Abstract
Ribonuclease MRP is an endonuclease, related to RNase P, which functions in eukaryotic pre-rRNA processing. In Saccharomyces cerevisiae, RNase MRP comprises an RNA subunit and ten proteins. To improve our understanding of subunit roles and enzyme architecture, we have examined protein-protein and protein–RNA interactions in vitro, complementing existing yeast two-hybrid data. In total, 31 direct protein–protein interactions were identified, each protein interacting with at least three others. Furthermore, seven proteins self-interact, four strongly, pointing to subunit multiplicity in the holoenzyme. Six protein subunits interact directly with MRP RNA and four with pre-rRNA. A comparative analysis with existing data for the yeast and human RNase P/MRP systems enables confident identification of Pop1p, Pop4p and Rpp1p as subunits that lie at the enzyme core, with probable addition of Pop5p and Pop3p. Rmp1p is confirmed as an integral subunit, presumably associating preferentially with RNase MRP, rather than RNase P, via interactions with Snm1p and MRP RNA. Snm1p and Rmp1p may act together to assist enzyme specificity, though roles in substrate binding are also indicated for Pop4p and Pop6p. The results provide further evidence of a conserved eukaryotic RNase P/MRP architecture and provide a strong basis for studies of enzyme assembly and subunit function.
INTRODUCTION
RNase MRP is an essential eukaryotic ribonucleoprotein endonuclease, first identified as having a role in mitochondrial DNA replication (1). Subsequent experiments have shown RNase MRP is primarily localized in the nucleolus (2,3), where it functions in pre-rRNA processing, cleaving at a specific site (A3) in the ITS1 of pre-rRNA, leading to the generation of the mature 5′ end of 5.8S rRNA (4,5). More recently, a critical role within the cell cycle has emerged, where RNase MRP promotes degradation of CL2B mRNA in Saccharomyces cerevisiae by cleavage within its 5′ UTR (6), possibly in cytoplasmic ‘processing bodies’ (7). In the human RNase MRP, mutations in the RNA component have been shown to cause the genetic disease cartilage hair hypoplasia (8).
Eukaryotic RNase MRP is structurally and functionally related to the ubiquitous ribonucleoprotein RNase P, which predominantly functions in the processing of pre-tRNAs. Recent evidence indicates RNase P also plays a role in pol III transcription of its RNA substrates (9), potentially providing a route for coordination of transcription with processing. In S. cerevisiae, RNase P and RNase MRP consist of 9 and 10 known protein subunits, respectively, and 1 distinct RNA molecule. Eight proteins are subunits of both RNase P and MRP (10), with each complex possessing one or more unique protein subunits; Snm1p and Rmp1p in RNase MRP and Rpr2p in RNase P (10,11). The conserved properties of both complexes suggest that they evolved from a common eukaryotic ancestor, the RNA subunit having diverged and unique protein subunits evolved accordingly (12). Indeed, phylogenetic analysis of the RNA subunits yields highly similar secondary structures despite limited sequence similarity (13–17).
In bacteria and some archaeal species, the RNA subunit of RNase P has been shown to cleave pre-tRNA in vitro in the absence of protein subunits (18,19). In S. cerevisiae and other eukaryotes, the protein–RNA ratio is much higher and the proteins appear to be a necessity for efficient enzymatic activity in vitro. Very recent studies on the human and Giardia lamblia P RNAs in the absence of proteins, however, have demonstrated low pre-tRNA RNA cleavage activity (20). Evidence is thus in favour of the RNA subunit providing the catalytic core of eukaryotic RNase P/MRP and the protein subunits having likely evolved to assume roles in RNA subunit folding and stabilization and/or to assist substrate binding and catalysis during one or more of the multiple functions of the respective enzymes.
Studies on the overall subunit composition and organization of the RNase MRP and RNase P complexes have proved challenging, primarily due to difficulty in obtaining biochemically purified native complexes and soluble individual purified protein subunits for reconstitution studies. However, yeast two-hybrid and yeast three-hybrid analyses of both the human and S. cerevisiae RNase P complexes (21–23), and GST pull-downs using the human RNase MRP subunits (24) have provided insights into mutual subunit interactions, and revealed that numerous protein-protein and RNA–protein interactions probably occur in both complexes (reviewed by Walker and Engelke, 2006 (25)). Exploration of direct subunit interactions through in vitro binding studies is lacking on the yeast RNase P and MRP enzymes. Here, we seek to redress this imbalance and to complement existing yeast two-hybrid data to obtain an improved understanding of the structural organization of yeast RNase MRP. This study reports the first successful soluble expression and purification of all 10 of the RNase MRP protein subunits and identifies one-to-one protein–protein and protein–RNA interactions. Comparative analysis with existing data on the yeast and human RNase P and/or RNase MRP systems (25), together with our novel data on protein-pre–rRNA interactions, provides new discussion of the enzyme architecture and protein subunit roles.
MATERIALS AND METHODS
Expression and purification of GST-fusion proteins
POP1, POP3, POP4, POP5, POP6, POP7, POP8 and RPP1 open reading frames were excised out of previously described p413Gal yeast expression vectors (10) and inserted at the SalI-XmaI sites of pGEX-6P-1 (GE Healthcare). Genes for Snm1p and Rmp1p were amplified from S. cerevisiae genomic DNA and cloned into pGEX-6P-1 between EcoR1 or BamH1 and Xho1 sites. A KS/pGEX construct allowing expression of a fragment of Snm1p (residues P124–S198) representing a lysine/serine (K/S)-rich domain of this protein was also created. All constructs were modified by insertion of a sequence at the BamH1 site that places a phosphorylation site (RRASV) for bovine heart protein kinase A (Sigma) at the N-terminus of the native protein sequence. The expression constructs were routinely transformed into E. coli BL21 RIL cells and grown with appropriate antibiotic selection. A subsequent overnight culture was diluted 1:200, and grown at 37°C to an OD600 = 0.8 at which stage expression was induced overnight at 16°C by addition 0.1 mM isopropyl β-d-galactopyranoside (IPTG). Pelleted cells were lysed (lysozyme and freeze-thaw cycles) in 25 mM Tris–Cl, pH 8.0, 1 M NaCl, supplemented with 1% Triton X-100, 10 mM dithiothreitol (DTT) and protease inhibitors (Complete, Roche). GST-fusion proteins were purified by affinity chromatography using glutathione Sepharose 4B, GS4B (GE Healthcare). The GS4B beads were prepared and washed with 1 × PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4), supplemented with 1% Triton X-100 + 10 mM DTT. The same buffer was used for wash and elution steps after application of GST-fusion protein. A GST-only vector (no insert sequence) was included as a control. The concentration and purity of GST-fusion protein on the beads was visually determined by SDS-PAGE.
Radiolabelling of the GST-fusion proteins and cleavage from GST
GS4B beads bearing bound fusion protein (bed-volume 1 ml) were equilibrated in HMK buffer (20 mM Tris–HCl, pH 8.0, 100 mM NaCl, 12 mM MgCl2) and then incubated with 100 U bovine heart kinase solution and 50 μCi [γ-32P] ATP (30 min, 4°C). The reaction was terminated using 10 ml of stop solution (10 mM sodium phosphate, pH 8.0, 10 mM sodium pyrophosphate, 10 mM EDTA, 10 mg BSA) and the beads subsequently washed with 5 × 10 ml of 1 × PBS + 1% Triton X-100 + 10 mM DTT. Bead samples were analysed by SDS-PAGE, followed by exposure to PhosphoImager screens. The efficiency and specificity of radiolabel incorporation was determined by autoradiography analysis (Typhoon scanner; AP Biotech). Radiolabelled GST-fusion proteins were subsequently cleaved overnight at 4°C with PreScission protease (AP Biotech), following the manufacturers recommended protocol. The purity of the cleaved proteins, estimated by gel analysis (with Coomassie staining), was improved (∼80% pure on average) over that of their respective GST-fusions, though the yield is reduced due to incomplete cleavage from GST (data not shown). The concentration of cleaved, untagged proteins was determined using the Bio-Rad protein assay.
In vitro transcription of RNase MRP RNA and pre-rRNA
NME1, the coding sequence for yeast MRP RNA, has been cloned into a pUC based vector, immediately downstream of a T7 promotor, creating the construct pHST7/NME1 for run-off transcription [a version of pHST7*NME1* described previously (17), without flanking ribozymes]. A pHST7/RPR1 construct was also generated to enable transcription of yeast P RNA. A region encoding a 187 nt RNA fragment starting 73 nt upstream of the A3 site in yeast pre-rRNA ITS1 (at the A2 site ACAC sequence) and ending 114 nt downstream was amplified from yeast genomic DNA and cloned into the same pUC-derived plasmid, creating the construct pHST7/pre-rRNA for use as an in vitro transcription template. All plasmid template sequences were confirmed via automated DNA sequencing. RNA was transcribed (37°C, 2 h) from EcoRI-linearized plasmid template in 20 µl reactions containing 80 mM Tris–HCl (pH 8.0), 2 mM spermine, 40 mM DTT, 30 mM MgCl2, 40 U RNasin, 7 mM rNTPs, [alpha-32P] UTP, 5U yeast inorganic pyrophosphatase, 15 U T7 RNA polymerase. Radiolabelled UTP was omitted for preparation of unlabelled competitor RNA. After transcription, RNAs were purified by treatment with RNase-free DNase I followed by acid phenol/chloroform extraction and ethanol precipitation and then taken through a refolding procedure (80°C for 3 min, cooled to 48°C at 2°C/min then cooled to 20°C at 1°C/min) in 30 mM HEPES pH 7.5 with 100 mM KCl (∼1 µM RNA). Conformational homogeneity of RNAs was monitored by UV absorbance melting profiles and by native 4% PAGE.
GST pull-down assay
For the analysis of protein–protein interactions, 50 μl of the beads (washed with PBS and containing bound GST-fusion protein at equivalent loadings, as estimated by SDS-PAGE) were incubated with 50 μl of ∼1 μM cleaved, untagged radiolabelled protein plus 5 μg BSA for 3 h at room temperature, under continuous agitation. After incubation, the beads were pelleted and the supernatant (containing any unbound cleaved protein) was removed. The beads were then washed three times with 1 ml of PBS + 1% triton + 10 mM DTT and analysed by SDS-PAGE. The gels were vacuum dried to blotting paper, and exposed to PhosphoImager screens. Protein–protein interactions were quantitated using a Typhoon scanner and ImageQuant software. Radiolabelled untagged protein retained on beads bearing a GST fusion protein was quantitated as percentage bound relative to total amount of input radiolabelled protein. In later experiments, NaCl concentration in the final wash was increased to 300 mM or 1 M, from the normal concentration of 150 mM.
Filter binding assay
For the analysis of RNA–protein interactions, 10 µM of each cleaved, untagged, protein subunit was incubated with 2 nM of annealed, in vitro transcribed 32P-labelled RNA, ∼1 μM competitor RNA (poly(IC) or poly(U) (Amersham)) and 5 mM MgCl2, on ice, for 20 min in binding buffer (50 mM Tris–HCl, pH7, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.01% Triton-X100). Experiments with poly(IC) and poly(U) were repeated four times in total. Pre-wetted nitrocellulose filter discs (Millipore, 0.45 μm) were placed onto the vacuum filter apparatus, and 0.45 μm filter discs placed on top of these. Both filters were pre-washed with 1 ml of binding buffer, before the sample was pipetted on to the top filter and vacuum applied. Filters were washed with 1 ml of 1× PBS then scintillation counted and the data analysed using Excel. The top 0.45 μm filters are used to remove aggregates commonly observed in RNA–protein interaction studies and to ensure RNA trapped on the nitrocellulose is part of a low ratio (preferably 1:1 or similar) complex(es). In our studies, at least 50% of the radiolabel is retained on the top filter for all proteins. Similar levels of aggregation are observed if fusion protein is used. The percentage bound values reported exclude RNA aggregates and are a percentage of the soluble RNA bound [counts per minute (c.p.m.) retained on nitrocellulose filter/(total input c.p.m. − c.p.m. on 0.45 μm filter]. For those subunits observed to bind MRP RNA in the filter binding screens, relative competition by poly(U), MRP RNA and P RNA was assessed by incubation of 32P-MRP RNA bound protein complexes with 1–1000 nM unlabelled RNA competitor.
RESULTS
Preparation of soluble protein subunits of S. cerevisiae RNase MRP
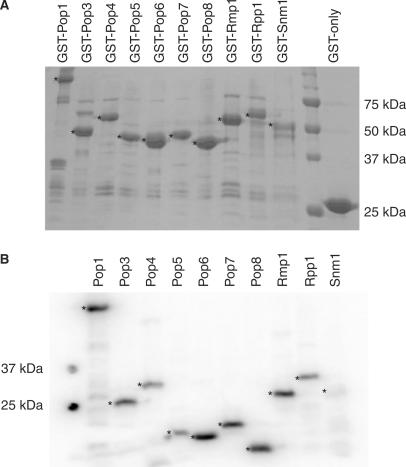
S. cerevisiae RNase MRP comprises one RNA molecule and at least 10 protein subunits, but little is known about the overall architecture of the complex. The approach taken here was to obtain each protein subunit in a recombinant form in order to screen for direct subunit interactions in vitro using the GST pull-down approach and thus lay the foundations for assembly of a recombinant enzyme. All 10 GST-fusion proteins (GST-Pop1p, GST-Pop3p, GST-Pop4p, GST-Pop5p, GST-Pop6p, GST-Pop7p, GST-Pop8p, GST-Rmp1p, GST-Rpp1p and GST-Snm1p), plus GST-alone (to be used as a negative control for the protein–protein interaction studies) were expressed in E. coli and purified by affinity chromatography using glutathione Sepharose (Figure 1A). This is the first report of successful soluble expression and purification all 10 of the yeast RNase MRP subunits, with yields of 15–25 mg protein per 1 l culture. Although the GST-fusion protein is the predominant product in all cases, all lanes show low levels of faster migrating polypeptides that probably represent degradation products or truncations of the recombinant proteins. Less prevalent slower migrating polypeptides are also observed, often common to all preparations, and most likely represent contaminating E. coli proteins. All of the GST-fusion proteins were stable for approximately 1 week when bound to the glutathione Sepharose beads and stored at 4°C in the presence of protease inhibitors.
Figure 1.
Preparation of RNase MRP protein subunits. (A) Expressed GST-fusion proteins bound to glutathione Sepharose 4B beads. The expression and purity of GST-fusion protein preparations were determined by SDS-PAGE analysis and Coomassie staining. The asterisks (*) indicate the full-length GST-(fusion) proteins. The bands seen beneath the full-length GST-(fusion) proteins probably represent truncated versions of the full-length recombinant proteins. The sizes of the molecular weight markers are shown on the right. (B) Radiolabelled, cleaved proteins. Whilst bound to glutathione Sepharose, GST fusions were treated with bovine heart kinase in the presence of γ-32P-ATP to achieve radiolabelling, followed by removal of the GST-tag by overnight cleavage with PreScission protease. The efficiency of radiolabelling and the purity of the cleaved proteins were assessed by SDS-PAGE analysis followed by exposure to PhosphoImager screens and analysis using a Typhoon scanner. The asterisks (*) indicate the radiolabelled, untagged proteins. The sizes of the molecular weight markers are shown on the left.
Radiolabelling of the GST-fusion protein was highly specific (Figure 1B), with the majority of the radiolabel (60–80%) being incorporated into the GST-fusion protein. Cleavage at the PreScission protease site enabled recovery of radiolabelled protein subunits with the GST-tag removed (Figure 1B). These untagged radiolabelled proteins were then combined with unlabelled GST-fusion protein samples bound to glutathione Sepharose beads to investigate direct one-to-one protein–protein interactions. With the exception of cleaved Snm1p, all RNase MRP protein subunits were stable as both cleaved subunits and as GST-fusions. Cleaved Snm1p was less stable; we were able to use it in protein–RNA interaction screens but were unable to use it to study protein–protein interactions, due to the prolonged incubation times and possible exposure to proteases.
Protein–protein interactions
Interactions between the bound GST-fusion proteins and the radiolabelled, untagged proteins were analysed by SDS-PAGE and exposure to PhosphoImager screens. Protein–protein interactions were quantitated, relative to the input radiolabelled protein, using a Typhoon scanner and ImageQuant™ software (GE Healthcare). GST-only was included as a negative control, in order to assess the specificity of the interactions.
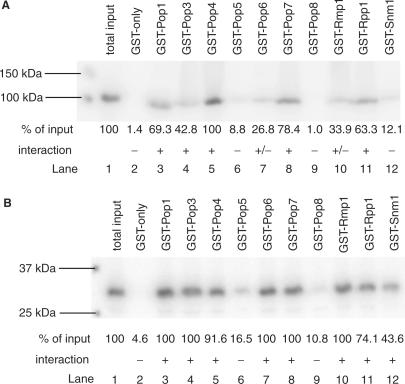
Interactions were assigned as being strong (‘+’; at least 40% of input radiolabelled protein bound to the GST-fusion protein), weak (‘+/−’; between 20 and 40% of input protein co-precipitated), or none (‘−’; <20% of input protein co-precipitated). Figure 2 shows a typical set of results for the Pop1p and Rmp1p protein subunits, obtained upon incubation of radiolabelled, untagged protein with each of the bound GST-fusion proteins at 150 mM NaCl concentration. In the case of Pop1p, e.g. Lane 1 shows the amount of input radiolabelled, cleaved Pop1p. Lanes 3, 4, 5, 8 and 11 show strong interactions, where over 40% of the input protein binds to the GST-fusion protein. Lanes 7 and 10 show weak interactions, and lanes 6, 9 and 12 are examples of what was observed when no interaction had occurred. As well as such non-interacting internal negative controls, none of the proteins should interact with GST-alone. However, subunits of protein complexes are often ‘sticky’ and may interact non-specifically. Indeed, such properties have been reported for human RNase MRP subunits (24). Figure 2 shows that there is no non-specific interaction between Pop1p, or Rmp1p, and GST-alone (1.4 and 4.6% of the input, respectively).
Figure 2.
Protein–protein interactions. The figure shows a typical set of interaction data for two protein subunits. The protein subunit in untagged radiolabelled form was screened for interaction with each RNase MRP GST-fusion protein immobilized on glutathione Sepharose beads in a phosphate buffer containing 150 mM NaCl. GST was included as a control. The first lane in each case contains untagged radiolabelled protein only, indicating the total (100%) amount of input. (A) Interactions of Pop1p, (B) interactions of Rmp1p. Protein–protein interactions were quantitated using a Typhoon scanner and ImageQuant software. Interactions were assigned as strong (‘+’; at least 40% of input protein co-precipitated), weak (‘+’/−; between 20 and 40% of input protein co-precipitated) or none (‘−’; <20% of input protein co-precipitated).
By conducting such assays for all protein subunits, we identified numerous binary protein–protein interactions, which are detailed in Figure 3 (top panel). Interaction screens were also carried out at higher salt concentrations, 300 mM (see Figure 3 lower panel) and 1 M NaCl (data not shown) in order to derive more information about the nature and strength of protein–protein interactions. The combination of these observations contributed to the final assignment of the number (13 strong and 18 weak) of protein–protein interactions and their strength, summarized in Figure 4A. In order for an interaction to be classified as ‘strong’ it was required to be both a mutual interaction (observed as ‘strong’ in both directions) and also maintained (at least weakly) or enhanced at elevated salt conditions (300 mM NaCl). Interactions that were not mutual, or interactions not mutually maintained at normal or elevated salt conditions were usually more conservatively assigned as ‘weak’. The following detailed examination of interactions illustrates this assignment procedure.
Figure 3.
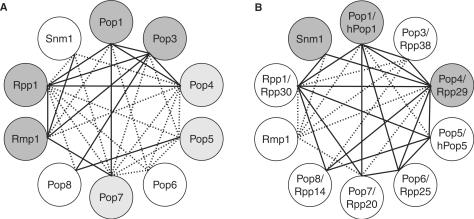
Protein–protein interactions. Top panel: pairwise protein–protein interactions observed in the presence of 150 mM NaCl. Dark grey squares indicate strong protein-protein interactions, light grey squares indicate weak interactions and white squares indicate that no significant interaction occurred. A ‘y’ within a box indicates a protein–protein interaction also seen by Houser-Scott et al. (22) using the yeast two-hybrid system. An ‘h’ (in bold) indicates a protein–protein interaction observed by Welting et al., (24) using GST-pulldown experiments on the human homologues of yeast RNase MRP subunits [these being hPop1, Rpp38 (Pop3p), Rpp29 (Pop4p), hPop5, Rpp25 (Pop6p), Rpp20 (Pop7p), Rpp14 (Pop8p), Rpp30 (Rpp1p)]. An ‘h’ denotes a protein–protein interaction observed by Jiang et al. (22) in a yeast two-hybrid study of human RNase P. Bottom panel: protein–protein interactions observed in the presence of 300 mM NaCl. Thick borders indicate interactions that are conserved in at least 2 of the other 3 major studies of yeast and human subunits. The bordering is dashed if the interaction is conserved but under-represented by other studies.
Figure 4.
A summary of the extensive network of direct protein–protein interactions between RNase MRP subunits. (A) Interactions (1:1) between yeast RNase MRP subunits observed in this report using GST pull-down assays. Solid and dashed lines represent strong and weak interaction, respectively. In total, 31 inter-subunit interactions are observed (13 strong, 18 weak). Dark and light grey circles indicate protein subunits that display strong and weak self-association, respectively. (B) Conserved protein–protein interactions. Solid lines represent 18 protein–protein interactions that are conserved in at least three of the four studies of eukaryal (yeast and human) RNase P/MRP (this report, 22–24), or the majority if a subunit is not represented in all four studies (e.g. Rpp1p, Pop5p, Snm1p). Dashed lines represent a further four interactions that are observed in any two out of the four studies, or Rmp1p interactions (six), reported for the first time here. Grey circles indicate those subunits for which there is convincing evidence of self-interaction. Of the 25 yeast inter-subunit interactions observed by the GST pull-down analysis here (excluding the Rmp1p subunit), 17 are strongly conserved (all solid lines, except that linking Pop4p and Pop8p).
The majority of the interactions were mutual at 150 mM NaCl, detected in ‘two directions’ at the same intensity, i.e. with either one or the other interacting protein fused to GST. Six of the interactions (Rmp1p + Pop1p, Pop4p + Rmp1p, Pop4p + Pop7p, Rpp1p + Pop6p, Pop6p + Rmp1p and Pop7p + Rpp1p) were detected as having a strong interaction in one direction and a weak interaction in the other. For the purposes of the diagram (Figure 4A), they were therefore classified as weak interactions. This difference in the strength of the interactions may be due to the influence of the GST-tag, either interfering with protein–protein association, or enhancing subunit stability and, therefore, interaction. For example, Rmp1p and Rpp1p display a number of weak interactions as GST fusions that are strong in the other direction (using untagged protein). In cases where two such factors combine, a particularly conservative assignment is usually made for an interaction. For example, we actually assign the interaction between GST-Pop6 and untagged Rmp1p as a false positive since, not only is the interaction not reciprocal, Rmp1p is clearly ‘stickier’ in its untagged form, whilst Pop6p demonstrates interactions of greater strength and number as a GST fusion.
Figure 3 (lower panel) shows that several strong interactions (notably Pop1p with Pop3p, Pop4p, Pop7p, Rpp1p and Rmp1p; Pop3p with Pop1p, Pop8p and Rmp1p; Pop4p with Pop1p, Pop3p, Pop5p, and Rpp1p; Pop5p with Pop4p and Pop8p; Pop7p with Pop1p and Rmp1p; Pop8p with Pop3p and Pop5p; Rpp1p with Pop1p, Pop4p and Rmp1p; Rmp1p with Pop1p, Pop3p, Pop7p and Rpp1p; Snm1p with Rmp1p) are maintained at 300 mM salt, confirming their categorization as ‘strong’ interactions. Self-association of Pop1p, Pop3p and Rmp1p is not affected by raising NaCl concentration to 300 mM. Other strong interactions (Pop3p with Pop5p, Pop7p and Rpp1p; Pop4p with Pop7p, Pop7p with Pop3p) become weaker or are abolished on increasing salt concentration, suggesting a specific electrostatic component to their association. The same applies to self-association of the Rpp1p and Pop4p subunits. In test experiments, 1 M NaCl was found to have no influence on GST binding and retention on the glutathione Sepharose beads (data not shown), demonstrating that loss of observed interaction on increasing salt is not due to a reduction in bound GST-fusion. It is possible that interactions weakened at higher salt concentration are in fact bridged by contaminating nucleic acid. However, during protein purification, 1 M NaCl was added to the initial freeze-thaw buffer in order to minimize the levels of contaminating nucleic acids, and spectrophotometer readings at OD260 and OD280 did not reveal significant levels of contaminants.
Weaker interactions (at 150 mM NaCl, Figure 3) for which an increase in strength is noted at 300 mM (Figure 3) and/or 1M NaCl (Pop1p with Rmp1p; Rpp1p with Pop8p) could be the result of genuine hydrophobic interaction, or could be due to protein aggregation and precipitation with the glutathione sepharose beads. The latter possibility is unlikely given no interaction with the GST control under the same conditions (data not shown) but was also tested by incubation of untagged proteins with unloaded glutathione Sepharose beads under the same high salt wash conditions and no protein precipitation on the beads was observed for any subunit. The only interactions to remain consistent regardless of ionic strength are the self-association of subunits Pop5p and Rmp1p.
The combination of these observations with the presence or not of mutuality to a binary interaction contributed to the final assignment of number and strength of interactions, summarized in Figure 4A. For example, the Pop3p-Pop5 interaction is classified as ‘weak’, despite detection as a strong interaction in both directions at 150 mM NaCl as a result of its abrogation in one direction (GST-Pop5 with Pop3p) and weakening in the other when assayed at 300 mM NaCl. Likewise, a strong interaction between GST-Pop4p and untagged Pop5p is maintained up to 1 M NaCl but is not observed in the opposite direction (GST-Pop5 with untagged Pop4p), hence overall we conservatively classify this interaction as weak.
Pop1p, Pop3p, Pop4p, Rpp1p and Rmp1p exhibit the highest number of strong interactions, suggesting that they may play a more central role in RNase MRP structure. Our data confirm Rmp1p as a new member of the RNase MRP complex (11), identifying strong interactions with four other subunits (Pop3p, Pop7p, Rpp1p and Snm1p) and weaker interactions with two others (Pop1p and Pop4p). Close association between Rmp1p and Snm1p, indicated by their strong interaction here, is interesting in the light of their possible role in distinguishing RNase MRP function from that of RNase P. Interactions identified here that are conserved in other studies of yeast and human RNase P/MRP are highlighted in Figure 3 by thicker bordering. A full comparative analysis is now possible, leading to a number of conclusions regarding subunit organization in RNase P/MRP (see Discussion section).
Interactions of protein subunits with MRP RNA
The RNA component of RNase MRP is expected to play a major role in the assembly and function of the enzyme complex. Here, we report the first screen of direct interactions of yeast MRP RNA (340 nt) with individual protein subunits and, as such, provide novel data that will be useful as a platform for future detailed examination of particular RNA–protein interactions. Further to the 10 protein subunits, a fragment of Snm1p representing its C-terminal Lys/Ser-rich region, named K/S, was included in binding screens to gain insight into the role of this region of the protein.
Interactions with MRP RNA were screened using a filter binding assay at excess protein concentrations (10 μM), likely representative of near saturation binding conditions (see Methods section). An interaction was identified as positive where at least 15% of radiolabelled RNA bound to the protein in the presence of one or other competitor RNA (poly(IC) double-stranded or poly(U) single-stranded competitor). Negative controls involved the addition of water or BSA (10 μM) to RNA samples. Note that formation of protein–RNA aggregates, RNA degradation and proper RNA folding can influence the extent of RNA binding observed. The percentage RNA bound values are thus calculated after exclusion of RNA in aggregates (see Methods section). The integrity of RNA during the binding reaction was also monitored (see Supplementary Data) and no significant decrease observed with any protein preparation. Conformational homogeneity of the RNA is more difficult to fully assess, although it was prepared as previously described (17) in a study that demonstrated the majority of the RNA adopts a single conformation.
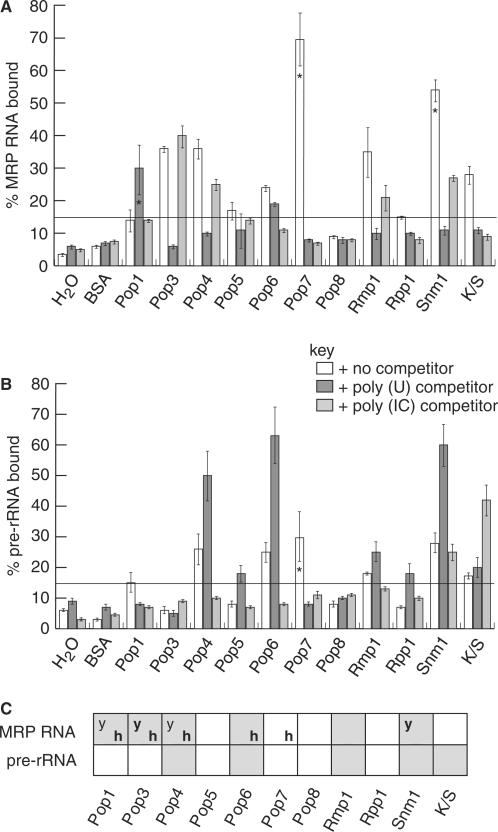
Figure 5A shows the results of the filter binding assays between each protein and MRP RNA. We identified six interactions between untagged protein subunits (Pop1p, Pop3p, Pop4p, Pop6p, Rmp1p and Snm1p) and MRP RNA, which are shown in Figure 5A. None of the subunits interact with MRP RNA in a truly specific manner as independent proteins since one or other RNA competitor, poly(U) or poly(IC), reduces binding. Abrogation of binding is, however, complete in all cases upon addition of excess unlabelled MRP RNA (see later).
Figure 5.
RNA–protein interaction data. Filter-binding RNA interaction data. The bar graphs show the percentage of RNA that interacted with each protein subunit, determined by scintillation counting of 32P-labelled RNA retained on nitrocellulose filters. (A) Interactions between the protein subunits and MRP RNA. (B) Interactions between the protein subunits and pre-rRNA substrate RNA. In (A) and (B), the horizontal line at 15% of RNA incorporation indicates the cutoff point for a positive interaction. Bar shading indicates which competitor RNA was present, described by the key. Note that the percentage RNA bound values were calculated after exclusion of RNA in aggregates from the total available RNA (see Methods section). Interaction mixtures that are particularly affected by formation of aggregates are indicated with an asterisk within the bar. (C) Summary of protein–RNA interactions of yeast RNase MRP subunits. A light grey square indicates an interaction was observed and a white square indicates that no interaction occurred. A bold ‘h’ indicates those interactions also observed for human subunits in vitro (24). A ‘y’ indicates those interactions detected by yeast three-hybrid analysis of yeast RNase P subunits (23,44). A bold ‘y’ indicates interactions detected by other in vitro binding analyses (28,56).
Pop3p provides convincing MRP RNA filter binding data (Figure 5A). This protein–RNA interaction is maintained in the presence of poly(IC) competitor but lost in the presence of poly(U) competitor, suggesting that the affinity for MRP RNA involves interaction with a single-stranded region. This would concur with previous analysis of the RNA binding properties of Pop3p (28). Pop4p, Rmp1p and Snm1p were also observed to interact with MRP RNA in the presence of poly(IC) competitor, but these interactions were again lost in the presence of poly(U) competitor, suggesting a structural preference for single-stranded RNA by these subunits. The same applies to Pop5p, although the percentage input RNA bound by Pop5p is only just above our chosen cutoff for an interaction [note that the Pop5p sample induces no obvious RNA degradation (see Supplementary Data), which hence cannot be the reason for low binding activity for this protein]. Competition by poly(U), however, may underestimate the degree of specificity of certain interactions given that inhibition of the RNase P holoenzyme by poly(U) has been reported (29). Pop6p interacts with MRP RNA in the presence of poly(U) but the interaction is clearly blocked by excess poly(IC), indicative of non-specific binding to double-stranded RNA. Pop1p may also have a preference for a double-stranded region of MRP RNA. Note that the levels of RNA binding by Pop1p are probably an underestimation of its RNA binding activity since its purity is lower and the protein has relatively poor stability.
In a further attempt to address specificity, competitor titration experiments were carried out to assess the relative ability of unlabelled MRP RNA, P RNA and poly(U) to compete off the 32P-MRP RNA. For proteins shown to bind 32P-MRP RNA less effectively in the presence of poly(IC) (Figure 5A), difficulties in accurately determining the molar amounts of poly(IC) due to heterogeneity, precluded an accurate comparison. In summary, for those proteins that possess apparent preference for single-stranded RNA (Pop3p, Pop4p, Pop5p, Rmp1p and Snm1p), only Snm1p displayed clear evidence of specificity for MRP RNA over poly(U) and P RNA (Figure 6). Pop4p would appear to have greater specificity for MRP RNA than P RNA, though this is complicated since addition of cold P RNA to a Pop4p-32P-MRP RNA complex reproducibly resulted in an initial decrease in retained radiolabel followed by an actual increase to above initial levels prior to complete competition (Figure 6). This anomaly will be further explored. Specificity of 1:1 protein–RNA interactions will require further study for full evaluation (see Discussion section).
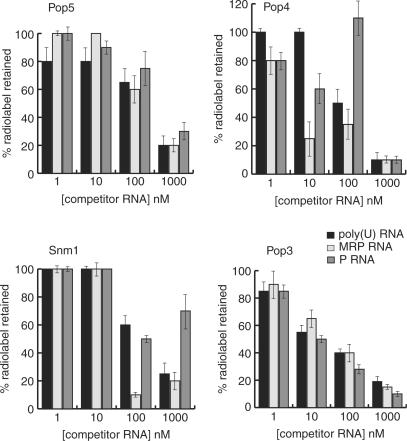
Figure 6.
Competition binding analysis of MRP RNA–protein interactions. Protein complexes with 32P-MRP RNA were challenged with increasing concentrations of unlabelled competitor RNA as indicated and the percentage of retained radiolabelled complex recorded. In general, protein subunits did not display clear specificity for MRP RNA. Data are shown for a subset of the protein–MRP RNA interactions; Pop5p, Pop4p, Snm1p and Pop3p. The Pop5p and Pop3p data clearly show no evidence of a specific interaction since the MRP RNA is competed at similar rates by non-specific poly(U) and potentially specific RNAs (MRP and P RNA). Pop4p data lacks clarity with respect to specificity. MRP appears to compete a little more effectively (∼3-fold) than poly(U). Competition by P RNA is complex, appearing to transiently enhance binding to labelled MRP-RNA prior to effectively displacing it. Uniquely, Snm1p competition binding analysis indicates a degree of specificity of Snm1p for MRP RNA, with effective displacement of labelled MRP RNA achieved at ∼5-fold lower concentrations of unlabelled MRP RNA relative to poly(U) and P RNA.
Note that a GST pull-down approach for screening protein–RNA interactions, similar to that described for studies of human RNase MRP subunits (24), was also adopted. Although the same subunits were observed to bind MRP RNA, with the addition of Pop7p, these results are not included here since a greater degree of non-specific association of RNA with immobilized proteins was observed. The same applies to screening of protein interactions with pre-rRNA (see Subsequently).
Interactions of protein subunits with the pre-rRNA substrate
Screening for interaction with the pre-rRNA substrate was carried out using a filter binding assay, as described in the preceding section for MRP RNA. Four protein subunits were identified as binding pre-rRNA; Pop4p, Pop6p, Rmp1p and Snm1p, see Figure 5B. The Snm1p K/S region also interacts with pre-rRNA. Although just above the chosen cutoff for an interaction, the case for an interaction of Pop5p with pre-rRNA is very marginal and it is not included as a positive result in our discussions. Tests were also again performed to assess the integrity of the pre-rRNA during incubation with proteins, for which the data are shown in the Supplementary Data.
No protein demonstrated truly specific pre-rRNA binding since one or other competitor reduced binding to background levels. Pop4p and Pop6p were observed to interact strongly with pre-rRNA in the presence of poly(U) competitor, but this interaction was lost in the presence of poly(IC) competitor. Rmp1p and Snm1p were observed to interact weakly with pre-rRNA in the presence of poly(U) competitor, but these interactions were again lost in the presence of poly(IC) competitor. These results indicate some preference of the proteins for double-stranded regions of pre-rRNA. The K/S tail of Snm1p was the only protein seen to interact weakly with pre-rRNA RNA in the presence of poly(IC) competitor, but this interaction was lost in the presence of poly(U) competitor, indicating some preference for single-stranded RNA. A GST pull-down approach confirmed interaction of Pop4p, Rmp1p, Snm1p and the Snm1p K/S tail with pre-rRNA (data not shown).
DISCUSSION
In the first steps towards assembly of a recombinant yeast RNase MRP complex for structure-function analysis, we have performed a systematic investigation of direct intermolecular interactions between individual protein and RNA subunits. These data are discussed in the context of existing data on the yeast and human RNase P/MRP systems (25) and support the evolutionary conservation of common core elements to the structure of the RNP enzymes.
Protein–protein interactions and implications for RNase MRP architecture
In summary, we have shown that each subunit can interact with at least three other subunits. Of the 31 protein–protein interactions, 13 were deemed strong and 18 weak (Figure 4A). The sheer number of interactions that the majority of the protein subunits can make indicates a highly integrated (and possibly dynamic) complex structure. All interactions in Figure 4 are reciprocal, with the exception of Pop4p-Pop5 and Pop6p-Pop7. Other non-reciprocal interactions were observed (Figure 3) but, on the basis of weak strength and the effects of higher salt concentrations, were excluded as being significant.
It is important to compare the data to previous observations in the yeast and human RNase P/MRP systems, since each study suffers its own experimental limitations. As such, where differences occur, it is difficult to conclude whether these are real differences between the possible enzyme architectures or a reflection of experimental artefacts leading to false positive or negative results. Hence, in assessing subunit interactions, we place most confidence in those reported here and also observed between RNase P/MRP subunits from the different organisms.
The foremost comparison is to previous studies of yeast RNase P/MRP subunits conducted by yeast two-hybrid analysis (23). We detect almost all of the 19 yeast two-hybrid reported pairwise interactions, adding weight to their validity as direct interactions. Exceptions to this are the observed Pop1p-Snm1p and Pop6p-Snm1p yeast two-hybrid interactions, which are notably rather weak. Inclusion of Rmp1p in our study contributes to the additional interactions we observe, in addition to a greater number of interactions observed for the Pop3p, Pop7p and Rpp1p subunits. Pop3p is the most notable, giving interactions with seven other subunits (five of these strong) versus only two also observed in two-hybrid analysis (Pop3p-Pop1p and Pop3p-Pop4p interactions). The authors mention possible Pop3p misfolding problems upon fusion in the yeast two-hybrid system (23). Here, Pop3p gives very similar data as either a GST fusion or in its cleaved, untagged form, suggesting it folds independently of the tag and also that the tag does not affect its interactions.
Human RNase MRP/P subunit interactions have also been studied (22,24). The human enzyme(s) also possess 10 subunits, of which 6 share low to moderate sequence identity with yeast RNase MRP protein subunit sequences (Pop1p/hPop1, Pop3p/Rpp38, Pop4p/Rpp29, Pop5p/hPop5, Pop7p/Rpp20, Rpp1p/Rpp30). In addition, Snm1p shares some weak sequence similarity to Rpr2p (the unique RNase P subunit) and to human Rpp21 (discuss later). A very recent sequence analysis (30) of RNase MRP/P subunits, has proposed that Pop6p may be the yeast homologue of metazoan Rpp25, placing Pop6p in the Alba superfamily, of which both the human Rpp25 and Rpp20 (Pop7) subunits are members (31). The same study suggests a relationship between Pop8p and Rpp14. Comparison of the yeast and human GST pull-down analyses (this work and ref. 24), reveals that yeast subunits with human homologues (assuming that Rpp14 and Rpp25 are homologues of Pop8p and Pop6p, respectively) make almost twice as many interactions in total as their human counterparts. The difference is again largely due to discrepancy in the number of interactions made by Pop3p/Rpp38, Pop7p/Rpp20 and Rpp1p/Rpp30; the human subunits exhibiting fewer interactions. A yeast two-hybrid analysis of human subunit interactions (22), however, detects more binding partners for these subunits, many of which are observed in our study (Figure 3).
One would expect subunits that lie more to the heart of the holoenzyme structure to make more contacts with other subunits. Considering strong interactions only (Figure 4A), this would place Pop1p, Pop4p, Rpp1p, Pop3p and Rmp1p at the core of yeast RNase MRP. Significantly, of these subunits, Pop1p, Pop4p and Rpp1p make the largest number of conserved interactions in all studies so far (six, eight and seven, respectively, including self-interactions), Figure 4B, supporting their central role in a conserved eukaryotic RNase MRP/P core structure. A central role for Pop3p is less well supported due to the difficulties encountered in yeast two-hybrid analysis of yeast RNase P alluded to earlier, although human studies identify six of the interactions we observe. Rmp1p is unique to RNase MRP and its interactions are discussed separately.
Given recent sequence analysis (30), the yeast RNase MRP/P subunits, Pop6p and Pop8p, are here considered homologues of human Rpp25 and Rpp14, respectively, and it is interesting to note that their interactions are very similar to those reported for their human counterparts (refer to Figure 3). Pop8p makes the fewest interactions of all subunits in both yeast studies, its interaction with the enzyme core possibly bridged by Pop5p. The lower strength and number of their interactions (in both yeast and human studies) place both Pop6p and Pop8p on the periphery of the RNase MRP/P structure, in keeping with roles such as regulation of substrate binding (directly or via the RNA subunit) or formation of higher order complexes, such as are known to occur in the ribosome biogenesis pathway (38). Peripheral subunits might also influence RNase MRP/P involvement in functions other than pre-rRNA/pre-tRNA processing (6,9). Notably, Pop8p, the only acidic yeast RNase MRP/P protein, is also the only subunit that can be depleted without a discernable effect on pre-rRNA processing (34–37).
Protein interactions of Snm1p and Rmp1p, subunits ‘unique’ to RNase MRP
In the absence of significant contrary evidence, the observations we have thus far made can be equally attributed to RNase MRP and RNase P and support a common ‘core’ structure to these related enzymes. RNase MRP differs from RNase P, however, with respect to the subunits Rmp1p (11) and Snm1p (55,56). This is the first report of interactions made by Rmp1p, strongly supportive of a key role within RNase MRP. An interaction with Snm1p is of particular note, suggesting that these two ‘unique’ subunits may work in close association, presumably in some way defining RNase MRP function, perhaps through directly or indirectly affecting substrate selection and/or cleavage. The absence of Rmp1p in RNase P is perhaps surprising given it interacts with subunits common to both RNase P and RNase MRP. Relatively few of its strong interactions are, however, with ‘core’ subunits and, crucially, interactions with Snm1p and MRP RNA, unique components of RNase MRP, are observed.
Snm1p makes relatively few interactions (to Pop4p, Rpp1p, Pop7p and Rmp1p) consistent with a position peripheral to the core enzyme structure. Although interactions of Snm1p are hard to corroborate given its identity as a ‘unique’ member of the yeast RNase MRP complex, Snm1p is a member of the Rpr2 Pfam alignment (Pfam accession PF04032), suggesting a structural and possibly functional link between the proteins. Rpr2p, however, has only been observed to interact with Pop4p and, perhaps, Pop6p (23). In pairwise sequence alignments using ClustalW and T-COFFEE (data not shown), although Rpr2p and Rpp21 share the highest similarity, Snm1p is clearly more closely related to Rpp21, both in the Cys-rich Zn-binding region and in its Lys/Ser-rich C-terminal region (the latter is absent in Rpr2p). In the absence of a clear human homologue to Snm1p, it is possible that Rpp21 has evolved to perform a role in both human RNase P and RNase MRP. Rpp21 has reported interactions with six proteins (Pop1p/hPop1, Pop4p/Rpp29, Pop3p/Rpp38, Pop8p/Rpp14, Rpp1p/Rpp30 and Pop7p/Rpp20) (22), of which Snm1p binds three, possibly utilizing similar recognition mechanisms.
Self-association of protein subunits indicates multiplicity within RNase P/MRP
Further to inter-subunit interactions, seven subunits self-associate (Pop1p, Pop3p, Rpp1p, Rmp1p, Pop4p, Pop5p and Pop7p), the first four of which do so strongly. Such self-association, together with the numerous protein–protein interactions would suggest the presence of these subunits in more than one copy in an assembled holoenzyme. Self-association has been previously observed for yeast Pop4p and Snm1p subunits (two-hybrid analysis) (23), the human Pop1 (hPop1), Pop3p (Rpp38), Pop4p (Rpp29) and Rpp1p (Rpp30) proteins (22,24) and archaeal Pop4p (Ph1771/Mth11) (33). Indeed, the only study to estimate stoichiometry using natively purified yeast RNase MRP (11) indicated most subunits are present in at least two copies with the exception of Pop1p. However, it would be unprecedented for a very large complex to have this number of multimeric subunits and the undoubtedly most accurate way to analyse stoichiometry would be through RNP assembly studies.
RNA–protein interactions
RNA binding subunits may provide RNA-chaperone and stabilization activities or play roles in substrate selection by the enzyme. Identification of RNA binding activities thus gives the first clues to possible protein subunit function. Here, we present novel RNA interaction data for many of the yeast RNase P/MRP subunits, including the first report of interactions with a pre-rRNA substrate.
In summary, six untagged protein subunits were observed to directly interact with MRP RNA and four with the pre-rRNA substrate (Figure 5). It is notable that none of the protein–RNA interactions are highly specific, although a preference for a double- or single-stranded context to the RNA bound is indicated in most cases. This observation is not altogether surprising given that the subunits integrate into a large particle for which specificity of RNA recognition (either MRP RNA or substrate pre-rRNA) may be conferred by the concerted interactions of multiple subunits. Synergistic binding to human MRP RNA has indeed been recently demonstrated for the human Rpp20 and Rpp25 proteins (39). Weak, non-specific RNA binding affinity may also simply assist with guiding substrate to the catalytic site. The binding of multiple subunits to single-stranded RNA may reflect the fact that the RNase MRP and RNase P enzymes cut their RNA substrates in single-stranded regions (40–42,29). In this respect, it is interesting that RNA homopolymers inhibit the yeast nuclear RNase P holoenzyme (29).
Detection of MRP RNA binding by Pop1p is not wholly unexpected since previous mutational analysis indicates interaction with the P3 domain of the yeast RNase P RNA subunit (23,44), a domain conserved in MRP RNA. Data on human RNase P/MRP is contradictory; yeast three-hybrid analysis of human subunits does not detect P RNA binding by hPop1 but GST-pull down analysis identifies hPop1 as one of six subunits to bind MRP RNA (24). We further detect interaction of Pop3p with MRP RNA, with clear preference for a single-stranded region. Although not detected as an RNA binding subunit in yeast three-hybrid analysis (23), recombinant yeast Pop3p has a predicted RNA binding fold (27) and was found to possess high affinity RNA-binding properties in vitro, with preferences for ssRNA (28). RNA binding by the human Pop3p homologue, Rpp38, has also been demonstrated (21,24,43). Although Pop3p is the only yeast RNase P subunit for which direct binding to pre-tRNA has been demonstrated (28), we do not observe interaction of Pop3p with substrate pre-rRNA.
Pop4p is the only yeast RNase MRP/P subunit besides Pop1p for which interaction with P RNA has been indicated through three-hybrid analysis (23). Evidence on the human (45,46,24) RNase MRP/P and archaeal (32) RNase P enzymes place this subunit in a central functional role, binding the RNA subunit and promoting RNA-based catalysis. Here, we provide the first evidence of direct RNA binding by yeast Pop4p, the protein displaying affinity for MRP RNA and the pre-rRNA substrate. Specificity for MRP/P RNA is not evident, though there is a possible preference for ssRNA. Conversely, the preference appears to be for double-stranded RNA in binding pre-rRNA. The crystal structures of archaeal Pop4p homologues (47,48) reveal a β-barrel structure highly similar to the Sm fold (known to bind U-rich single-stranded RNA), flanked by α-helices. Two possible sites for RNA binding are proposed (47), which would be in keeping with binding to both the RNase P/MRP RNA subunit and the RNA substrate. The yeast and human Pop4p proteins possess additional N-terminal regions that might also contribute to binding RNA or protein.
For Pop6p, we observe non-specific double-stranded binding activity to both the MRP RNA and pre-rRNA substrate. Binding to the latter would fit with the more peripheral position of this subunit in the RNP complex, discussed earlier. This is the first indication of RNA binding by this yeast subunit. The possible aforementioned similarity of Pop6p to alba superfamily member, human Rpp25 (30), is again interesting in this respect since GST pulldown analysis detects binding of Rpp25 to P RNA at two possible sites. Structures of the alba domain (53,54) indicate possibilities for specific and non-specific interaction sites on the domain. Interestingly, an extended β-hairpin makes contact with a DNA minor groove (non-specific). When we align Pop6p to the Alba superfamily, this hairpin region between the third and fourth β-strands is particularly extended and includes a number of basic residues.
Of the remaining subunits shared between RNase P and RNase MRP, we do not observe significant RNA binding by Pop5p, Pop7p, Pop8p or Rpp1p. Pop8p is the only acidic protein subunit, suggestive of non-involvement in RNA interactions. The Rpp1p result is in agreement with yeast three-hybrid interaction screens (23) for yeast RNase P/MRP subunits but contradicts previous data for the human (21,43) Rpp1p homologue in RNase P. However, differences between the yeast and human enzyme architectures may explain this inconsistency. Alternatively, Rpp1p in yeast RNase P/MRP may require association with another subunit for full RNA binding activity. Relatively, weak RNA binding by Pop7p (the human Rpp20 homologue) has been detected in one study only (24). Given that Pop7p is also a member of the alba superfamily, it might be expected to show at least non-specific RNA binding. Figure 5 does indicate binding to MRP RNA in the absence of any competitor. We observe significant formation of Pop7p-MRP aggregate in this experiment. Heparin, in addition to poly(IC) and poly(U), blocks the interaction (and aggregate formation) indicating a general ‘stickiness’ of Pop7p for nucleic acid that may suggest no more than a counter-ion role upon assembly into the RNase MRP complex. Interestingly, RNA binding by human Rpp20 is enhanced when complexed with Rpp25 (39). Pop5p has not emerged as a strong candidate for RNA binding in any of the other aforementioned screens conducted for eukaryal RNase P/MRP subunits, despite likely possessing an RRM-like fold (49). Even classical RRM domains, however, are diverse in their recognition mechanisms (50); some require association with another protein before binding RNA specifically (51) or may instead bind protein (52).
It is likely that the subunits Rmp1p and Snm1p will play important roles in the specific function of RNase MRP, whether in substrate recognition or in RNA processing. It is therefore interesting that we observe direct binding of each subunit to both MRP RNA and the pre-rRNA RNA, further to having observed the proteins interacting with each other. Future work to examine the RNA binding properties of both proteins could shed light on key aspects of RNase MRP versus RNase P function. Previous Snm1p mutagenesis studies in yeast (55) suggest that the protein has two functionally independent domains; an N-terminal zinc binding region probably required for association with the MRP RNA, and a C-terminal domain (the Lys/Ser-rich tail) not required for binding MRP RNA but necessary for RNA processing, indicating some involvement in substrate selection or cleavage. In accordance with these suggestions, we demonstrate direct interaction of Snm1p with both the MRP RNA and the pre-rRNA substrate RNA, while the Lys/Ser-rich C-terminal domain is only seen to interact with the substrate.
All screens of binary 1:1 interactions suffer from the same drawback that they may not fully reflect the affinity and specificity of interactions within the context of a holoenzyme where synergistic interactions are possible. Furthermore, a subunit may also attain conformational stability only on integration into the holoenzyme. Although proteins susceptible to misfolding, degradation or poor expression will be underrepresented in a comparative data analysis, we have established that a significant majority of the interactions identified here are conserved in other studies, providing further evidence of a conserved eukaryotic RNase MRP/P architecture and a strong basis from which to assemble enzyme complexes, a critical step in the study of RNase MRP/P architecture and the identification of subunit functions.
ACKNOWLEDGEMENTS
We are grateful to Emma Hollingsworth and Mark Timms for their early contributions to this project, and to Debbie Johns and Felicia Scott for plasmid DNA. Funding for the research was provided by Medical Research Council (UK) Career Establishment Grant (G9900092/69277 to J.M.A.); National Institutes of Health (R01 GM34869 to D.R.E.) and Biotechnology and Biological Sciences Research Council, UK (studentships to J.M.B.G. and H.J.B.). Funding to pay the Open Access publication charges for this article was provided by the University of Manchester (UK).
Conflict of interest Statement. None declared
REFERENCES
- 1.Chang DD, Clayton DA. A novel endoribonuclease cleaves at a priming site of mouse mitochondrial DNA replication. EMBO J. 1987;6:409–417. doi: 10.1002/j.1460-2075.1987.tb04770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reimer G, Raska I, Sheer U, Tan EM. Immunolocalization of 7-2-ribonucleoprotein in the granular components of the nucleolus. Exp. Cell Res. 1986;176:117–128. doi: 10.1016/0014-4827(88)90126-7. [DOI] [PubMed] [Google Scholar]
- 3.Li K, Smagula CS, Parsons WJ, Richardson JA, Gonzalez M, Hagler HK, Williams RS. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J. Cell Biol. 1994;124:871–882. doi: 10.1083/jcb.124.6.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lygerou Z, Allmang C, Tollervey D, Seraphin B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science. 1996;272:268–270. doi: 10.1126/science.272.5259.268. [DOI] [PubMed] [Google Scholar]
- 5.Tollervey D. Genetic and biochemical analyses of yeast RNase MRP. Mol. Biol. Rep. 1996;22:75–79. doi: 10.1007/BF00988709. [DOI] [PubMed] [Google Scholar]
- 6.Gill T, Cai T, Aulds J, Wierzbicki S, Schmitt ME. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: novel method of mRNA degradation. Mol. Cell. Biol. 2004;24:945–953. doi: 10.1128/MCB.24.3.945-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gill T, Aulds J, Schmitt ME. A specialized processing body that is temporally and asymmetrically regulated during the cell cycle in Saccharomyces cerevisiae. J. Cell Biol. 2006;173:35–45. doi: 10.1083/jcb.200512025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridanpaa M, Van Eenennaam H, Pelin K, Chadwick R, Johnson C, Yuan B, vanVenrooij W, Pruijn G, Salmela R, et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell. 2001;104:195–203. doi: 10.1016/s0092-8674(01)00205-7. [DOI] [PubMed] [Google Scholar]
- 9.Reiner R, Ben-Asouli Y, Krilovetzky I, Jarrous N. A role for the catalytic ribonucleoprotein RNase P in RNA polymerase III transcription. Genes Dev. 2006;20:1621–1635. doi: 10.1101/gad.386706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chamberlain JR, Lee Y, Lane WS, Engelke DR. Purification and characterization of the nuclear RNase P holoenzyme complex reveals extensive subunit overlap with RNase MRP. Genes Dev. 1998;12:1678–1690. doi: 10.1101/gad.12.11.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salinas K, Wierzbicki S, Zhou L, Schmitt ME. Characterization and purification of Saccharomyces cerevisiae RNase MRP reveals a new unique protein component. J. Biol. Chem. 2005;280:11352–11360. doi: 10.1074/jbc.M409568200. [DOI] [PubMed] [Google Scholar]
- 12.Collins LJ, Moulton V, Penny D. Use of RNA secondary structure for studying the architecture of the RNA component of yeast RNase MRP. J. Mol. Biol. 2000;292:827–836. [Google Scholar]
- 13.Forster AC, Altman S. Similar cage-shaped structures for the RNA components of all ribonuclease P and ribonuclease MRP enzymes. Cell. 1990;62:407–409. doi: 10.1016/0092-8674(90)90003-w. [DOI] [PubMed] [Google Scholar]
- 14.Frank DN, Adamidi C, Ehringer MA, Pitulle C, Pace NR. Phylogenetic-comparative analysis of the eukaryl Ribonuclease P RNA. RNA. 2000;6:1895–1904. doi: 10.1017/s1355838200001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindahl L, Fretz S, Epps N, Zengel JM. Functional equivalence of hairpins in the RNA subunit of RNase MRP and RNase P in Saccharomyces cerevisiae. RNA. 2000;6:653–658. doi: 10.1017/s1355838200992574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, Frank DN, Pace N, Zengel JM, Lindahl L. Phylogenetic analysis of the structure of RNase MRP RNA in yeasts. RNA. 2002;8:740–751. doi: 10.1017/s1355838202022082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walker SC, Avis JM. A conserved element in the yeast RNase MRP RNA subunit can participate in a long-range base-pairing interaction. J. Mol. Biol. 2004;341:375–388. doi: 10.1016/j.jmb.2004.05.076. [DOI] [PubMed] [Google Scholar]
- 18.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of Ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 19.Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some archaea are catalytically active. Proc. Natl Acad. Sci. USA. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kikovska E, Svârd SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl Acad. Sci. USA. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang T, Guerrier-Takada C, Altman S. Protein-RNA interactions in the subunits of human nuclear RNase P. RNA. 2001;7:937–941. doi: 10.1017/s1355838201010299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang T, Altman S. Protein-protein interactions with subunits of human nuclear RNase P. Proc. Natl Acad. Sci. USA. 2001;98:920–925. doi: 10.1073/pnas.021561498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houser-Scott F, Xiao S, Millikin CE, Zengel JM, Lindahl L, Engelke DR. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proc. Natl Acad. Sci. USA. 2002;99:2684–2689. doi: 10.1073/pnas.052586299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welting TJ, van Venrooij WJ, Pruijn GJ. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32:2138–2146. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Battle DJ, Doudna JA. The stem-loop binding protein forms a highly stable and specific complex with the 3′ stem-loop of histone mRNAs. RNA. 2001;7:123–132. doi: 10.1017/s1355838201001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dlakic M. 3D models of yeast RNase P/MRP proteins Rpp1 and Pop3. RNA. 2005;11:123–127. doi: 10.1261/rna.7128905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brusca EM, True HL, Celander DW. Novel RNA-binding properties of Pop3p supports a role for eukaryotic RNase P protein subunits in substrate recognition. J. Biol. Chem. 2001;276:42543–42548. doi: 10.1074/jbc.M107293200. [DOI] [PubMed] [Google Scholar]
- 29.Ziehler WA, Day JJ, Fierke CA, Engelke DR. Effects of 5′ leader and 3′ trailer structures on pre-tRNA processing by nuclear RNase P. Biochemistry. 2000;39:9909–9916. doi: 10.1021/bi000603n. [DOI] [PubMed] [Google Scholar]
- 30.Rosenblad MA, Lopez MD, Piccinelli P, Samuelsson T. Inventory and analysis of the protein subunits of ribonucleases P and MRP provides further evidence of homology between the yeast and human enzymes. Nucleic Acids Res. 2006;34:5145–5156. doi: 10.1093/nar/gkl626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aravind L, Iyer LM, Anantharaman V. The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003;4:R64. doi: 10.1186/gb-2003-4-10-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kouzuma Y, Mizoguchi M, Takagi H, Fukuhara H, Tsukamoto M, Numata T, Kimura M. Reconstitution of archaeal Ribonuclease P from RNA and four protein components. Biochem. Biophys. Res. Commun. 2003;306:2684–2689. doi: 10.1016/s0006-291x(03)01034-9. [DOI] [PubMed] [Google Scholar]
- 33.Boomershine WP, McElroy CA, Tsai HY, Wilson RC, Gopalan V, Foster MP. Structure of Mth11/MthRpp29, an essential protein subunit of archaeal and eukaryotic RNase P. Proc. Natl Acad. Sci. USA. 2003;100:15398–15403. doi: 10.1073/pnas.2535887100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dichtl B, Tollervey D. Pop3p is essential for the activity of the RNase MRP and RNase P ribonucleoproteins in vivo. EMBO J. 1997;16:417–429. doi: 10.1093/emboj/16.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lygerou Z, Mitchell P, Petfalski E, Seraphin B, Tollervey D. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 1994;8:1423–1433. doi: 10.1101/gad.8.12.1423. [DOI] [PubMed] [Google Scholar]
- 36.Chu S, Zengel JM, Lindahl L. A novel protein shared by RNase MRP and RNase P. RNA. 1997;3:382–391. [PMC free article] [PubMed] [Google Scholar]
- 37.Stolc V, Altman S. Rpp1, an essential protein subunit of nuclear RNase P required for processing of precursor tRNA and 35 S precursor rRNA in Saccharomyces cerevisiae. Genes Dev. 1997;11:2414–2425. doi: 10.1101/gad.11.18.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fatica A, Tollervey D. Making ribosomes. Curr. Opin. Cell Biol. 2002;14:313–318. doi: 10.1016/s0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 39.Welting TJM, Peters FMA, Hensen SMM, van Doorn NL, Kikkert BJ, Raats JMH, van Venrooij WJ, Pruijn GJM. Heterodimerization regulates RNase MRP/RNase P association, localization, and expression of Rpp20 and Rpp25. RNA. 2007;13:65–75. doi: 10.1261/rna.237807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitt ME, Clayton DA. Nuclear RNase MRP is required for correct processing of pre-5.8S ribosomal RNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1993;13:7935–7941. doi: 10.1128/mcb.13.12.7935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Nues RW, Rientjes JMJ, Vandersande CAFM, Zerp SF, Sluiter C, Venema J, Planta RJ, Raue HA. Separate structural elements within internal transcribed spacer-1 of Saccharomyces cerevisiae precursor ribosomal-RNA direct the formation of 17S and 26S ribosomal RNA. Nucleic Acids Res. 1994;22:912–919. doi: 10.1093/nar/22.6.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chamberlain JR, Paganramos E, Kindelberger DW, Engelke DR. An RNase P RNA subunit mutation affects ribosomal RNA processing. Nucleic Acids Res. 1996;24:3158–3166. doi: 10.1093/nar/24.16.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pluk H, vanEenennaam H, Rutjes SA, Pruijn JM, vanVenrooij WJ. RNA-protein interactions in the human RNase MRP ribonucleoprotein complex. RNA. 1999;5:512–524. doi: 10.1017/s1355838299982079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ziehler WA, Morris J, Scott FH, Millikin C, Engelke DR. An essential protein-binding domain of nuclear RNase P RNA. RNA. 2001;7:565–575. doi: 10.1017/s1355838201001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mann H, BenAsouli Y, Schein A, Moussa S, Jarrous N. Eukaryotic RNase P: role of RNA and protein subunits of a primordial catalytic ribonucleoprotein in RNA-based catalysis. Mol. Cell. 2003;12:925–935. doi: 10.1016/s1097-2765(03)00357-5. [DOI] [PubMed] [Google Scholar]
- 46.Sharin E, Schein A, Mann H, Ben-Asouli Y, Jarrous N. RNase P: role of distinct protein cofactors in tRNA substrate recognition and RNA-based catalysis. Nucleic Acids Res. 2005;33:5120–5132. doi: 10.1093/nar/gki828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Numata T, Ishimatsu I, Kakuta Y, Tanaka I, Kimura M. Crystal structure of archaeal ribonuclease P protein Ph1771 p from Pyrococcus horikoshii OT3: an archaeal homolog of eukaryotic ribonuclease P protein Rpp29. RNA. 2004;10:1423–1432. doi: 10.1261/rna.7560904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sidote DJ, Heideker J, Hoffman DW. Crystal structure of archaeal ribonuclease P protein aRpp29 from Archaeoglobus fulgidus. Biochemistry. 2004;43:14128–14138. doi: 10.1021/bi048578z. [DOI] [PubMed] [Google Scholar]
- 49.Wilson RC, Bohlen CJ, Foster MP, Bell CE. Structure of Pfu Pop5, an archaeal RNase P protein. Proc. Natl Acad. Sci.USA. 2006;103:873–878. doi: 10.1073/pnas.0508004103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maris C, Dominguez C, Allain FHT. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;9:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 51.Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B''-U2A' protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 52.Fribourg S, Gatfield D, Izaurralde E, Conti E. A novel mode of RBD-protein recognition in the Y14-Mago complex. Nat. Struct. Biol. 2003;10:433–439. doi: 10.1038/nsb926. [DOI] [PubMed] [Google Scholar]
- 53.Wardleworth BN, Russell RJM, Bell SD, Taylor GL, White MF. Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 2002;21:4654–4662. doi: 10.1093/emboj/cdf465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Biyani K, Kahsai MA, Clark AT, Armstrong TL, Edmondson SP, Shriver JW. Solution structure, stability, and nucleic acid binding of the hyperthermophile protein Sso10b2. Biochemistry. 2005;44:14217–14230. doi: 10.1021/bi051266r. [DOI] [PubMed] [Google Scholar]
- 55.Cai T, Reilly TR, Cerio M, Schmitt ME. Mutagenesis of SNM1, which encodes a protein component of the yeast RNase MRP, reveals a role for this ribonucleoprotein endoribonuclease in plasmid segregation. Mol. Cell. Biol. 1999;19:7857–7869. doi: 10.1128/mcb.19.11.7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmitt ME, Clayton DA. Characterization of a unique protein component of yeast RNase MRP - an RNA binding protein with a zinc-cluster domain. Genes Dev. 1994;8:2617–2628. doi: 10.1101/gad.8.21.2617. [DOI] [PubMed] [Google Scholar]