC axons travel through developing brain tissue devoid of other GFP-expressing domains, allowing us to trace optic projections with great certainty. Because GFP expression is intrinsic within the axons, specificity is greater than either anteriograde or retrograde RGC axon labeling techniques, and no surgical manipulations are needed. Finally, Math5-GFP expression in RGCs occurs in vivo during axonal outgrowth, guidance, and synaptogenesis, allowing for real-time visualization of these processes. In the future, Math5-GFP mice will be used to time-lapse image RGC outgrowth in retinal explants, retinal flat mount cultures and retinal-brain cocultures. We conclude that these transgenic mice are a valuable tool for understanding RGC axon pathfinding and visual system innervation of the developing brain.
What restricts Math5 expression to the developing retina and cochlear nucleus?
The upstream 2.1 Kb of Math5 noncoding DNA contains a retinal enhancer, but transgenic mice containing this DNA show no reporter expression in the cochlear nucleus, an endogenous expression domain of Math5 (Riesenberg et al., 2007; Saul et al., 2007). Therefore the cochlear nucleus enhancer is located in more distal Math5 regulatory DNA. Instead, we observed multiple ectopic Math5-GFP transgenic expression domains, at times and places where endogenous Math5 is not expressed. We did not anticipate that these upstream Math5 DNA sequences are capable of driving GFP reporter expression in the lower rhombic lip, spinal cord, inner ear, whisker barrels, pontine nucleus, molars, pineal gland, and neocortex. Importantly, none of these ectopic expression domains were found in multiple Math5 lineage studies (Yang et al., 2003; Brzezinski, 2005; Saul et al., 2007), in Math5LacZ targeted deletion mice (Brown et al., 2001; Wang et al., 2001), or by extensive in situ hybridization experiments analyzing Math5 mRNA expression from E10.5 to birth (Brown et al., 1998; Brown et al., 2001). Because both Math5-GFP transgenes display the same deregulation, the regulatory DNA responsible for suppressing endogenous Math5 must be more distal. Interestingly, the addition of the 1.6 Kb of 3′ Math5 noncoding DNA in Math5-GFP2 was sufficient to silence ectopic GFP expression in inner ear hair cells, suggesting that the elements that keep Math5 off in Math1 domains may reside 3′ to the Math5 coding exon. Future experiments will test additional Math5 5′ and 3′ DNA to identify the cochlear nucleus enhancer and those elements that normally mask Math1-like expression.
Regulatory conservation in the evolutionary divergence of bHLH factors
The bHLH protein domains of Math5 and Math1 are as closely related to each other as they are to that of Drosophila atonal (Brown et al., 1998; Hassan and Bellen, 2000). In the fly, atonal is expressed in the developing eye and specifies the first retinal neuron, R8 (Jarman et al., 1993). Both the fly R8 photoreceptor neuron and vertebrate RGC are the first retinal neurons to differentiate and project axons that innervate the brain. Atonal is also expressed in chordotonal organs, which process proprioceptive information during larval movement, and in Johnston’s organ that functions analogously to the mammalian auditory system (Jarman et al., 1993). Math1 is expressed in several components of the proprioceptive pathway in mouse, and in auditory hair cells of the inner ear (Bermingham et al., 1999; Bermingham et al., 2001). Here we report Math5LacZ expression in the cochlear nucleus, a component of the auditory system. In this manner, the functions of atonal appear to have been divided between Math5 and Math1, with Math5 in vision, Math1 in the proprioceptive system, and both Math5 and Math1 in auditory processing.
Our Math5 transgenes are expressed in multiple Math1 domains in the auditory and proprioceptive systems. Math5-GFP1 is expressed in Math1-lineages of the spinal cord, lower rhombic lip, inner ear hair cells, and whisker barrel Merkel cells. Moreover, Math5-GFP1 is regulated in a similar manner to Math1. Math1 is regulated by Pax6 in the lower rhombic lip and autoregulated in the dorsal neural tube (Helms et al., 2000; Landsberg et al., 2005). Similarly, Math5-GFP1 expression is absent in the lower rhombic lip of Pax6-null embryos, and coexpression of Math5-GFP and Math1LacZ is reduced in the lower rhombic lip and dorsal spinal cord of Math1-null embryos. These changes are unlikely caused by the loss of a progenitor cell population, as Math1-expressing cells likely remain undifferentiated or switch fates in the absence of Math1 in the rhombic lip and spinal cord, or Pax6 in the rhombic lip (Ben-Arie et al., 1997; Ben-Arie et al., 2000; Bermingham et al., 2001; Landsberg et al., 2005; Machold and Fishell, 2005; Wang et al., 2005). Alternatively, the effects of Pax6 loss on Math5-GFP lower rhombic lip expression might be due to the loss of Math1. However, unlike Math5-GFP1, Math1 is reduced but not absent in the rhombic lip of Pax6-nulls, indicating that Pax6 regulates Math5-GFP1 in part via a Math1-independent mechanism. All together, regulatory similarities found between Math5-GFP1 and Math1 further indicate that Pax6 regulation and autoregulation are well-conserved features of the atonal gene family (Sun et al., 1998; Zhang et al., 2006).
Math1 expression is largely controlled by two 3′ enhancers, identified by their ability to direct LacZ reporter gene expression in vivo (Helms et al., 2000). Subsequently, a slightly smaller 3′ DNA fragment, containing most, but not all, of the Math1 enhancers was used to create a Math1-GFP transgene (Lumpkin et al., 2003). Intriguingly, Math1-GFP expression was reported in non-Math1 domains, including the retina, suggesting the unmasking of a retinal Math1 enhancer in this particular GFP transgenic construct. That both the Math5-GFP1 and Math1-GFP2 transgenes are capable of expression in the reciprocal gene’s pattern further suggests that the Math5 5′ and Math1 3′ regulatory DNA retain conserved regulatory motifs. By contrast, Math5-GFP1 is also expressed in regions not associated with endogenous Math1 expression, including the ventral spinal cord, forebrain, pineal gland, and molars. As such, the Math5 5′ DNA may contain pan-proneural activation elements in common with other bHLH factors, such as Mash1, Ngn1, Ngn2, or NeuroD. Interestingly, all of these genes, except NeuroD, genetically require Pax6 (Marquardt et al., 2001; Blader et al., 2004). The activation of Ngn1 and Ngn2 is directly regulated by Pax6 binding to particular CNS enhancers (Marquardt et al., 2001; Scardigli et al., 2003; Blader et al., 2004).
Our bioinformatic and EMSA analyses of these regulatory sequences yielded several interesting findings. First, the distal conserved region of Math5, which contains a retinal enhancer (Riesenberg et al., 2007) does not align to the Math1 enhancer sequence, suggesting that the retinal enhancer for Math5 may have been added after these genes duplicated and diverged, or it was subsequently lost from Math1. Second, the Math1 autoregulatory E-box, that maintains expression in the dorsal spinal cord, and the Ath5 E1 E-box, required for maintenance of retinal expression in frog and chick (Skowronska-Krawczyk et al., 2004; Hutcheson et al., 2005), align to one another, providing one explanation for Math1 cross-regulation of our transgenes. Finally, we demonstrate functional Pax6 binding sites for both Math5 and Math1, which could account for the similar regulation of Math5-GFP and Math1 by this factor. Although the other predicted Pax6 binding sites do not demonstrate Pax6 binding, some may yet turn out to be functional, but require tissue-specific cofactors absent from our in vitro experiments.
The duplication and divergence of Math5 and Math1 during vertebrate evolution resulted in tissue-specific, restricted expression of each gene that, when added together, recapitulates the expression domains of Drosophila atonal. As the vertebrate nervous system expanded and elaborated, these genes became segregated to mammalian visual, auditory, and proprioceptive systems. We propose one mechanism of evolutionary divergence may have occurred at the level of cis-regulation, where common enhancer elements within the Math1 and Math5 genes became silenced by an as yet unknown repressive mechanism. It is possible that these elements are also present in other vertebrate bHLH atonal gene homologues. Future elucidation of gene-specific repressor sequences, the factors that bind to them, and the overall mechanism by which semi-orthologous genes develop complementary expression patterns will contribute important information to the evolution of the vertebrate nervous system.
Experimental Methods
Generation of transgenic mice
Math5-GFP constructs and transgenic mice were generated as described (Hutcheson et al., 2005; Riesenberg et al., 2007). Transgenic mice are maintained in a CD-1 background. Math5-GFP1 and Math5-GFP2 were crossed with Math5LacZ/+, Math1LacZ/+, or Pax6Sey/+ mice to compare Math5-GFP expression to that of the Math5LacZ and Math1LacZ reporters and to assess changes in GFP expression in Math5, Math1, or Pax6 mutant embryos. Mouse embryos were harvested from timed pregnancies for GFP imaging, cryosectioning, and immunofluorescence (see below), with the observed plug date taken as E0.5. A minimum of three embryos from at least two litters was used for each experiment. Genotyping for Math5-GFP, Math5LacZ, Math1LacZ and Pax6Sey embryos or adult mice was performed by PCR as described (Brown et al., 1998; Ben-Arie et al., 2000; Brown et al., 2001; Hutcheson et al., 2005).
GFP imaging and immunofluorescence
Embryos were maintained in cold PBS for whole-mount imaging on a Leica MZ-FLIII dissecting microscope equipped with a GFP fluorescence lamp, digital camera, and Magnafire software. For immunofluorescence, embryos were fixed for 1–2 hours in 4% PFA/PBS at 4°C, cryoprotected in 5% and 15% sucrose/PBS, embedded in OCT, and cryosectioned in 10μm increments. Primary antibodies used include rabbit anti-GFP Alexa-Fluor 488 (1:500–1:1000, Molecular Probes), rabbit anti-βgal (1:5000, ICN), rabbit anti-Math1 (1:250, gift from Jane Johnson), rabbit anti-Pax6 (1:1000, Covance), rabbit anti-Pax2 (1:100, Covance), mouse anti-Islet1 (DSHB), and DAPI (1:500). Secondary antibodies used include goat anti-rabbit IgG Alexa-Fluor 594 (1:2000, Molecular Probes), biotinylated donkey anti-rabbit (1:200, Jackson), and streptavidin Texas Red (1:200, Jackson). For antibody experiments in which the direct conjugate rabbit anti-GFP antibody and another rabbit primary antibody were employed, potential cross-reactivity was eliminated by incubating the slides in 10% rabbit serum/TST for 2–3 hours. This step was performed after the final amplification of the rabbit primary antibody and before applying the anti-GFP antibody. Images were generated on a Zeiss Axioplan 2 fluorescent microscope with an Apotome deconvolution device and Axiovision software.
Sequence analysis
Noncoding nucleotide sequences for Math5 (NCBI accession #AAF418923) and Math1 (NCBI accession #AF218258) were obtained from NCBI. The 5′ 2.1 Kb sequence was extracted from a larger Math5 sequence. The 5′ 2.1 Kb sequence was aligned with the 3′ Math1 enhancer sequence using the VISTA program (http://genome.lbl.gov/vista). The VISTA alignment was assessed for consensus identity using a calculation window of 20 or 30 bp, and consensus identity defined as 70%. Potential Pax6 binding site sequences were predicted using the Transfac® (http://www.biobase-international.com) MATCH™ (Matrix Search for Transcription Factor Binding Sites) program version 10.3 and matrices M00979 (V$PAX6_Q2), M00097 (V$PAX6_01), and M00808 (V$PAX6_06). The black box in the Math5 distal conserved region represents a conserved binding site 5-J, initially tested in Riesenberg et al (2007). All other putative binding sites (shown in grey) were predicted using any of the three matrices, and selected based on their Core score and potential evolutionary conservation between Math5 and Math1 as shown in Figure 7. The E-box binding sites in the Math5 and Math1 sequences have been previously tested in vivo (Helms et al., 2000; Hutcheson et al., 2005).
Electrophoretic Mobility Shift Assay (EMSA)
GST and GST-Pax6 paired domain proteins (Epstein et al., 1994), were purified from BL21 bacterial lysates by incubation with glutathione agarose beads (Sigma) for 1 hour at 4°C, washed in PBS, eluted with 25 mM glutathione/0.1M Tris pH 8 and dialyzed into 50 mM Tris pH 7.5, 250 mM NaCl, 5 mM MgCl2, 2.5 mM EDTA, 20% glycerol. Gel-shift reactions used a 5X binding buffer (50 mM Tris pH 7.5, 250 mM NaCl, 5 mM MgCl2, 2.5 mM EDTA, 2.5 mM DTT, 0.25 mg/ml poly-dI-dC, and 20% glycerol). 20 μl reactions contained 4 μl of 5X binding buffer, 0.1, 0.5 or 1 μg of recombinant protein and 75 fmol of γ32P end-labeled, annealed, double-stranded oligonucleotides (400,000 Cerenkov counts per reaction). After DNA probe addition, reactions were incubated for 20 minutes at room temperature and run on a 4% polyacrylamide gel in 0.5X Tris borate-EDTA buffer, gels were then dried and exposed to x-ray film.
Acknowledgments
This work was supported by NIH Grant EY13612. We thank Jane Johnson for the gift of anti-Math1 polyclonal antibody; Huda Zoghbi, Kate Barald, and Tom Glaser for Math1LacZ mice and a GST-Pax6 paired domain construct; Tom Glaser and Brian Gebelein for advice regarding EMSA assays; Kenny Campbell, Richard Lang, and April Smith for technical advice; and Kenny Campbell, Tiffany Cook, and Masato Nakafuku for critical comments on this manuscript.
Abbreviations
- 4V
fourth ventricle
- cn
cochlear nucleus
- gcl
ganglion cell layer
- hc
inner ear hair cell
- lg
lateral geniculate nucleus
- ll
lateral lemniscus
- lrl
lower rhombic lip
- mc
Merkel cells
- on
optic nerve
- oc
optic chiasm
- ot
optic tract
- pn
pontine nucleus
- sc
superior colliculus
- tb
trapezoid body
- wb
whisker barrels
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akazawa C, Ishibashi M, Shimizu C, Nakanishi S, Kageyama R. A mammalian helix-loop-helix factor structurally related to the product of Drosophila proneural gene atonal is a positive transcriptional regulator expressed in the developing nervous system. J Biol Chem. 1995;270:8730–8738. doi: 10.1074/jbc.270.15.8730. [DOI] [PubMed] [Google Scholar]
- Alder J, Cho NK, Hatten ME. Embryonic precursor cells from the rhombic lip are specified to a cerebellar granule neuron identity. Neuron. 1996;17:389–399. doi: 10.1016/s0896-6273(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, McCall AE, Berkman S, Eichele G, Bellen HJ, Zoghbi HY. Evolutionary conservation of sequence and expression of the bHLH protein Atonal suggests a conserved role in neurogenesis. Hum Mol Genet. 1996;5:1207–1216. doi: 10.1093/hmg/5.9.1207. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Ben-Arie N, Hassan BA, Bermingham NA, Malicki DM, Armstrong D, Matzuk M, Bellen HJ, Zoghbi HY. Functional conservation of atonal and Math1 in the CNS and PNS. Development. 2000;127:1039–1048. doi: 10.1242/dev.127.5.1039. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Wang VY, Fernandez M, Banfi S, Bellen HJ, Fritzsch B, Zoghbi HY. Proprioceptor pathway development is dependent on Math1. Neuron. 2001;30:411–422. doi: 10.1016/s0896-6273(01)00305-1. [DOI] [PubMed] [Google Scholar]
- Bermingham NA, Hassan BA, Price SD, Vollrath MA, Ben-Arie N, Eatock RA, Bellen HJ, Lysakowski A, Zoghbi HY. Math1: an essential gene for the generation of inner ear hair cells. Science. 1999;284:1837–1841. doi: 10.1126/science.284.5421.1837. [DOI] [PubMed] [Google Scholar]
- Blader P, Lam CS, Rastegar S, Scardigli R, Nicod JC, Simplicio N, Plessy C, Fischer N, Schuurmans C, Guillemot F, Strahle U. Conserved and acquired features of neurogenin1 regulation. Development. 2004;131:5627–5637. doi: 10.1242/dev.01455. [DOI] [PubMed] [Google Scholar]
- Brown NL, Patel S, Brzezinski J, Glaser T. Math5 is required for retinal ganglion cell and optic nerve formation. Development. 2001;128:2497–2508. doi: 10.1242/dev.128.13.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Dagenais SL, Chen CM, Glaser T. Molecular characterization and mapping of ATOH7, a human atonal homolog with a predicted role in retinal ganglion cell development. Mamm Genome. 2002;13:95–101. doi: 10.1007/s00335-001-2101-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Kanekar S, Vetter ML, Tucker PK, Gemza DL, Glaser T. Math5 encodes a murine basic helix-loop-helix transcription factor expressed during early stages of retinal neurogenesis. Development. 1998;125:4821–4833. doi: 10.1242/dev.125.23.4821. [DOI] [PubMed] [Google Scholar]
- Brzezinski JA. Human Genetics. Ann Arbor, MI: University of Michigan; 2005. The Role of Math5 in Retinal Development. [Google Scholar]
- Burrill JD, Moran L, Goulding MD, Saueressig H. PAX2 is expressed in multiple spinal cord interneurons, including a population of EN1+ interneurons that require PAX6 for their development. Development. 1997;124:4493–4503. doi: 10.1242/dev.124.22.4493. [DOI] [PubMed] [Google Scholar]
- Cant NB, Benson CG. Parallel auditory pathways: projection patterns of the different neuronal populations in the dorsal and ventral cochlear nuclei. Brain Res Bull. 2003;60:457–474. doi: 10.1016/s0361-9230(03)00050-9. [DOI] [PubMed] [Google Scholar]
- Cepko CL. The roles of intrinsic and extrinsic cues and bHLH genes in the determination of retinal cell fates. Curr Opin Neurobiol. 1999;9:37–46. doi: 10.1016/s0959-4388(99)80005-1. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelkamp D, Rashbass P, Seawright A, van Heyningen V. Role of Pax6 in development of the cerebellar system. Development. 1999;126:3585–3596. doi: 10.1242/dev.126.16.3585. [DOI] [PubMed] [Google Scholar]
- Epstein J, Cai J, Glaser T, Jepeal L, Maas R. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994;269:8355–8361. [PubMed] [Google Scholar]
- Farago AF, Awatramani RB, Dymecki SM. Assembly of the brainstem cochlear nuclear complex is revealed by intersectional and subtractive genetic fate maps. Neuron. 2006;50:205–218. doi: 10.1016/j.neuron.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Hassan BA, Bellen HJ. Doing the MATH: is the mouse a good model for fly development? Genes Dev. 2000;14:1852–1865. [PubMed] [Google Scholar]
- Helms AW, Abney AL, Ben-Arie N, Zoghbi HY, Johnson JE. Autoregulation and multiple enhancers control Math1 expression in the developing nervous system. Development. 2000;127:1185–1196. doi: 10.1242/dev.127.6.1185. [DOI] [PubMed] [Google Scholar]
- Hutcheson DA, Hanson MI, Moore KB, Le TT, Brown NL, Vetter ML. bHLH-dependent and -independent modes of Ath5 gene regulation during retinal development. Development. 2005;132:829–839. doi: 10.1242/dev.01653. [DOI] [PubMed] [Google Scholar]
- Jarman AP, Grau Y, Jan LY, Jan YN. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell. 1993;73:1307–1321. doi: 10.1016/0092-8674(93)90358-w. [DOI] [PubMed] [Google Scholar]
- Landsberg RL, Awatramani RB, Hunter NL, Farago AF, DiPietrantonio HJ, Rodriguez CI, Dymecki SM. Hindbrain rhombic lip is comprised of discrete progenitor cell populations allocated by Pax6. Neuron. 2005;48:933–947. doi: 10.1016/j.neuron.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Le TT, Wroblewski E, Patel S, Riesenberg AN, Brown NL. Math5 is required for both early retinal neuron differentiation and cell cycle progression. Dev Biol. 2006 doi: 10.1016/j.ydbio.2006.03.055. [DOI] [PubMed] [Google Scholar]
- Lee HY, Wroblewski E, Philips GT, Stair CN, Conley K, Reedy M, Mastick GS, Brown NL. Multiple requirements for Hes 1 during early eye formation. Dev Biol. 2005;284:464–478. doi: 10.1016/j.ydbio.2005.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Lumpkin EA, Collisson T, Parab P, Omer-Abdalla A, Haeberle H, Chen P, Doetzlhofer A, White P, Groves A, Segil N, Johnson JE. Math1-driven GFP expression in the developing nervous system of transgenic mice. Gene Expr Patterns. 2003;3:389–395. doi: 10.1016/s1567-133x(03)00089-9. [DOI] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB, et al. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Riesenberg AN, Le TT, Willardsen MI, Blackburn DC, Spencer ML, Vetter ML, Brown NL. Initiation of mouse retinal neurogenesis via Pax6 regulation of Math5. 2007 doi: 10.1002/dvg.20479. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodieck RW. The First Steps in Seeing. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- Saul SM, Altschuler RA, Shore SE, Kabara LL, Halsey KE, Glaser T. Math5 expression and function in the central auditory system. 2007 doi: 10.1016/j.mcn.2007.09.006. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scardigli R, Baumer N, Gruss P, Guillemot F, Le Roux I. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development. 2003;130:3269–3281. doi: 10.1242/dev.00539. [DOI] [PubMed] [Google Scholar]
- Schambra UB, Lauder JM, Silver J. Atlas of the Prenatal Mouse Brain. San Diego, California: Acadmic Press, Inc; 1992. [Google Scholar]
- Skowronska-Krawczyk D, Ballivet M, Dynlacht BD, Matter JM. Highly specific interactions between bHLH transcription factors and chromatin during retina development. Development. 2004;131:4447–4454. doi: 10.1242/dev.01302. [DOI] [PubMed] [Google Scholar]
- Sun Y, Jan LY, Jan YN. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development. 1998;125:3731–3740. doi: 10.1242/dev.125.18.3731. [DOI] [PubMed] [Google Scholar]
- Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang VY, Rose MF, Zoghbi HY. Math1 expression redefines the rhombic lip derivatives and reveals novel lineages within the brainstem and cerebellum. Neuron. 2005;48:31–43. doi: 10.1016/j.neuron.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ding K, Pan L, Deng M, Gan L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev Biol. 2003;264:240–254. doi: 10.1016/j.ydbio.2003.08.005. [DOI] [PubMed] [Google Scholar]
- Zhang T, Ranade S, Cai CQ, Clouser C, Pignoni F. Direct control of neurogenesis by selector factors in the fly eye: regulation of atonal by Ey and So. Development. 2006;133:4881–4889. doi: 10.1242/dev.02669. [DOI] [PubMed] [Google Scholar]