Abstract
Nucleotide excision repair (NER) is a conserved DNA repair mechanism capable of removing a variety of helix-distorting DNA lesions. Rad26, a member of the Swi2/Snf2 superfamily of proteins, has been shown to be involved in a specialized NER process called transcription coupled NER. Rad16, another member of the same protein superfamily, has been shown to be required for genome-wide NER. Here we show that Rad16 and Rad26 play different roles in repairing repressed and actively transcribed genes in yeast. Rad16 is partially dispensable, and Rad26 plays a significant role in repairing certain regions of the repressed GAL1-10, PHO5 and ADH2 genes, especially in the core DNA of well positioned nucleosomes. Simultaneous elimination of Rad16 and Rad26 results in no detectable repair in these regions of the repressed genes. Transcriptional induction of the GAL1-10 genes abolishes the role of Rad26, but does not affect the role of Rad16 in repairing the nontranscribed strand of the genes. Interestingly, when the transcription activator Gal4 is eliminated from the cells, Rad16 becomes partially dispensable and Rad26 plays a significant role in repairing both strands of the GAL1-10 genes even under inducing conditions. Our results suggest that Rad16 and Rad26 play different and, to some extent, complementary roles in repairing both strands of repressed genes, although the relative contributions of the two proteins can be different from gene to gene, and from region to region of a gene. However, Rad16 is solely responsible for repairing the nontranscribed strand of actively transcribed genes.
Keywords: nucleosome, nucleotide excision repair, Rad16, Rad26, transcription coupled, Saccharomyces cerevisiae
1. Introduction
Nucleotide excision repair (NER) is a conserved DNA repair mechanism that removes a wide range of bulky DNA lesions, including UV induced cyclobutane pyrimidine dimers (CPDs) and 6-4 photoproducts [for a review see [1]]. NER is a multistep reaction and requires the coordinated action of about 30 proteins implicated in damage recognition, helix opening, lesion verification, dual incision of the damaged strand bracketing the lesion, removal of an oligonucleotide containing the lesion, gap-filling DNA synthesis, and ligation [2].
A specialized NER pathway, called transcription coupled NER (TC-NER), refers to preferential repair in the transcribed strand (TS) of an actively transcribed gene. A transcribing RNA polymerase complex stalled at a DNA lesion on the TS may serve as a signal for rapidly recruiting NER machinery [2]. The genome-wide NER process is sometimes termed as global genomic NER (GG-NER), to be distinguished from the process of TC-NER. Most NER proteins are shared by GG-NER and TC-NER. However, some factors have been shown to be specifically required for GG-NER or TC-NER. In Saccharomyces cerevisiae, Rad7 and Rad16, components of a complex that has DNA-dependent ATPase activity [3], have been shown to be essential for repairing the nontranscribed strand (NTS) of actively transcribed genes, but dispensable for TC-NER [4]. Rad16 is a member of the Swi2/Snf2 superfamily of proteins, many of which are involved in remodeling of chromatin structure [5,6]. Rad7 and Rad16 appear to be present in different protein complexes. A stable heterotrimeric complex of Rad7/Rad16/Abf1 (replicating sequence binding factor 1) was isolated from yeast [7,8]. This heterotrimeric complex generates superhelicity in DNA through the catalytic activity of the Rad16 component. It was proposed that the torsion generated in the DNA by this complex is necessary to remove the damage-containing oligonucleotide during NER [7,8]. Recently, it was found that Rad7 and Rad16 are also components of an E3 ubiquitin ligase complex [9,10].
In mammalian cells, Cockayne syndrome (CS) group A (CSA) and B (CSB) proteins have been shown to be specifically required for TC-NER, but not for GG-NER [11–14]. In S. cerevisiae, Rad26, the yeast homologue of the mammalian CSB, and Rpb9, a nonessential subunit of RNA polymerase II (Pol II), appear to be involved in two subpathways of TC-NER [15–17]. Rad26 [18] and CSB [19] are also DNA-dependent ATPases of the Swi2/Snf2 superfamily of proteins, and both factors enhance transcription elongation [20,21]. Our recent data suggests that the roles of both Rad26 and Rpb9 in TC-NER may be indirect, as elimination of a transcription elongation/repression factor reinstates TC-NER in cells lacking both Rad26 and Rpb9 [22].
Chromatin structure may significantly affect NER in living cells [23]. In the nuclei of eukaryotic cells, DNA is packaged into a nucleoprotein complex known as chromatin [24]. This complex provides the compaction and structural organization of DNA for processes such as replication, transcription, recombination, and repair. The fundamental subunits of chromatin are nucleosome cores, where 147 bp of DNA is wrapped in 1.65 left-handed superhelical turns around a histone octamer [25]. The DNA between adjacent nucleosome cores is called linker DNA; it varies in length from about 20 to 90 bp in different organisms and tissues or even between individual nucleosome cores in the same cell [26].
In this paper, we present evidence that, in repressed genes, Rad16 is partially dispensable, and Rad26 plays a significant role in repairing both DNA strands, especially in the core regions of well positioned nucleosomes. Active transcription abolishes the role of Rad26 but does not affect the role of Rad16 in repairing the NTS.
2. Materials and Methods
2.1. Yeast strains
The wild-type yeast strain Y452 (MATα, ura3-52, his3–1, leu2-3, leu2-112, cir°) and its isogenic rad16, rpb9 and rad26 deletion mutants were created as described previously [16]. Deletions of RAD1, RAD2, RAD4, RAD7 and RAD14 were made by replacing sequences between nucleotides (with respect to the start codon ATG) −212 and +3853, +129 and +2911, +75 and 2885, +214 to +1454, and +202 and +1114 with the yeast URA3 gene, respectively. Deletion of GAL4 was achieved by replacing the sequence between nucleotides +8 and +2126 with the LEU2 gene.
2.2. UV irradiation, repair incubation and DNA isolation
Yeast cells were grown at 30°C in minimal medium, containing 2% glucose, 2% galactose, or a combination of 2% galactose, 3% glycerol and 2% ethanol to late log phase (OD600 ≈ 1.0), washed and resuspended in 2% glucose, 2% galactose, or a combination of 2% galactose, 3% glycerol and 2% ethanol, respectively. The resuspended cells were irradiated with 50 J/m2 UV light. One-tenth volume of a stock solution containing 10% yeast extract and 20% peptone was added immediately to the irradiated cell suspension, and the cells were incubated for various times in the dark at 28°C before being pelleted. Total DNA was isolated from the pelleted cells using a glass beads method [16].
2.3. Mapping of NER
DNA fragments of interest in the yeast genomic GAL1-10 genes were strand specifically end labeled using the procedure described previously [27,28]. Briefly, ~ 1 μg of total DNA was digested with restriction enzyme(s) to release the fragments of interest and incised at CPD sites with an excess amount of purified T4 endonuclease V (Epicentre). Excess copies of biotinylated oligonucleotides, which are complementary to the 3′ end of the fragments to be labeled, were mixed with the sample. The mixture was heated at 95°C for 5 minutes to denature the DNA and then cooled to an annealing temperature of around 50°C. The annealed fragments were attached to streptavidin magnetic beads (Invitrogen) and the other fragments were removed by washing the beads at the annealing temperature. The attached fragments were labeled with [α-32P]dATP (Perkin Elmer), resolved on sequencing gels. The gels were dried and exposed to a Phosphorimager screen (GE Healthcare or Bio-Rad).
2.4. Northern blot analysis
Yeast cells were cultured to late log phase under the same conditions as those used for NER analysis. Total RNA was isolated using a hot acidic phenol method, as described [29]. The RNA was fractionated by electrophoresis on formaldehyde-agarose gels [30], transferred to Hybond-N+ membranes (GE Healthcare), and hybridized with radioactive probes generated using the Prime-It® II Random Primer Labeling Kit (Stratagene). The probe for the GAL1-10 transcripts was generated from a 2 kb GAL1-10 DNA fragment encompassing the UAS and 5′ portions (0.7 kb) of each of the genes. The probes for ACT1 and 25S rRNA (used as loading controls) were made from 1 kb ACT1 and 2.9 kb 25S rDNA fragments.
3. Results
3.1. Rad16 is partially dispensable for repairing both strands of the repressed GAL1-10 genes
The divergent yeast GAL1 and GAL10 genes, which share a common upstream activating sequence (UAS), are regulated identically [31,32]. Extensive studies have shown that these genes are completely repressed in glucose media and highly induced by galactose. In agreement with previous reports, essentially no transcription was detected in either strand of the GAL1-10 genes in glucose cultured yeast cells (Fig. 1, lanes 1 – 4). In contrast, a high level of transcription occurred in these genes when cells were grown in galactose media (Fig. 1, lanes 5 – 8). These results suggest that transcription in the glucose repressed GAL1-10 genes is either absent or too low to be detected.
Fig. 1. Northern blot showing transcription in the GAL1-10 genes.
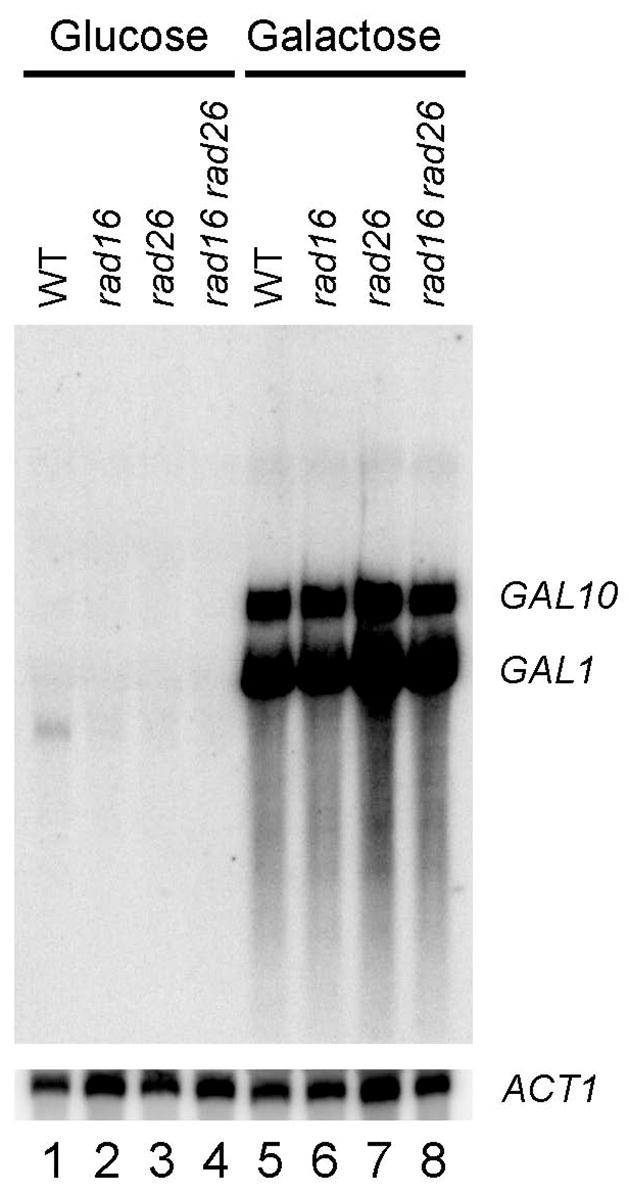
Total RNA was isolated from late log phase cells cultured in minimal medium containing 2% glucose or 2% galactose. The RNA was resolved on a formaldehyde-agarose gel, transferred to a membrane, and hybridized with radioactive probes. The probes were generated by random primer extension, using a 2 kb GAL1-10 fragment, encompassing the shared UAS and ~ 700 bp of each of the genes, as template. ACT1 mRNA serves as an internal loading control.
Repair of CPDs was measured with a high resolution method, which uses streptavidin magnetic beads and biotinylated oligonucleotides to facilitate end-labeling of DNA fragments of interest [27,28]. Yeast cells were cultured to late log phase (OD600 ≈ 1.0), UV irradiated and incubated in a repair medium for different lengths of time. Total genomic DNA was isolated, digested with restriction enzymes to release the fragments of interest, and incised at the UV induced CPDs with excess amount of T4 endonuclease V [33]. The incised fragments were strand specifically end-labeled, resolved on DNA sequencing gels and exposed to a phosphorimager screen [27,28]. To specifically locate the damaged sites along a fragment of interest, sequencing ladders of the same fragment were generated through the Maxam-Gilbert method and resolved on sequencing gels along with UV irradiated samples. The band intensities in the gel lane of “0” time repair reflect the yields of CPDs at different sites. A decrease in band intensities at respective sites indicates that CPDs at these sites are repaired.
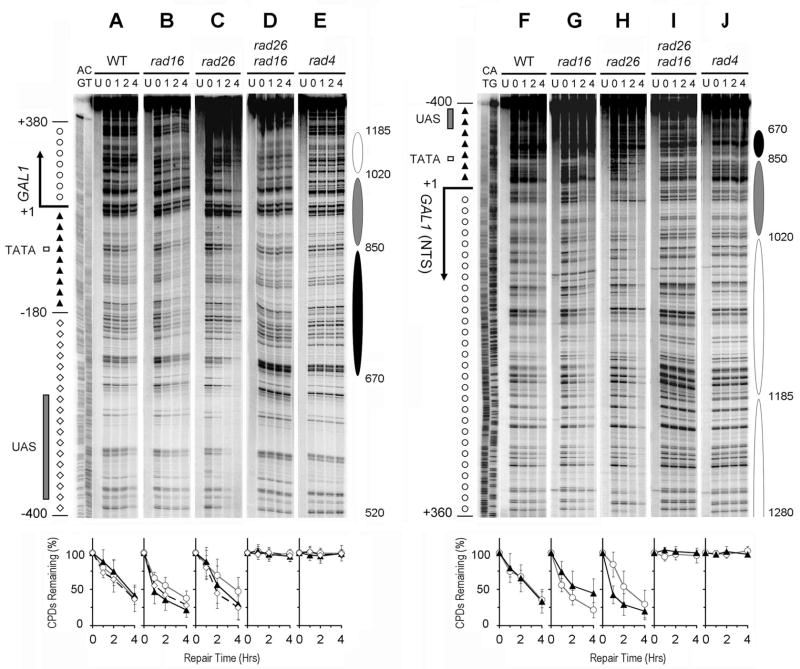
NER occurred in both strands of the GAL1 gene in glucose cultured wild type cells (Fig. 2A and F; Fig. 3, black squares and lines). Surprisingly, quite efficient repair also occurred at most sites in both strands of the glucose repressed GAL1 gene in cells lacking Rad16 (Fig. 2B and G, Fig. 3 blue triangles and lines), which has been shown to be required for GG-NER [4]. Further analysis showed that, in glucose cultured rad16 cells, this repair was not limited to the GAL1 gene but occurred in both strands of the whole GAL1-10 region (Fig. 3, blue triangles and lines). Deletion of genes that are essential for NER (e.g., RAD1, RAD2, RAD4 and RAD14) eliminated repair in the glucose repressed GAL1-10 genes (Fig. 2E and J, and data not shown), indicating that the repair was accomplished by a Rad16-independent NER mechanism.
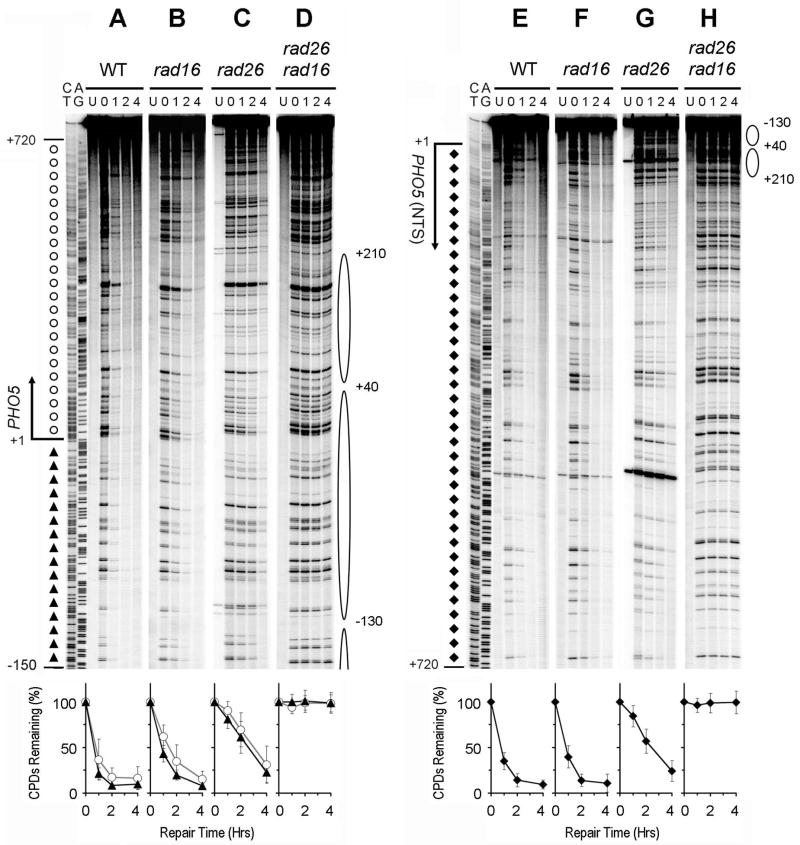
Fig. 2. NER in the glucose repressed GAL1 gene.
Panels A – E and F – J show NER in the TS and NTS of the gene, respectively. Lanes AG and CT are Maxam-Gilbert sequencing ladders. Lanes U are unirradiated samples. Other lanes are samples of different times (in hours) of repair incubation. The solid arrows on the left of the gels indicate the major transcription start site (when the gene is induced). Open box and shaded bar on the left of the gels mark the TATA box and UAS, respectively. Nucleotide positions marked on the left of the gels are relative to the major transcription start site of GAL1. Open circles and solid triangles on the left of panel A mark the transcribed and upstream regions of the TS, respectively, where TCR operates when the gene is induced by galactose. Open diamonds on the left of panel A indicate the upstream region of the TS where TCR does not operate. Solid triangles and open circles on the left of panel F indicate the upstream and the coding regions of the NTS, respectively. Ovals on the right of the gels indicate positions of nucleosomes in the repressed gene, with the extent of shading reflecting the variability of positioning (darkest, least variable; lightest, most variable) [34]. Nucleotide positions marked on the right of the gels are numbered from a site that is 810 nucleotides downstream of the major transcription start site of the GAL10 gene (see Fig. 3 for a schematic of the GAL1-10 genes). Plots underneath each of the panels show percent of CPDs remaining (mean ± SD) for the respective strains. The symbols (open circles, solid triangles and open diamonds) in the plots represent percent of CPD remaining in different regions (as marked on the left of panels A, for the TS, and F, for the NTS) of the gene. WT, wild type.
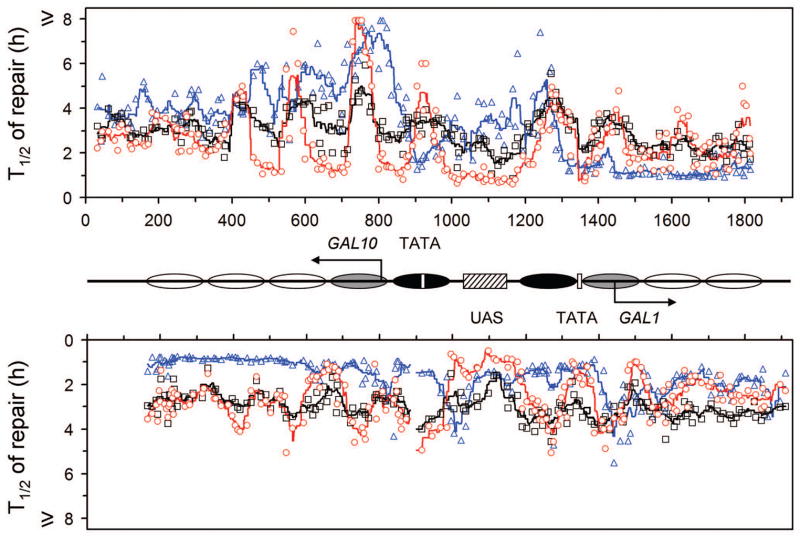
Fig. 3. Plot showing NER in the glucose repressed GAL1-10 genes.
The upper and lower panels represent the top (TS for GAL10 and NTS for GAL1 genes) and bottom strands, respectively. Between the panels is a schematic diagram of the GAL1-10 region, where ovals denote the locations of nucleosomes, with the extent of shading reflecting the variability of positioning (darkest, least variable; lightest, most variable) [34]. Abscissa values are nucleotide positions numbered from a site that is 810 nucleotides downstream of the major transcription start site of the GAL10 gene. Arrows indicate the major transcription start sites when the genes are induced. Open vertical bars denote TATA boxes of the GAL10 and GAL1 genes, and hatched box denotes the common UAS of the two genes. Individual symbols in the plot represent time (in hours) required for repairing 50% (T1/2) CPDs at individual sites of the GAL1-10 genes. Black squares, blue triangles and red circles represent wild type, rad16 and rad26 cells, respectively. Black, blue and red lines in the plot are smoothed T1/2 values for wild type, rad16 and rad26 cells, respectively. Smoothing was carried out by averaging the individual T1/2 values at continuous intervals of 40 nucleotides, where the 40 nucleotide brackets were ramped along the DNA by one nucleotide.
We showed previously that, in glucose repressed GAL1-10 genes of intact cells, well positioned nucleosomes exist in regions adjacent to the UAS, and the variability of nucleosome positioning sharply increases with increasing distance from the UAS [34]. In rad16 cells, the repair rates in the glucose repressed GAL1-10 genes did not seem to be correlated with the positioning of nucleosomes, even in the most well positioned ones close to the UAS (Fig. 2B and G; Fig. 3, blue triangles and lines). Therefore, nucleosome structure may not pose (or indirectly impose) an inhibitory effect on the Rad16-independent NER.
3.2. Rad26 is responsible for Rad16-independent NER in both strands of the repressed GAL1-10 genes
We searched for the factor(s) that is/are responsible for the Rad16-independent NER in both strands of the repressed GAL1-10 genes. Deletion of RPB9, which mediates a subpathway of TC-NER [16], did not affect repair in the repressed GAL1-10 genes of rad16 cells (not shown). Surprisingly, deletion of RAD26 in rad16 cells eliminated repair in the glucose repressed genes (Fig. 2D and I), indicating that the Rad16-independent NER is dependent on Rad26.
To dissect the relative contributions of Rad16 and Rad26 to repair of the repressed GAL1-10 genes, we analyzed rad26 cells, where the NER should be solely dependent on Rad16 (as deletions of both RAD16 and RAD26 completely abolish NER in the repressed genes). In rad26 cells, repair rates in the repressed GAL1-10 genes seemed to be correlated tightly with the nucleosomes that are close to the UAS, being fast in the nucleosome linker and slow in the nucleosome core regions (Fig. 2C and H; Fig. 3, red circles and lines). This correlation fades off in nucleosomes that are more distant from the UAS, in agreement with the observations that the variability of nucleosome positioning increases with distance from the UAS [34]. This indicates that Rad16-dependent NER in the repressed genes is effective in nucleosome linker regions, but not efficient in the core regions of well positioned nucleosomes.
As mentioned above, in the repressed GAL1-10 genes of rad16 cells, where NER should be solely dependent on Rad26, the repair rates were not correlated with positioning of nucleosomes (Fig. 2B and G; Fig. 3, blue triangles and lines). This indicates that Rad26-dependent NER in the repressed genes is effective in nucleosome core regions. Taken together, our results indicate that the roles of Rad16 and Rad26 in NER of the repressed genes are not entirely redundant, but can be complementary to a certain extent.
3.3. Active transcription abolishes the role of Rad26 but does not affect the role of Rad16 in NER in the NTS of the GAL1-10 genes
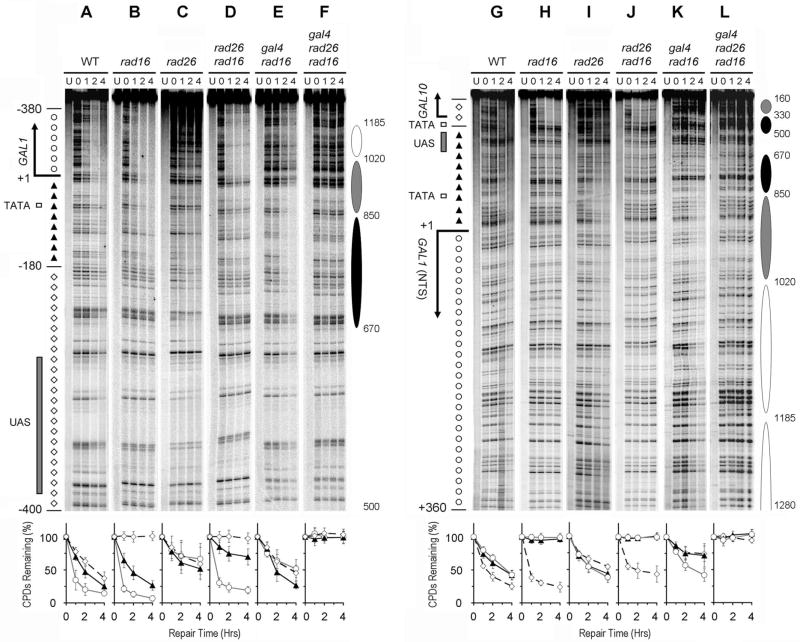
Galactose induction of wild type and rad16 cells caused rapid repair in the TS of the GAL1-10 genes (Fig. 4A, B, G and H; Fig. 5, black and blue symbols). This rapid repair is accomplished by Rad26 and Rpb9 mediated TC-NER [15,16]. In agreement with our previous results [15,16], TC-NER initiates ~ 180 and 120 bp upstream of the transcription start sites of the GAL1 and GAL10 genes, respectively (Fig. 4A, B, G and H; Fig. 5, blue and black symbols). Galactose induction also caused very rapid repair in the TS of the GAL1-10 genes in rad16 rad26 cells (Fig. 4D and J), agreeing with our previous results that Rad26 is almost dispensable for TC-NER in the highly induced genes [15,16]. However, in rad26 cells, most sites in the TS of the induced GAL1-10 genes were repaired quite slowly (Fig. 4C and I; Fig. 5, red circles and lines in the TS regions of the genes), indicating that, in the presence of Rad16, Rad26 may play a larger role in TC-NER of the highly induced GAL1-10 genes. At present, it is unclear why Rad26 is required to different degrees for TC-NER in the induced genes in rad16 and RAD16+ cells. One possibility is that GG-NER mediated by Rad16 competes with TC-NER. Elimination of Rad16 makes more NER factors available for Rpb9 mediated TC-NER, making Rad26 almost dispensable for TC-NER. We also consistently noticed that TC-NER rates are more or less faster in rad16 or rad7 cells than in wild type cells (e.g., Fig. 4, compare panels A and B, and G and H), indicating that a competition of NER factors between GG-NER and TC-NER may indeed exist in the cells.
Fig. 4. NER in the GAL1-10 genes of cells cultured in minimal medium containing 2% galactose, 3% glycerol and 2% ethanol.
Panels A – F and G – L show NER in the TS and NTS of the GAL1 gene, respectively. Lanes U are unirradiated samples. Other lanes are samples of different times (in hours) of repair incubation. The solid arrows on the left of the gels indicate the major transcription start sites. Open box and shaded bar on the left of the gels mark the TATA boxes and UAS, respectively. Nucleotide positions marked on the left of the gels are relative to the major transcription start site of the GAL1 gene. Open circles and solid triangles on the left of panel A mark the transcribed and upstream regions of the GAL1 TS, respectively, where TCR operates. Open diamonds on the left of panel A indicate the upstream region where TCR does not operate. Open diamonds, open circles and solid triangles on the left of panel G indicate the GAL10 TS, GAL1 NTS and the upstream regions (where TCR does not operate), respectively. Ovals on the right of the gels indicate positions of nucleosomes in the repressed genes, with the extent of shading reflecting the variability of positioning (darkest, least variable; lightest, most variable) [34]. Nucleotide positions marked on the right of the gels are numbered from a site that is 810 nucleotides downstream of the major transcription start site of the GAL10 gene (see Fig. 3 for a schematic of the GAL1-10 genes). Plots underneath each of the panels show percent of CPDs remaining (mean ± SD) for the respective strains. The symbols (open circles, solid triangles and open diamonds) in the plots represent percent of CPD remaining in different regions (as marked on the left of panels A, for the GAL1 TS, and G, for the GAL1 NTS) of the genes. WT, wild type.
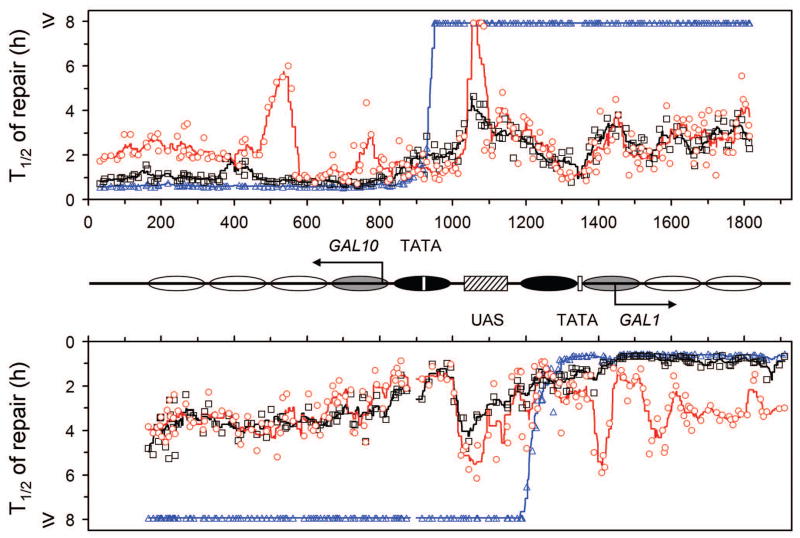
Fig. 5. Plot showing NER in the galactose induced GAL1-10 genes.
The upper and lower panels represent the top (TS for GAL10 and NTS for GAL1 genes) and bottom strands, respectively. Between the panels is a schematic diagram of the GAL1-10 region, where ovals denote the locations of nucleosomes, with the extent of shading reflecting the variability of positioning (darkest, least variable; lightest, most variable) [34]. Abscissa values are nucleotide positions numbered from a site that is 810 nucleotides downstream of the major transcription start site of the GAL10 gene. Arrows indicate the major transcription start sites. Open vertical bars denote TATA boxes of the GAL10 and GAL1 genes, and hatched box denotes the common UAS of the two genes. Individual symbols in the plot represent time (in hours) required for repairing 50% (T1/2) CPDs at individual sites of the GAL1-10 genes. Black squares, blue triangles and red circles represent wild type, rad16 and rad26 cells, respectively. Black, blue and red lines in the plot are smoothed T1/2 values for wild type, rad16 and rad26 cells, respectively. Smoothing was carried out by averaging the individual T1/2 values at continuous intervals of 40 nucleotides, where the 40 nucleotide brackets were ramped along the DNA by one nucleotide.
No repair can be seen in the NTS of the induced GAL1-10 genes in rad16 cells (Fig. 4H; Fig. 5, blue symbols in the NTS of the genes). Furthermore, in agreement with previous reports [4,35], no repair occurred in the NTS of the constitutively expressed RPB2 gene in either glucose- or galactose-cultured rad16 cells (not shown). However, essentially normal repair occurred in the NTS of the induced genes in rad26 cells (Fig. 5, compare black and red symbols in the NTS of the GAL1-10 genes). These results indicate that active transcription abolishes the role of Rad26, but does not affect the role of Rad16 in NER of the NTS.
3.4. Both Rad16 and Rad26 play roles in repairing both strands of the GAL1-10 genes in cells lacking Gal4
The experiments described above suggest that Rad26 plays a role in NER in both strands of a gene only in the absence of detectable transcription. To further examine the notion, we analyzed repair in cells lacking Gal4, which activates transcription of the GAL1-10 genes [31,32]. A medium containing galactose, ethanol and glycerol supports growth of cells with or without Gal4, and induces transcription of the GAL1-10 genes when Gal4 is present [36]. Deletion of the GAL4 gene resulted in essentially no detectable transcription from the GAL1-10 genes in all the cells tested (Fig. 6, compare lanes 1 – 3 and 4 – 6; data not shown). However, apparent repair can be seen in both strands of the GAL1-10 genes in rad16 gal4 cells cultured in the galactose medium (Fig. 4E and K). However, no repair can be seen in both strands of the GAL1-10 genes in rad16 rad26 gal4 cells cultured in the same galactose medium (Fig. 4F and L). These results indicate that Rad16 becomes partially dispensable and Rad26 plays a role in repairing both strands of the GAL1-10 genes when transcription is absent or critically low, through either elimination of the transcription activator Gal4 or glucose repression (see above).
Fig. 6. Northern blot showing transcription in the GAL1-10 genes in GAL4+ and gal4 cells.
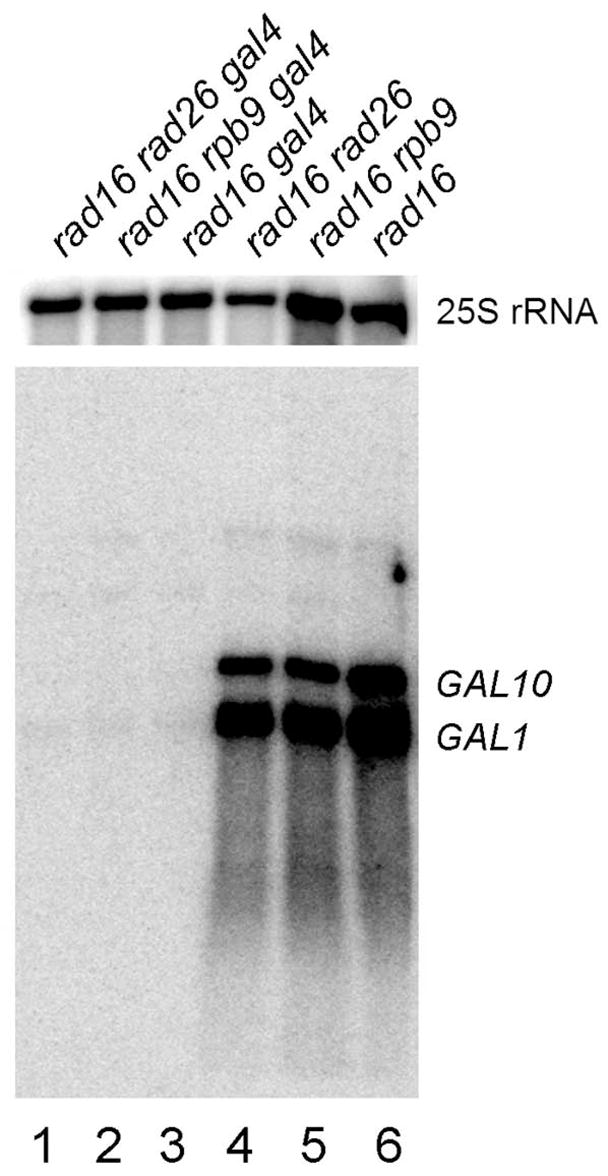
Total RNA was isolated from late log phase cells cultured in minimal medium containing 2% galactose, 3% glycerol and 2% ethanol. The RNA was resolved on a formaldehyde-agarose gel, transferred to a membrane, and hybridized with radioactive probes. The probes were generated by random primer extension, using a 2 kb GAL1-10 fragment, encompassing the shared UAS and ~ 700 bp of each of the genes, as a template. 25S rRNA serves as an internal loading control.
3.5. Both Rad16 and Rad26 play roles in repairing both strands of the repressed PHO5 and ADH2 genes
The GAL1-10 genes are unique in that they share the same UAS, and are transcribed divergently. One question we asked is whether Rad26 plays a general role in repairing repressed genes. To address this question, we analyzed NER in the repressed PHO5 and ADH2 genes, which are regulated differently from each other and from the GAL1-10 genes.
PHO5, which encodes the major phosphate-regulated secreted acid phosphatase, is repressed when phosphate is abundant and highly expressed when phosphate is depleted [37,38]. Efficient NER occurred in both strands of the PHO5 gene in wild type (Fig. 7A and E) and rad16 (Fig. 7B and F) cells cultured in normal minimal medium, which contains a high level of inorganic phosphate. This indicates that Rad16-independent repair also occurs in the repressed PHO5 gene. However, no repair can be seen in the repressed gene in rad16 rad26 cells (Fig. 7D and H), indicating that the Rad16-independent repair is dependent on Rad26.
Fig. 7. NER in the repressed PHO5 gene.
Panels A – D and E – H show NER in the TS and NTS of the gene, respectively. Lanes CT and AG are Maxam-Gilbert sequencing ladders. Lanes U are unirradiated samples. Other lanes are samples of different times (in hours) of repair incubation. Open circles and solid triangles on the left of panel A mark the transcribed and upstream regions of the gene, respectively. Solid diamonds on the left of panel E indicate the coding region of the NTS. Ovals on the right of the gels indicate positions of nucleosomes present in the repressed gene [39]. The rest of the gene is not marked with nucleosomes, as nucleosome positioning is irregular in the regions that are distant from the promoter [39]. Nucleotide positions are relative to the transcription start site of the gene. Plots underneath each of the panels show percent of CPDs remaining (mean ± SD) for the respective strains. The symbols (open circles, solid triangles and diamonds) in the plots represent percent of CPD remaining in different regions (as marked on the left of panels A, for the TS, and E, for the NTS) of the gene. WT, wild type.
It has been shown that, in the PHO5 promoter region, certain nuclease-hypersensitive sequences serve as boundary for nucleosome positioning [39]. At a greater distance from these sequences, nucleosome positioning becomes more irregular [39]. Therefore, in the repressed PHO5 gene, well positioned nucleosomes have been detected only in the promoter-proximal regions. Deletion of RAD26 slowed down NER in both strands of the gene (Fig. 7, compare panels A and C, and E and G). Furthermore, in rad26 cells, there appears to be a mild correlation between repair rates and nucleosome positioning in the regions around the transcription start site, being slower in the core regions and faster in the linker DNA (Fig. 7, compare panels C and G with nucleosome positions marked on the right of the gels). However, repair in other regions of the repressed gene showed no apparent periodicity of the size of a nucleosome (Fig. 7). Presumably, irregularity of nucleosome positioning masked the effect of nucleosome structure on Rad26-independent repair.
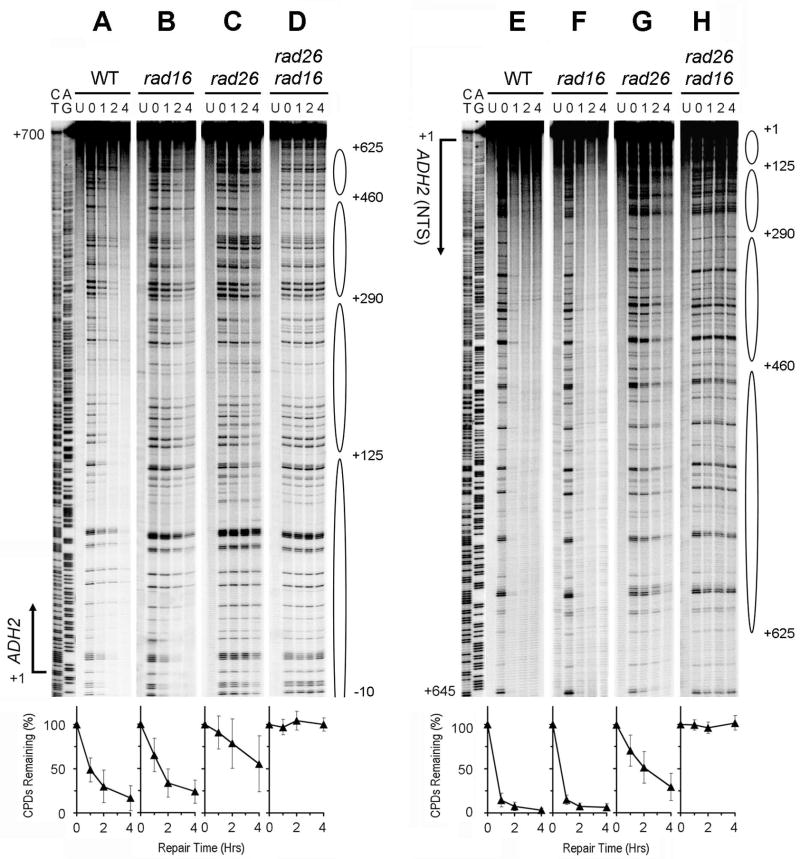
ADH2 encodes an alcohol dehydrogenase, which is responsible for catalyzing the initial step in the utilization of ethanol as a carbon source [40]. This gene is also repressed by glucose [41,42]. Efficient repair can be seen in both strands of the gene in wild type (Fig. 8A and E) and rad16 (Fig. 8B and F) cells cultured in minimal medium containing 2% glucose. For unknown reason(s), the repair was faster in the NTS than in the TS (Fig. 8, compare panels A and E, and B and F). This indicates that Rad16-independent repair also occurs in the repressed ADH2 gene. However, no repair can be seen in the repressed gene in rad16 rad26 cells (Fig. 8D and H), indicating that the Rad16-independent repair is dependent on Rad26.
Fig. 8. NER in the repressed ADH2 gene.
Panels A – D and E – H show NER in the TS and NTS of the gene, respectively. Lanes CT and AG are Maxam-Gilbert sequencing ladders. Lanes U are unirradiated samples. Other lanes are samples of different times (in hours) of repair incubation. Ovals on the right of the gels indicate positions of nucleosomes in the repressed gene [43]. Nucleotide positions are relative to the transcription start site of the gene. Plots underneath each of the panels show percent of CPDs remaining (mean ± SD) for the entire fragments analyzed in the respective strains. WT, wild type.
Well positioned nucleosomes have been detected in the repressed ADH2 gene [43]. Deletion of RAD26 slowed down repair in both strands of the repressed ADH2 gene (Fig. 8, compare panels A and C, and E and G; Fig. 9, red symbols). Furthermore, in rad26 cells, repair rates in the repressed ADH2 gene appear to be tightly correlated with nucleosomes, being slow in the nucleosome cores and fast in the nucleosome linkers (Fig. 8C and G; Fig. 9, redsymbols).
Fig. 9. Plot showing NER in the repressed ADH2 gene.
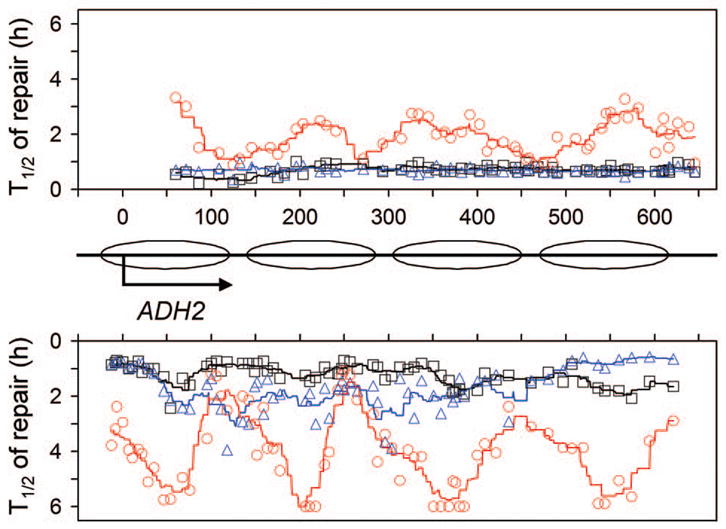
Upper and lower panels represent NTS and TS, respectively. Between the panels is a schematic diagram of the ADH2 gene, where ovals denote locations of nucleosomes [43]. Abscissa values are nucleotide positions relative to the transcription start site. Individual symbols in the plot represent time (in hours) required for repairing 50% (T1/2) CPDs at individual sites of the gene. Black squares, blue triangles and red circles represent wild type, rad16 and rad26 cells, respectively. Black, blue and red lines in the plot are smoothed T1/2 values for wild type, rad16 and rad26 cells, respectively. Smoothing was carried out by averaging the individual T1/2 values at continuous intervals of 40 nucleotides, where the 40 nucleotide brackets were ramped along the DNA by one nucleotide.
4. Discussion
In this study, we present evidence that Rad16 and Rad26 play different and, to some extent, complementary roles in repairing both strands of the repressed GAL1-10, PHO5 and ADH2 genes in yeast. It was shown previously that some repair also occurs in the repressed GAL7 gene in rad7 and rad16 cells [35]. Also, the Rad7/Rad16-independent repair in the repressed GAL7 gene appears to be dependent on Rad26, as essentially no repair was seen in rad7 rad26 and rad16 rad26 cells [35]. Therefore, Rad26 may play a common role in repairing both strands of repressed genes.
A heterotrimeric complex of Rad7/Rad16/Abf1 can generate superhelicity in naked DNA, through the catalytic activity of the Rad16 component [7,8]. It was proposed that the torsion generated in the DNA by this complex is necessary to remove the damage-containing oligonucleotide during NER [7,8]. Although Rad16 is a DNA-dependent ATPase [3], and shares homology with Snf2 protein [5], the catalytic subunit of the Swi2/Snf2 chromatin remodeling complex, there is no evidence that Rad16 plays a role in nucleosome remodeling during NER in the cells. We observed that, in rad26 cells, a strong correlation exists between repair rates and well positioned nucleosomes in the repressed GAL1-10 and ADH2 genes (Figs. 3 and 9, red symbols). A mild correlation can also be seen in the region of the repressed PHO5 gene that is close to the promoter (Fig. 7). These results indicate that nucleosome structure may inhibit Rad26-independent repair, which we show here is dependent on Rad16. Therefore, nucleosome structure may hinder the action of Rad16 for inducing torsional change in the nucleosome core DNA, resulting in inefficient removal of the damage-containing oligonucleotide in the core region.
The mechanism as to how Rad26 plays a role in repairing both strands of repressed genes remains to be understood. One possibility is that the Rad26-dependent repair is simply a form of TC-NER, due to “noise” transcription in both strands of the repressed genes. Recent studies suggest that “noise” transcription, which can not be detected by traditional ways, is quite common in eukaryotic cells [44]. In the absence of Rad26, a well positioned nucleosome may pose a halt to a Pol II complex engaged in “noise” transcription, resulting in inefficient repair in the nucleosome core regions. Induced or constitutive transcription from the TS of a gene may abolish “noise” transcription from the NTS, resulting in confinement of Rad26-dependent repair to the TS. However, the “noise” transcription may not be fully responsible for Rad26-dependent repair in both strands of a repressed gene. First, it was found recently that an unstable, uncoding, antisense RNA is transcribed from the NTS of the repressed PHO5 gene [45]. This antisense transcription may explain the slightly faster repair in the NTS than in the TS of the repressed PHO5 gene in rad16 cells (Fig. 7, compare panels B and F). However, this can not explain why Rad26 also plays a role in repairing the TS of the repressed gene (Fig. 7, compare panels B, C and D). Second, many chromatin immunoprecipitation studies [e.g. [46,47]] have shown that no association between Pol II and GAL1-10 genes exists in glucose cultured yeast cells. Third, we recently observed that deletion of the GAL1 TATA element caused Rad26 mediated repair to occur in both DNA strands downstream of the GAL1 promoter [48]. However, the deletion also resulted in no detectable association of Pol II with the sequence downstream of the TATA-deleted GAL1 promoter [48].
Alternatively, Rad26 may play a role in repairing both strands of a repressed gene through a mechanism that is independent of Pol II. Like Rad16, Rad26 and its human homolog CSB are members of the Swi2/Snf2 superfamily of ATPases, and both of these proteins have DNA-dependent ATPase activity [18,19]. In an in vitro system, it was shown that CSB can remodel reconstituted mononucleosome cores and disarrange an array of nucleosomes regularly spaced on plasmid DNA [49]. Furthermore, CSB interacts not only with double-stranded DNA but also directly with core histones, and intact histone tails play an important role in CSB remodeling [49]. It was proposed that the capability of CSB to modulate DNA double helix conformation may directly facilitate TC-NER [49]. We observed that, in rad26 cells, repair in nucleosome core regions is inefficient. It is possible that Rad26, and its human homolog CSB, may facilitate NER by remodeling nucleosome structure in the cells.
The contributions of Rad26 to NER in different repressed genes, or in different regions or strands of the same repressed gene can vary significantly. Rad26-dependent (Rad16-independent) repair appears to be faster in the TS than in the NTS of the repressed GAL7 gene [35]. In contrast, Rad26-dependent repair seems to be more or less faster in the NTS than in the TS of the GAL1-10, PHO5, and ADH2 genes (Fig. 2, compare panels B and G; Fig. 3, compare blue symbols in the top and bottom panels; Fig. 7, compare panels B and F; Fig. 8, compare panels B and F; Fig. 9, compare blue symbols in the top and bottom panels). The variations may be caused by different levels of “noise” transcription, and/or differences in local chromatin structure.
In view of the fact that the yeast Rad26 and the human CSB are highly conserved [17], it is reasonable to speculate that CSB may play a similar role in repairing both strands of repressed genes (or genomic regions) in human cells. Yeast rad26 single mutants are not UV sensitive [16,17], indicating that the role of Rad26 in NER of repressed genes may be limited to a small portion of the yeast genome. However, human CSB cells are UV sensitive [50], which may be partially due to a role for CSB in repairing a larger fraction of the human genome — the repressed genes or genomic regions.
Acknowledgments
This study was primarily supported by NIH grant ES012718 (to SL) from the National Institute of Environmental Health Sciences (NIEHS), and initiated while SL wassupported by NIH grant ES04106 (to MJS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reardon JT, Sancar A. Nucleotide excision repair. Prog Nucleic Acid Res Mol Biol. 2005;79:183–235. doi: 10.1016/S0079-6603(04)79004-2. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. ASM Press; Washington D.C: 2006. [Google Scholar]
- 3.Guzder SN, Sung P, Prakash L, Prakash S. The DNA-dependent ATPase activity of yeast nucleotide excision repair factor 4 and its role in DNA damage recognition. J Biol Chem. 1998;273:6292–6296. doi: 10.1074/jbc.273.11.6292. [DOI] [PubMed] [Google Scholar]
- 4.Verhage R, Zeeman AM, de Groot N, Gleig F, Bang DD, van de Putte P, Brouwer J. The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6135–6142. doi: 10.1128/mcb.14.9.6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bang DD, Verhage R, Goosen N, Brouwer J, van de Putte P. Molecular cloning of RAD16, a gene involved in differential repair in Saccharomyces cerevisiae. Nucleic Acids Res. 1992;20:3925–3931. doi: 10.1093/nar/20.15.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reed SH, Akiyama M, Stillman B, Friedberg EC. Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes Dev. 1999;13:3052–3058. doi: 10.1101/gad.13.23.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu S, Owen-Hughes T, Friedberg EC, Waters R, Reed SH. The yeast Rad7/Rad16/Abf1 complex generates superhelical torsion in DNA that is required for nucleotide excision repair. DNA Repair (Amst) 2004;3:277–287. doi: 10.1016/j.dnarep.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 9.Ramsey KL, Smith JJ, Dasgupta A, Maqani N, Grant P, Auble DT. The NEF4 complex regulates Rad4 levels and utilizes Snf2/Swi2-related ATPase activity for nucleotide excision repair. Mol Cell Biol. 2004;24:6362–6378. doi: 10.1128/MCB.24.14.6362-6378.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ribar B, Prakash L, Prakash S. Requirement of ELC1 for RNA polymerase II polyubiquitylation and degradation in response to DNA damage in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:3999–4005. doi: 10.1128/MCB.00293-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lommel L, Hanawalt PC. The genetic defect in the Chinese hamster ovary cell mutant UV61 permits moderate selective repair of cyclobutane pyrimidine dimers in an expressed gene. Mutat Res. 1991;255:183–191. doi: 10.1016/0921-8777(91)90052-q. [DOI] [PubMed] [Google Scholar]
- 12.Troelstra C, van Gool A, de Wit J, Vermeulen W, Bootsma D, Hoeijmakers JH. ERCC6, a member of a subfamily of putative helicases, is involved in Cockayne’s syndrome and preferential repair of active genes. Cell. 1992;71:939–953. doi: 10.1016/0092-8674(92)90390-x. [DOI] [PubMed] [Google Scholar]
- 13.van Hoffen A, Natarajan AT, Mayne LV, van Zeeland AA, Mullenders LH, Venema J. Deficient repair of the transcribed strand of active genes in Cockayne’s syndrome cells. Nucleic Acids Res. 1993;21:5890–5895. doi: 10.1093/nar/21.25.5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venema J, Mullenders LH, Natarajan AT, van Zeeland AA, Mayne LV. The genetic defect in Cockayne syndrome is associated with a defect in repair of UV-induced DNA damage in transcriptionally active DNA. Proc Natl Acad Sci U S A. 1990;87:4707–4711. doi: 10.1073/pnas.87.12.4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li S, Smerdon MJ. Dissecting transcription-coupled and global genomic repair in the chromatin of yeast GAL1-10 genes. J Biol Chem. 2004;279:14418–14426. doi: 10.1074/jbc.M312004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li S, Smerdon MJ. Rpb4 and Rpb9 mediate subpathways of transcription-coupled DNA repair in Saccharomyces cerevisiae. EMBO J. 2002;21:5921–5929. doi: 10.1093/emboj/cdf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Gool AJ, Verhage R, Swagemakers SM, van de Putte P, Brouwer J, Troelstra C, Bootsma D, Hoeijmakers JH. RAD26, the functional S. cerevisiae homolog of the Cockayne syndrome B gene ERCC6. EMBO J. 1994;13:5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzder SN, Habraken Y, Sung P, Prakash L, Prakash S. RAD26, the yeast homolog of human Cockayne’s syndrome group B gene, encodes a DNA-dependent ATPase. J Biol Chem. 1996;271:18314–18317. doi: 10.1074/jbc.271.31.18314. [DOI] [PubMed] [Google Scholar]
- 19.Selby CP, Sancar A. Human transcription-repair coupling factor CSB/ERCC6 is a DNA-stimulated ATPase but is not a helicase and does not disrupt the ternary transcription complex of stalled RNA polymerase II. J Biol Chem. 1997;272:1885–1890. doi: 10.1074/jbc.272.3.1885. [DOI] [PubMed] [Google Scholar]
- 20.Lee SK, Yu SL, Prakash L, Prakash S. Requirement for yeast RAD26, a homolog of the human CSB gene, in elongation by RNA polymerase II. Mol Cell Biol. 2001;21:8651–8656. doi: 10.1128/MCB.21.24.8651-8656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selby CP, Sancar A. Cockayne syndrome group B protein enhances elongation by RNA polymerase II. Proc Natl Acad Sci U S A. 1997;94:11205–11209. doi: 10.1073/pnas.94.21.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S, Ding B, Chen R, Ruggiero C, Chen X. Evidence that the transcription elongation function of Rpb9 is involved in transcription-coupled DNA repair in Saccharomyces cerevisiae. Mol Cell Biol. 2006;26:9430–9441. doi: 10.1128/MCB.01656-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong F, Kwon Y, Smerdon MJ. Nucleotide excision repair in chromatin and the right of entry. DNA Repair (Amst) 2005;4:884–896. doi: 10.1016/j.dnarep.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Wolffe Chromatin AP. Structure and Function. Academic Press; London and New York: 1999. [Google Scholar]
- 25.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 26.van Holde KE. Chromatin. Springer-Verlag KG; Berlin: 1989. [Google Scholar]
- 27.Li S, Waters R. Nucleotide level detection of cyclobutane pyrimidine dimers using oligonucleotides and magnetic beads to facilitate labelling of DNA fragments incised at the dimers and chemical sequencing reference ladders. Carcinogenesis. 1996;17:1549–1552. doi: 10.1093/carcin/17.8.1549. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Waters R, Smerdon MJ. Low- and high-resolution mapping of DNA damage at specific sites. Methods. 2000;22:170–179. doi: 10.1006/meth.2000.1058. [DOI] [PubMed] [Google Scholar]
- 29.Collart MA, Oliviero S. Preparation of yeast RNA. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. John Wiley & Sons, Inc; New York: 2004. pp. 13.12.11–13.12.15. [Google Scholar]
- 30.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2001. [Google Scholar]
- 31.Bash R, Lohr D. Yeast chromatin structure and regulation of GAL gene expression. Prog Nucleic Acid Res Mol Biol. 2001;65:197–259. doi: 10.1016/s0079-6603(00)65006-7. [DOI] [PubMed] [Google Scholar]
- 32.Lohr D, Venkov P, Zlatanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 33.Lloyd RS. Investigations of pyrimidine dimer glycosylases--a paradigm for DNA base excision repair enzymology. Mutat Res. 2005;577:77–91. doi: 10.1016/j.mrfmmm.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Li S, Smerdon MJ. Nucleosome structure and repair of N-methylpurines in the GAL1-10 genes of Saccharomyces cerevisiae. J Biol Chem. 2002;277:44651–44659. doi: 10.1074/jbc.M206623200. [DOI] [PubMed] [Google Scholar]
- 35.Verhage RA, van Gool AJ, de Groot N, Hoeijmakers JH, van de Putte P, Brouwer J. Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol Cell Biol. 1996;16:496–502. doi: 10.1128/mcb.16.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohr D, Lopez J. GAL4/GAL80-dependent nucleosome disruption/deposition on the upstream regions of the yeast GAL1-10 and GAL80 genes. J Biol Chem. 1995;270:27671–27678. doi: 10.1074/jbc.270.46.27671. [DOI] [PubMed] [Google Scholar]
- 37.Lenburg ME, O’Shea EK. Signaling phosphate starvation. Trends Biochem Sci. 1996;21:383–387. [PubMed] [Google Scholar]
- 38.Oshima Y. The phosphatase system in Saccharomyces cerevisiae. Genes Genet Syst. 1997;72:323–334. doi: 10.1266/ggs.72.323. [DOI] [PubMed] [Google Scholar]
- 39.Almer A, Horz W. Nuclease hypersensitive regions with adjacent positioned nucleosomes mark the gene boundaries of the PHO5/PHO3 locus in yeast. EMBO J. 1986;5:2681–2687. doi: 10.1002/j.1460-2075.1986.tb04551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciriacy M. Genetics of alcohol dehydrogenase in Saccharomyces cerevisiae. II. Two loci controlling synthesis of the glucose-repressible ADH II. Mol Gen Genet. 1975;138:157–164. doi: 10.1007/BF02428119. [DOI] [PubMed] [Google Scholar]
- 41.Blumberg H, Hartshorne TA, Young ET. Regulation of expression and activity of the yeast transcription factor ADR1. Mol Cell Biol. 1988;8:1868–1876. doi: 10.1128/mcb.8.5.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hedges D, Proft M, Entian KD. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1915–1922. doi: 10.1128/mcb.15.4.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verdone L, Camilloni G, Di Mauro E, Caserta M. Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Mol Cell Biol. 1996;16:1978–1988. doi: 10.1128/mcb.16.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- 45.Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kotovic KM, Lockshon D, Boric L, Neugebauer KM. Cotranscriptional recruitment of the U1 snRNP to intron-containing genes in yeast. Mol Cell Biol. 2003;23:5768–5779. doi: 10.1128/MCB.23.16.5768-5779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li S, Chen X, Ruggiero C, Ding B, Smerdon MJ. Modulation of Rad26- and Rpb9-mediated DNA repair by different promoter elements. J Biol Chem. 2006;281:36643–36651. doi: 10.1074/jbc.M604885200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Citterio E, Van Den Boom V, Schnitzler G, Kanaar R, Bonte E, Kingston RE, Hoeijmakers JH, Vermeulen W. ATP-dependent chromatin remodeling by the Cockayne syndrome B DNA repair-transcription-coupling factor. Mol Cell Biol. 2000;20:7643–7653. doi: 10.1128/mcb.20.20.7643-7653.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nance MA, Berry SA. Cockayne syndrome: review of 140 cases. Am J Med Genet. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]