Abstract
The length of the poly(A) tail of an mRNA plays an important role in translational efficiency, mRNA stability and mRNA degradation. Regulated polyadenylation and deadenylation of specific mRNAs is involved in oogenesis, embryonic development, spermatogenesis, cell cycle progression and synaptic plasticity. Here we report a new technique to analyse the length of poly(A) tails and to separate a mixed population of mRNAs into fractions dependent on the length of their poly(A) tails. The method can be performed on crude lysate or total RNA, is fast, highly reproducible and minor changes in poly(A) tail length distribution are easily detected. We validated the method by analysing mRNAs known to undergo cytoplasmic polyadenylation during Xenopus laevis oocyte maturation. We then separated RNA from NIH3T3 cells into two fractions with short and long poly(A) tails and compared them by microarray analysis. In combination with the validation experiments, the results indicate that ∼25% of the expressed genes have a poly(A) tail of less than 30 residues in a significant percentage of their transcripts.
INTRODUCTION
The poly(A) tail of an mRNA plays an important role during mRNA metabolism. After transcription the mRNA is capped, spliced and the 3′ end undergoes processing. After recognition of the hexanucleotide and a downstream GU-rich sequence the mRNA is cleaved and a poly(A) tail of ∼100–250 nt is added (1). After export to the cytoplasm the poly(A) tail usually gets gradually shortened. However, in some specific cases poly(A) tail length is strictly regulated and therefore dramatic changes of the poly(A) tail length can occur. The poly(A) tail recruits the cytoplasmic poly(A)-binding protein (PABP), which interacts with the cap-binding translation initiation factor eIF4E via the scaffolding protein eIF4G (2).
mRNA stability and degradation are essential for the regulation of gene expression. In general mRNA decay is thought to be dependent on deadenylation (3). In somatic cells, mRNA degradation and deadenylation of specific mRNAs have been shown to be strongly linked. AU-rich elements (AREs) promote rapid deadenylation-mediated decay and are found in some mRNAs-encoding cytokines, proto-oncogenes and growth factors (4). Several microRNAs (miRNAs) direct rapid deadenylation of mRNAs with sequence elements, which have imperfect complementarity, which correlates with the degradation of these mRNAs (5,6).
The regulation of poly(A) tail length has also been implicated in translational control. In general a long poly(A) tail correlates with active translation while a short poly(A) tail is linked with translational repression (7). The best characterized role of poly(A) tail length in translation has been in oocytes and embryos of Xenopus laevis (1,8,9). During oocyte maturation and early embryogenesis transcription is silent and therefore translational control is the main regulator of gene expression. Many mRNAs with short poly(A) tails are stored in an inactive state during oogenesis to be translationally activated by cytoplasmic polyadenylation, while others are inactivated by deadenylation (8,9). These regulatory mechanisms are not just restricted to X. laevis, but are very well conserved in many species including Drosophila and mouse (10). It is also important for spermatogenesis (11,12), cell cycle progression (9) and synaptic plasticity (13–15). Several factors implicated in polyadenylation and deadenylation have been reported to play a role in human diseases such as cancer, myotonic dystrophy and rheumatoid arthritis (16–22).
Although several techniques are available to study poly(A) tail length, none have proved generally applicable, sensitive and reliable in estimating the absolute length of the poly(A) tail. One of the major problems is that the poly(A) tail length of a specific mRNA usually has a wide distribution and in some cases deadenylated and polyadenylated mRNAs may be present in the same mRNA preparation, for instance when studying cell cycle mRNAs in non-synchronized cells (23). Longer poly(A) tails tend to be more diverse in size and it is therefore easy to underestimate the fraction of mRNA with a long poly(A) tail in methods that visualize the size of the poly(A) tail of an mRNA by gel electrophoresis, because the signal spreads over a considerable area and the intensity is soon too close to background for reliable quantification.
Most mRNAs are too long for the change in poly(A) tail length to be detectable by direct northern blotting. The classical method to determine the poly(A) tail length of an endogenous mRNA is to cleave the mRNA at a specific point with an oligonucleotide and RNAse H to obtain a 600–300 nt fragment which can be resolved sufficiently on an agarose gel and detected by northern blotting. Using markers and a control in which oligo dT is added to the RNase H treatment to remove the poly(A) tail, an estimate of tail length can be obtained (24–26). This technique is limited to relatively abundant mRNAs and is very prone to the underestimation of the long poly(A) tail fraction, as outlined above.
The introduction of PCR-based methods has improved the sensitivity of the analysis. The RACE-PAT (rapid amplification of cDNA ends poly(A) test) and the LM-PAT (ligation-mediated poly(A) test) are both based on the use of an oligo(dT) linked to an anchor sequence for the RT reaction. For the RACE-PAT the oligo can anneal anywhere in the poly(A) tail while in the LM-PAT the oligo binds at the 3′end of the poly(A) tail (27). A modification of these protocols is the RNA-ligation coupled RT-PCR (RL-PAT) where an oligo is ligated directly to the 3′end of the mRNA and the RT is performed using an oligo annealing to the first oligo (28). For the cRT-PCR (circularization RT-PCR) method, the cap is removed from the mRNA after which the RNA is circularized using T4 RNA ligase. The RT-PCR is the performed over the joint to determine the length of the poly(A) tail (29). For all PCR-based methods oligo(dT)/RNaseH treatment can be used first to remove the poly(A) tail to obtain a size estimate. All PAT methods are prone to the usual problems associated with RT-PCR, it is not uncommon for unexplained bands to appear and often three to four primers have to be tested before a suitable assay can be designed, making the analysis of a large number of mRNAs very tedious. In addition, the underestimation of the long poly(A) tail fraction also occurs using these methods, with the RACE-PAT exacerbating the problem by priming over the full-length of the poly(A) tail. The estimate of the poly(A) tail lengths can be improved significantly if the PCR is performed with a radioactive label to increase the signal-to-noise ratio.
By using reporter RNAs, the length of the transcript studied can be changed at will. In Xenopus oocytes and embryos radiolabelled RNAs can be injected to analyse changes in poly(A) tail length of reporter RNAs. Alternatively radiolabelled RNAs can be incubated in extracts. This is a powerful technique that has revealed much on the roles of deadenylation and cytoplasmic polyadenylation in translational control (30). Because the radioactive probe can be very short and can be detected with a high sensitivity and low background, the underestimation of long poly(A) tails is not as problematic as with RNaseH cleavage and PAT. It is however not applicable to most other experimental systems.
In the study of the role of deadenylation in mRNA stability, rapid on/off inducible reporter genes with short transcripts are often used (31). In this system, poly(A) changes can be easily detected by northern blotting. Because the transcripts are the same age, their poly(A) tail length is usually more uniform. Unfortunately, the restrictions on size of the mRNA exclude the study of long 3′ UTRs in their entirety, unless a combination with RNase H mapping is employed. As with all reporter assays, it is necessary to check that the endogenous mRNA behaves in the same way as the reporter.
All the methods described above study individual mRNAs, which is what has informed most of our current understanding of poly(A) tail metabolism. To analyse the global distribution of poly(A) tail length by microarray it is necessary to select RNA with a certain poly(A) tail length. Graindorge et al. (32) simply used an oligo(dT) primer to synthesize the cDNA required for the array. This method included all RNAs with a poly(A) tail or 10 nt or more with equal efficiency, while it did not bind any of the RNAs with a poly(A) tail of 5 nt or less. This technique therefore does not distinguish between oligoadenylated and polyadenylated mRNAs. Du and Richter (33) employed poly(U) agarose and eluted poly(A) RNA at 60°C with a buffer containing 25% formamide, resulting in elution of all RNAs with a poly(A) tail of at least 55 As. While this technique has potential, it is labour-intensive and difficult to operate reproducibly. In addition, in systems other than Xenopus oocytes and embryos, the unbound fraction will contain mRNA degradation products in addition to full-length oligoadenylated mRNAs, making it unsuitable for the identification of oligoadenylated mRNAs.
Here we describe a technique that allows for the isolation of mRNAs with different poly(A) tails in multiple fractions. Poly(A) RNA is bound to biotinylated oligo(dT) which in turn is bound to paramagnetic beads. The mRNAs are then eluted using decreasing salt concentration, resulting in the separation of mRNAs based on the length of their poly(A) tail. This method can be used on crude lysate or total RNA, is fast, highly reproducible and detects small changes in poly(A) tail length. We validated the method by analysing mRNAs known to undergo cytoplasmic polyadenylation during X. laevis oocyte maturation. We then separated RNA from NIH3T3 cells into two fractions of short and long poly(A) tails and subjected these fractions to microarray analysis. Validation of the microarray results by RL-PAT and northern analysis demonstrated that there is a remarkably high number of mRNA species with 25% or more of transcripts with an oligoadenylate tail of less than 30 residues.
MATERIALS AND METHODS
Oocytes and tissue culture cells, RNA isolation and RNA 3′end-labelling
Ovaries of adult female X. laevis were obtained and cultured as described previously (34). Some of the stage six oocytes were treated with progesterone (100 nM) to induce maturation.
NIH3T3 fibroblasts were cultured in DMEM with 10% fetal bovine serum. The day before RNA isolation, 3 million cells were seeded in a 15 cm culture dish. RNA isolation was performed according to Chomczynski and Sacchi (35).
RNA was 3′end-labelled with 32P as described by Minvielle-Sebastia et al. (36).
Probe synthesis and polyadenylation
pBluescript II SK- (Stratagene) was digested with EcoRV and used to synthesize a radioactive RNA using T3 RNA polymerase (Promega). The RNA was extracted with phenol/chloroform and purified over a Sephadex G50 spin column. The marker was synthesized in a similar way using a mixture of different templates.
The probe was polyadenylated using a poly(A) tailing kit (Ambion) according to manufacturer's instructions with the following alterations: several dilutions of E-PAP were used to obtain the desired poly(A) tail length and the reaction time was exactly 30 min. The reaction was stopped by the addition of an equal volume of TE and the RNA was extracted using phenol/chloroform. After analysis of a fraction of the probe and polyadenylated products by urea—PAGE, they were mixed to get a sample in which poly(A) tails of 0–200 adenylate residues were represented.
Poly(A) fractionation
Ten X. laevis oocytes or 80 μg of total RNA isolated from NIH3T3 cells were resuspended in 400 μl GTC buffer (4 M guanidine thiocyanate, 25 mM sodium citrate, pH 7.1, 2% β-mercaptoethanol). Five microlitres polyadenylated probe mix (see above) and 10 μl (for oocytes) or 15 μl (for RNA from NIH3T3 cells) biotinylated oligo(dT) (50 pmol/μl, Promega) were added. The biotin is covalently attached to the 25-mer DNA oligo via an 18-carbon spacer arm (37). Attempts to replace this oligo with a LNA or DNA oligo with the biotin directly linked to the oligo were not successful (data not shown). 816 μl dilution buffer (6× SSC, 10 mM Tris–HCl, pH 7.4, 1 mM EDTA, 0.25% SDS, 1% β-mercaptoethanol) was added, the mixture was incubated for 5 min at 70°C and centrifuged at 12 000g for 10 min at room temperature. Per sample 600 μl of paramagnetic beads (Promega) were washed 3× with 0.5× SSC. The supernatant was added to the beads and the oligo(dT) was allowed to bind for 15 min at room temperature while rotating. Room temperature was usually 18°C, but no significant differences were detected at temperatures up to 25°C. The unbound fraction was collected and kept on ice. After three washes with 0.5× SSC at room temperature, the polyadenylated RNA was eluted using elution buffers containing decreasing concentrations of SSC at room temperature. Between the elution steps the beads were kept rotating at room temperature for at least 5 min. All fractions were precipitated with 10 μg yeast tRNA. Part (10–20%) of all fractions was analysed on urea–PAGE to determine the poly(A) tail length in each fraction. The remainder of the fractions was analysed by microarray, RT-PCR or northern blotting.
Microarray analysis
The microarrays consisted of a set of mouse ESTs derived from the IMAGE clone collection and the NIA mouse 15K set. The average insert size is 1000 bp. The set was prepared by PCR amplification using vector-specific primers on a 384-well basis. Amplification was assessed using gel electrophoresis on a 96-well format, and the clones resuspended in 1.5 M betaine with 3× SSC for printing. The arrays were printed on poly(L) lysine-coated glass using a pin deposition method on a Stanford type arrayer.
The samples for the microarray were generated by poly(A) fractionation. The RNA was eluted with 0.075× SSC (oligoadenylated RNA) and H2O (polyadenylated RNA). cDNA was primed with random hexamers and transcribed using Superscript III (Invitrogen) in the presence of 0.3 mM aminoallyl dUTP, 0.2 mM dTTP, 0.5 mM dATP, 0.5 mM dCTP, 0.5 mM dGTP and labelled by coupling Cy5 (oligo adenylated RNA) or Cy3 (polyadenylated RNA) (GE Healthcare) to the aminoallyl dUTP in the presence of 0.1 M Na2CO3 pH 9 for 1 h at room temperature The reaction was stopped by the addition of hydroxylamine to a concentration of 1.2 M. The labelled cDNA was mixed with polyA, tRNA and mouse Cot 1 DNA, placed on the arrays and hybridized overnight at 50°C.
The arrays were scanned on an Axon Genepix 4000b scanner and quantified with Genepix Pro 6. Probes with a signal below or close to background and probes obscured by speckles or splodges were removed from analysis, leaving 2308 probes out of 10 944. The LogRatio was calculated as follows: LogRatio = 2log[(Mred − LBGred)/(Mgreen − LBGgreen)], in which M are the median values for red and green fluorescence in a probe spot and LBG is the local background. The corrected LogRatio was obtained by subtracting the LogRatio median value of all probes with a detectable signal, setting the median LogRatio value at 0. To generate Supplementary Table 2, the average corrected LogRatio and its SD were calculated. Probes with a standard deviation of 0.7 or higher were removed, leaving 2246 probes. To generate Table 2 and Supplementary Table 3, EST sequences for the 220 probes with the highest LogRatio were used for a BLAST homology search using the protocol for somewhat similar sequences (blastn, NCBI). Probes from ESTs that cross-hybridized to mRNAs from related genes over more than 100 nt were discarded, with the exception of products from duplicate genes of such high homology that they can be expected to be regulated identically (90% identity or more). The only probe that gave consistently negative results in the validation experiments, Sae2/Uble1b, was also removed from the lists in Table 2 and Supplementary Table 3 to reduce confusion. This left 148 probes, which were placed manually in the functional categories.
Table 2.
Functional classification of the top 10% deadenylated mRNAs
| Initiation factors (7) |
| Eif2s2, Eif3s4, Eif3s7, Eif3s9, Eif4b, Eif4e2, Eif4g2 |
| Initiation factor associated proteins(2) |
| Eif4ebp2, Eif3s6ip |
| Aminoacyl-tRNA-synthetases (3) |
| Aars, Eprs, Yars |
| Other RNP proteins (18) |
| Cdc5l, Csda, Cugbp2, Ddx21, Ddx24, Eif2c2, Hdlbp, Heatr1, Larp5, Mrto4, Nol5, Npm3, Serbp1, Sf3b2, Srp72, Tcof1, Tnpo3, Tsn |
| Cell adhesion and actin cytoskeleton (13) |
| AA536749 (p116Rip), Actn4, Adrm1, Basp1, C920006C10Rik, Cald1, Capzb, Cdc42, Cdh2, Mapkapk2, Mapre1, Tm4sf1,Tpbg |
| Cell cycle (10) |
| Aurkb, Calm1, Ccnd2, Ccnd3, Cdc42, Cdc5l, Mad2l1bp, Nubp1, Nuf2, Tbrg1 |
| Chromatin components and transcription factors (19) |
| Actl6a, Cebpa, Ercc1, Ercc5, Gins4, Hist1h3e, Hist1h4c, Hist1h4d, Hmga1, Msh6, Nfic, Pold2, Rnf4, Rpo1-1, Snd1, Sssca1, Tcf25, Tsn, Whsc1 |
| Ubiquitination, proteasome and associated proteins (11) |
| Adrm1, Cct5, Cops4, Psmc5, Psmd1, Psmd11, Rnf26, Rnf4, Sqstm1, Trip12, Ube2r2 |
| Endoplasmatic reticulum, golgi and lysosomes (14) |
| Ap3b1, Ccdc115, Cdipt, Elovl6, Foxred2, Gorasp2, Ift20, Mtmr4, Pigc, Sec63, Srp72, Stt3b, Tmed5, Tmed8 |
| Other mRNAs (47) |
| 1200011I18Rik, 1600014C10Rik, 2210412D01Rik, 2510006D16Rik, 2810026P18Rik, 4921533L14Rik, Acaa2, Alad, Armcx2, Asnsd1, Aven, C030045D06Rik, Comt, Csnk2b, D15Wsu75e, Ddah1, Drg1, Dtl, EG622339, Ext1, Frrs1, Gcat, Gdap1, Gpiap1, Grina, Hdgf, |
| Hsp90aa1, Il6st, Irgq, Lap3, Lrrc17, Mark2, Muc5ac, Neat2, Neat2, Pex13, Sh3d19, Spred1, Thop1, Tmprss2, Tomm22, Tspan31, Ttc1, Txndc9, Wbp5, Zfp606 |
The non-cross-hybridizing top 10% of deadenylated mRNAs (148 probes) were put into functional categories as indicated. This table is an extract of Supplementary Table 3.
RL-PAT, RT-PCR and northern blotting
RNA ligation-mediated polyadenylation assays (RL-PAT) were performed as described (38). For the analysis of the X. laevis genes, RTs were performed using the 1st Strand Synthesis Kit for RT-PCR, AMV (Roche) according to protocol with the use of a mix of specific primers (Supplementary Table 1). For the mouse RNAs the RT was performed using Superscript III (Invitrogen) and random hexamers. If required the samples were treated with Dnase before the RT. PCRs were performed using Taq DNA polymerase (Roche). For primers see Supplementary Table 1. Both the RL-PAT and PCR products were analysed by agarose gel electrophoresis.
Northerns were performed as described before (39). For the northerns on Xenopus samples the equivalent of five oocytes was used for the poly(A) fractions and the equivalent of 1.5 oocyte was used for total RNA. Plasmids required for templates for Xenopus probes were described previously: CyclinB1 (40); or obtained from the Image clone collection: RPS6 (Image clone 4056253) and GAPDH (Image clone 4404458). Templates for probes were created by digesting plasmids with the appropriate enzymes and isolating the fragment from gel: CyclinB1 with BamHI and EcoRI, GAPDH and RPS6 with NotI and SalI. For the northerns on mouse RNA, 4 μg of total RNA was loaded and for each of the poly(A) fractions (unbound, short, long) the equivalent of 20 μg total RNA was loaded. The probes for the Northern blots on mouse mRNA were generated by RT-PCR with the primers listed in Supplementary Table 1, except for the probes for polypyrimidine tract-binding protein (Ptb) which were the 333 and 500 bp Nco1 fragments from human PTB from pCMV-PTB (41), the full coding region from human YBX1 cloned from an RT-PCR product (Cobbold et al., manuscript submitted for publication) and an 800 nt Xho1 fragment representing the 3′ end of a mouse Tacc3 cDNA clone including the coiled-coil domain coding region and the full 3′ UTR.
RESULTS
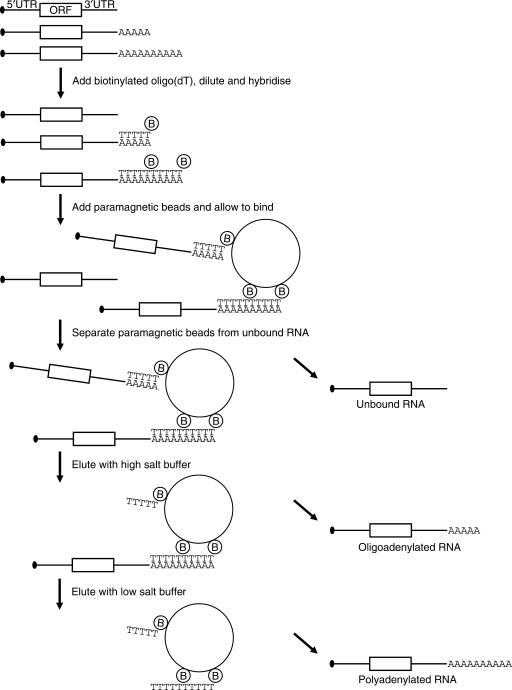
To analyse the length of poly(A) tails in detail we adapted the oligo(dT) affinity chromatography protocol developed by Promega (37) (Figure 1). In contrast to the hybridization properties of oligo(dT) coupled to a support, we expected that the solution hybridization of saturating concentrations of T25 oligonucleotides would lead to a continuous nicked duplex, in which the oligonucleotides are stacked to provide increased thermodynamic stability, as has been described for nicked DNA duplexes (42). In addition, under conditions where only a portion of the duplexes is melted, the presence of more oligo(dT) molecules should significantly increase the chances of an mRNA to stay on the beads. Cells or total RNA were resuspended in a GTC buffer, the lysate was mixed with biotinylated oligo(dT), diluted and bound to paramagnetic beads. The poly(A) RNA was then eluted in fractions using buffers of decreasing salt concentration. To calibrate the eluted poly(A) tail lengths a radiolabelled and polyadenylated probe with a variety of adenylate tails of up to 120 nt or more was added to each sample. Comparison of the distribution of this probe with a marker enables the determination of the length of the poly(A) tail in each fraction (For details on synthesis, see Supplementary Figure 1).
Figure 1.
Poly(A) fractionation method. Schematic representation of the poly(A) fractionation protocol. Cells or total RNA are resuspended in a GTC buffer, mixed with a radiolabelled polyadenylated probe and biotinylated oligo(dT) and diluted. The oligo(dT) is then hybridized with the RNA, resulting in stacking of the oligonucleotides on longer poly(A) tails and bound to paramagnetic beads. The beads are washed with 0.5 × SSC and the RNA is eluted using decreasing salt concentrations, eluting the RNAs with short poly(A) tails first.
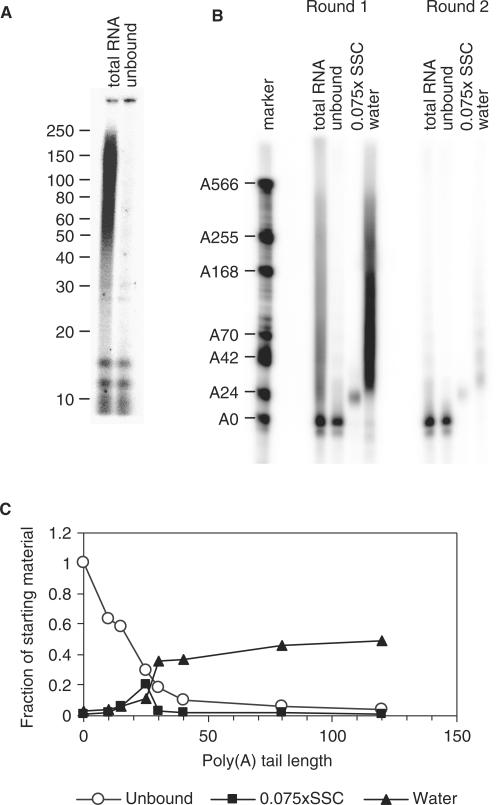
For maximal recovery of the polyadenylated RNAs it indeed proved important that an excess of oligo(dT) is used (Supplementary Figure 2). To evaluate whether the capture of short poly(A) tails can be achieved with reasonable efficiency using this system, we needed to determine which sizes of poly(A) tail are retained on the paramagnetic beads. For this purpose, we 3′ end-labelled the total RNA from NIH3T3 cells and digested the RNA with RNases A and T1, which only will leave labelled poly(A) tails. Figure 2A shows that the poly(A) tails in these cells generally range from 25 to 250 residues, with discrete bands at around 12 and 15–16 nt. Following the same procedure, the unbound fraction (U) obtained after oligo(dT) capture was analysed, and all poly(A) tail sizes above 15 nt were found to be significantly depleted (Figure 2A). In a second experiment, the radioactively labelled probe was mixed with the same total RNA and the beads eluted with 0.075 × SSC (short poly(A) tails) and water (long poly(A) tails). Again, RNAs with poly(A) tails of A25 and longer were depleted efficiently from the unbound fraction (Figure 2B). The 0.075 × SSC elution recovered RNAs with poly(A) tails of ∼25 nt, while longer tails were eluted with water, indicating that separation can be achieved with this method. We quantified the signal in this figure for different sizes of poly(A) tail and carried out corrections for the amounts loaded on the gel. The numbers obtained indicate that RNA with a poly(A) tail of 25 was 70% depleted in the unbound fraction and longer poly(A) tails were over 80% depleted (Figure 2C). Approximately 20% of the starting material with a 25 nt oligoadenylate tail was recovered in the 0.075 × SSC elution and 10% was eluted with water. For the RNAs with poly(A) tails of 30 residues and over, 40–50% of the starting material was recovered in the water elution. These data show that the 0.075× SSC elution selectively isolates mRNAs with a poly(A) tail of ∼25 residues. To ascertain that short poly(A) tails had been bound efficiently, a second round of poly(A) fractionation was performed on the unbound fractions. As can be seen in the second panel of Figure 2B, only small amounts of probe with poly(A) tails of 25 nt and 30–80 nt were recovered from this fraction, again indicating that 70% of poly(A) tails of 25 nt and over are bound in the first instance. From these data we can conclude that the unbound fraction contains only minimal amounts of RNA with tails of more than 30 adenylates.
Figure 2.
Characterization of the binding and elution properties of biotinylated oligo(dT) chromatography. (A) Total RNA from NIH3T3 cells was subjected to oligo(dT) affinity chromatography. Starting material and the unbound fraction were examined for poly(A) tail lengths by 3′ end-labelling followed by RNase A and T1 digestion. The markers are the indicated sizes in number of nucleotides. (B) Total RNA from NIH3T3 cells was mixed with radiolabelled polyadenylated probe with A0–500 and subjected to chromatography as above and eluted with 0.075 × SSC and water (Round 1). The unbound fraction was resubjected to the same procedure (Round 2). For total and unbound, equivalent amounts were loaded, while for the elutions, five times more was loaded. The marker is labelled with the number of nucleotides in excess over the unadenylated probe, which is 79 nt long. (C) Quantification of (B) for different lengths of poly(A) tails. The figure was quantified for each size using a phosphorimager and the background was subtracted. The numbers for the 0.075 × SSC and water elutions were divided by 5 to correct for the loading difference.
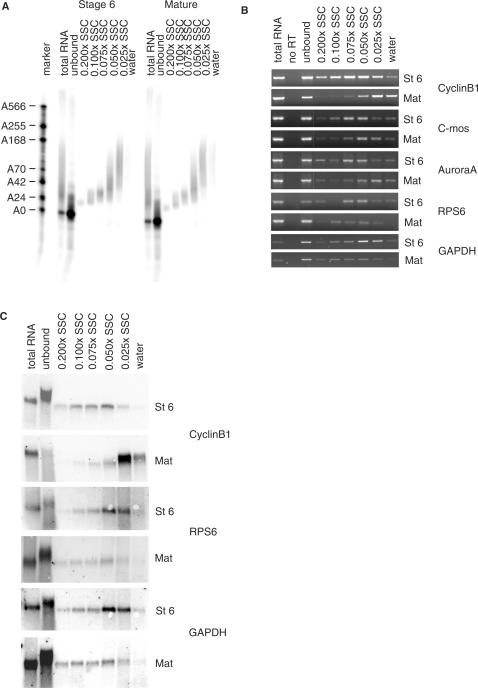
To determine whether the system could be refined to determine poly(A) lengths for specific endogenous mRNAs we used multiple elution steps to analyse some mRNAs which are known to be regulated during oocyte maturation in X. laevis (26,43,44). Cyclin B1, Aurora A (Eg2) and c-mos mRNAs have short tails in non-mature oocytes and are polyadenylated during maturation due to cytoplasmic polyadenylation elements in their 3′ UTRs, while mRNAs encoding ‘household’ proteins such as ribosomal protein S6 (RPS6) and glyceraldehydeposphate dehydrogenase (GAPDH) loose their poly(A) tails during maturation (8).
A portion of each fraction was used to analyse the distribution of the polyadenylated probe between the fractions (Figure 3A). As observed previously, RNA with poly(A) tails of 25 nt were depleted from the unbound fraction with 70% efficiency, while longer poly(A) tailed transcripts were bound quantitatively (Figure 3A, please note that in contrast to Figure 2B, equivalent amounts of the unbound and the eluted fractions were loaded). Buffers consisting of 0.2–0.075 × SSC eluted oligoadenylated RNA (15–30A) and 0.05 × SSC eluted RNAs with tails between 25 and 90 residues, 0.025 × SSC eluted A40–150, while water elution recovered even longer poly(A) tails (of which there were not very much in the probe used in this experiment). The fractions obtained from the assay were analysed by RT-PCR (Figure 3B). As can be seen by comparing the RT-panels for stage six and mature oocytes the mRNAs-encoding cyclin B1, and the kinases c-mos and Aurora A get polyadenylated during maturation and shift to the fractions with the longer poly(A) tails. The shift for cyclin B1 mRNA is clearly bigger than for c-mos and Aurora A, in agreement with the known poly(A) tail lengths of these mRNAs. In contrast, the mRNAs for ribosomal protein RPS6 and GAPDH become deadenylated during maturation and shift to the unbound fraction and the fractions with shorter poly(A) tails. The northern blots are even clearer in detecting shifts, due to their more quantitative nature (Figure 3C). Before maturation, most cyclin B1 mRNA was in the unbound fraction and upon maturation it moved to the more polyadenylated fractions while RPS6 and GAPDH mRNAs move back to the fractions with a short poly(A) tail. Please note that the apparent size change in the unbound lane is due to the very high levels of ribosomal RNA in this sample. These data confirm that the poly(A) fractionation method is working satisfactorily.
Figure 3.
Analysis of poly(A) tail length during Xenopus oogenesis. Stage 6 and mature X. laevis oocytes were lysed and mixed with radiolabelled polyadenylated probe with A0–150. Both samples were subjected to oligo(dT) affinity chromatography and eluted using decreasing SSC concentrations. (A) Ten percent of the resulting fractions was analysed by urea–PAGE and the polyadenylated probe detected using a phosphor imager. Note that the water lane has only a weak signal in this instance because there is little probe with a poly(A) tail of more than 150 adenylate residues. (B) RT-PCR analysis of endogenous mRNAs in the fractions shown in (A). Samples in the no RT lane were treated exactly the same as the samples in the total RNA lane except for omitting the AMV-RT. (C) Northern analysis on 50% of the fractions shown in (A). Lanes with total RNA contain 15% of starting material. St 6, stage 6 oocytes; Mat, mature oocytes.
Microarray analysis
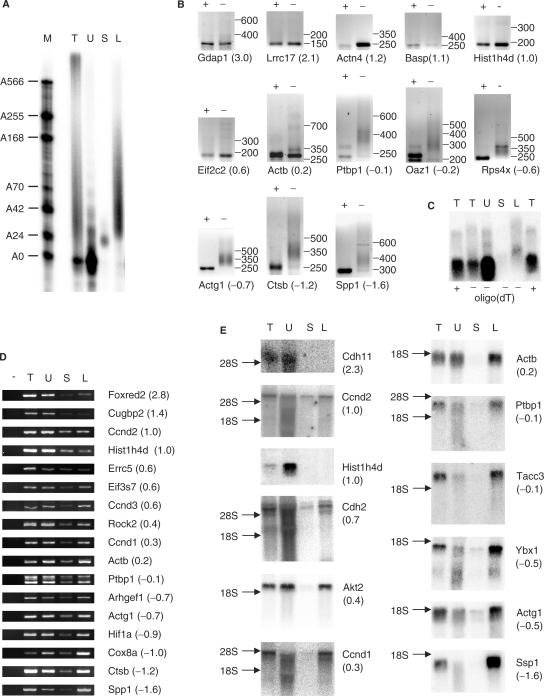
The poly(A) fractionation method described above should yield material to study changes in poly(A) tail length by microarray. Unbound RNA from oligo(dT) selections contains high levels of degradation products for some mRNAs, in addition to full–length mRNAs with short or no poly(A) tails, while oligo(dT) bound mRNAs tend to be full-length, as can be seen in northern blots of these fractions (Figure 4E). To identify mRNAs with short poly(A) tails, we therefore decided to compare mRNAs with oligoadenylate tails to mRNAs with long poly(A) tails. Total RNA isolated from NIH3T3 cells was fractionated into two fractions by elution of the RNA with 0.075× SSC (A25) and H2O (>A25) (Figure 4A). These fractions were then used to prepare cDNA for microarray analysis on mouse cDNA arrays with almost 11 000 probes per array. Of these some 2300 probes gave detectable signals with NIH3T3 RNA. The triplicate of independent microarray experiments showed good reproducibility and different probes for the same mRNA usually had similar LogRatio values (Supplementary Table 2). A high LogRatio indicates a relatively high amount of oligoadenylated RNA while a negative LogRatio indicates RNAs, which are predominantly polyadenylated. Thirty-one transcripts were validated using RL-PAT, RT-PCR and/or northern blotting of which 30 gave results consistent with their LogRatio (Table 1). The exception was SUMO1 activating enzyme subunit two (Uble1b), which appeared to have a long poly(A) tail in both RNA ligation-mediated poly(A) test (RL-PAT, see Introduction section) and northern blot of poly(A) fractions.
Figure 4.
Validation of poly(A) fractionation microarray. T, total RNA; U, unbound; S, short (oligoadenylated RNA eluted with 0.075× SSC); L, long (polyadenylated RNA eluted with H2O). Gene names are according to Genbank, numbers in brackets indicate the corrected LogRatio obtained for the mRNA in the microarray screen. A high LogRatio should correlate with a short poly(A) tail. (A) Total RNA from NIH3T3 cells was mixed with radiolabelled polyadenylated probe and biotinylated oligo(dT) and then fractionated using the poly(A) fractionation method. Ten percent of the resulting fractions was analysed by urea–PAGE as in Figure 3A. The lane with total RNA contains 2% of starting material. The remainder of the fractions with short and long poly(A) tails was used for microarray analysis (see Supplementary Table 2). (B) Total RNA from NIH3T3 was subjected to RNaseH treatment in the presence of oligo(dT) (+ lanes) or used untreated (− lanes) and subjected to RL-PAT using specific sense primers (Supplementary Table 1) for genes identified in the microarray analysis. The size difference in PCR products between RNaseH treated and untreated corresponds to the length of the poly(A) tail. The numbers after the gene name indicate the corrected LogRatio obtained in the microarray experiment. (C) Total RNA from NIH3T3 cells was treated with RNaseH in the presence of oligo(dT) and an antisense oligo specific for Actb (+) or the presence of the specific oligo only (−). The sample treated with the specific oligo only was subsequently fractionated using the poly(A) fractionation method (U, S, L). Actb RNA was detected by Northern analysis. (D) RNA was fractionated as described in (A). RT-PCR was performed on the resulting fractions. Samples in the no RT lane (−) were treated the same as samples in the total RNA lane (T) except for the omission of Superscript III. (E) RNA was fractionated as described in (A). Twenty-five percent of each fraction was then analysed by northern blotting. Total: 5% of starting material. The numbers in brackets behind the gene names refer to the average corrected LogRatio (see Supplementary Table 2).
Table 1.
mRNAs selected for the validation of the microarray experiment
| Rank | Gene | Refseq | Description | Validated | Corrected LogRatio | Standard deviation |
|---|---|---|---|---|---|---|
| 1 | Gdap1 | NM_010267 | Ganglioside-induced differentiation-associated-protein | + | 2.97 | 0.46 |
| 3 | Foxred2 | NM_001017983 | FAD-dependent oxidoreductase domain containing 2 | + | 2.76 | 0.59 |
| 4 | Sae2 | NM_016682 | SUMO1 activating enzyme subunit 2 (Uble1b) | − | 2.72 | 0.51 |
| 6 | Cdh11 | NM_009866 | Cadherin 11 | + | 2.32 | 0.58 |
| 9 | Lrrc17 | XM_919092 | Leucine-rich repeat containing 17 | + | 2.13 | 0.27 |
| 21 | Cugbp2 | NM_010160 | CUG triplet repeat, RNA-binding protein 2(ETR3, Napor) | + | 1.43 | 0.35 |
| 37 | Actn4 | NM_021895 | Actinin alpha 4 | + | 1.18 | 0.19 |
| 43 | Basp1 | NM_027395 | Brain abundant, membrane attached signal protein (CAP23) | + | 1.12 | 0.13 |
| 48 | Ccnd2 | NM_009829 | Cyclin D2 | + | 1.03 | 0.08 |
| 50 | Hist1h4d | NM_175654 | Histone cluster 1, H4d | + | 1.01 | 0.20 |
| 54 | Basp1 | NM_027395 | Brain abundant, membrane attached signal protein (CAP23) | + | 0.97 | 0.08 |
| 86 | Cdh2 | NM_007664 | Cadherin 2 (N-cadherin) | + | 0.73 | 0.22 |
| 112 | Ercc5 | NM_011729 | Excision repair cross-complementing rodent repair 5 | + | 0.64 | 0.08 |
| 116 | Eif3s7 | NM_018749 | Eukaryotic translation initiation factor 3, subunit 7 (zeta) | + | 0.62 | 0.38 |
| 135 | Ccnd3 | NM_007632 | Cyclin D3 | + | 0.59 | 0.11 |
| 142 | Eif2c2 | BC064741 | Argonaute 2 | + | 0.57 | 0.10 |
| 276 | Akt2 | NM_007434 | Thymoma viral proto-oncogene 2 | + | 0.42 | 0.20 |
| 314 | Rock2 | NM_009072 | Rho-associated coiled-coil containing protein kinase 2 | + | 0.38 | 0.28 |
| 425 | Ccnd1 | NM_007631 | Cyclin D1 | + | 0.30 | 0.43 |
| 589 | Actb | NM_007393 | Beta actin | + | 0.21 | 0.49 |
| 673 | Actb | NM_007393 | Beta actin | + | 0.17 | 0.47 |
| 690 | Actb | NM_007393 | Beta actin | + | 0.17 | 0.56 |
| 1402 | Ptbp1 | NM_008956 | Polypyrimidine tract binding protein 1 | + | −0.13 | 0.15 |
| 1438 | Tacc3 | NM_001040435 | Transforming, acidic coiled-coil containing protein 3 | + | −0.15 | 0.21 |
| 1569 | Oaz1 | NM_008753 | Ornithine decarboxylase antizyme 1 | + | −0.22 | 0.41 |
| 1921 | Ybx1 | NM_011732 | Y box protein 1 | + | −0.47 | 0.22 |
| 2023 | Rps4x | NM_009094 | Ribosomal protein S4, X-linked | + | −0.57 | 0.36 |
| 2110 | Arhgef1 | NM_008488 | RhoA GTP exchange factor 1 | + | −0.69 | 0.08 |
| 2139 | Actg1 | NM_009609 | Actin, gamma, cytoplasmic 1 | + | −0.75 | 0.07 |
| 2185 | Hif1a | NM_010431 | Hypoxia inducible factor alpha | + | −0.88 | 0.12 |
| 2216 | Cox8a | NM_007750 | Cytochrome c oxidase, subunit VIIIa | + | −1.00 | 0.40 |
| 2236 | Ctsb | NM_007798 | Cathepsin B | + | −1.13 | 0.13 |
| 2239 | Ctsb | NM_007798 | Cathepsin B | + | −1.28 | 0.26 |
| 2242 | Spp1 | NM_009263 | Secreted phosphoprotein 1 | + | −1.45 | 0.40 |
| 2243 | Spp1 | NM_009263 | Secreted phosphoprotein 1 | + | −1.75 | 0.20 |
Each probe on the microarray for the selected mRNAs is represented by a line in the listing, with multiple listings for each mRNA if it is represented by multiple probes. Probes cross-hybridizing to other mRNAs were excluded from this list. The rank indicates the number of the probe in the list of 2246 probes. The Genbank gene name is used. For unequivocal identification, the Refseq number from Genbank is cited, note that this is not the EST used on the array. For each probe, it is indicated if the validation was successful, what average corrected LogRatio was obtained and what the standard deviation over the three arrays was.
To validate the poly(A) fractionation by RL-PAT, we isolated total RNA from NIH3T3 cells and treated part of it with RNaseH in the presence of oligo(dT) to remove the poly(A) tail. Both treated and untreated RNA were then used for RL-PAT analysis (Figure 4B). The difference in product size between treated (+ lanes) and untreated (− lanes) samples represents the length of the poly(A) tail. The mRNAs at the top of the ranking (high LogRatio) all show no size difference between the two samples indicating that the poly(A) tail is too short to detect. This corresponds to RNAs, which are mostly oligoadenylated (<30 As). None of the RNAs with a negative LogRatio had a large amount of oligoadenylate mRNA judging by the LR-PAT assay. β Actin, (Actb, LogRatio 0.2) appears to represent the mRNAs which contain both oligo adenylated and polyadenylated transcripts.
We also validated the poly(A) fractionation coupled microarray by RT-PCR (Figure 4D) and northern blotting (Figure 4E). Both were performed on samples from poly(A) fractionations independent from those used for the microarrays. The RT-PCR analysis does not show big differences in the signal strength in the lanes with oligo adenylated RNA (lane 4, short) but there is a clear increase in the amount of polyadenylated RNA corresponding to a decreasing LogRatio. Unexpectedly, for all RNAs tested, a strong signal was obtained for RNAs that did not bind oligo(dT) (lane 3, unbound) suggesting that many RNAs consist of a large fraction of transcripts with a poly(A) tail of less than 25 residues. To test whether these unpolyadenylated RNAs were full length or represented degradation products, we analysed fractionated RNA from NIH3T3 cells by northern blotting (Figure 4E). As expected, no oligoadenylated of polyadenylated mRNAs were detected for histone H4. Surprisingly, cadherin 11 (Cdh11) shows a similar pattern to histone H4. For cadherin 2 (Cdh2) and cyclin D2 (Ccnd2), a relatively high percentage of the full-length mRNA was in the oligoadenylate lane (10–20%), in addition to significant levels of full-length mRNA in the unbound lane.
The mRNAs that are below the top 10% but still have a positive LogRatio, such as Akt2, cyclin D1 and beta actin, high percentages of full-length unbound mRNA are detected, indicating a large fraction of mRNA with a short or no adenylate tail. Quantification of several northern blots has shown that ∼25% of the β actin (Actb) mRNA is in the unbound fraction (corrected for the difference in recovery, as shown in Figure 2C). As the three non-cross-hybridizing Actb probes are ranked between 589 and 690 of 2246 probes, these data indicate that as much as 25% of all expressed genes have an oligoadenylate tail of less than 30 in 25% or more of their full-length mRNA transcripts. In contrast, mRNAs with negative LogRatios, representing 50% of all transcripts, have much lower levels of full-length transcripts in the unbound fraction. However, there are usually degradation products detectable in this fraction (Figure 4E), indicating that our choice of using the 0.075 × SSC eluate rather than the unbound fraction for the microarray screen was correct.
To further compare our new method with the classical method of RNase H treatment for the Actb transcript, total RNA from NIH3T3 cells was treated with RNaseH in the presence of a specific antisense Actb oligo resulting in the cleavage 459 nt upstream of the polyadenylation site. Oligo(dT) was added to half of the RNA to remove the poly(A) tail. The sample that was treated with the specific Actb oligo only was subsequently fractionated into three fractions: unbound, short (25 As) and long (>25 As) poly(A) tails. All samples were analysed by northern blotting (Figure 4C). When comparing the total RNA samples treated with RNaseH in the presence (lane 1) or absence (lane 2) of oligo(dT) the smear in lane 2 is slightly longer than in lane 1. The smear remaining in lane 1 is probably caused by a small amount of nuclear uncleaved transcript, as these bands move with the size change of the mRNA after RNase H cleavage. The smear in lane 2 represents the polyadenylated transcripts (and uncleaved RNA). When comparing the fractionated samples (lanes 3–5) the polyadenylated transcripts are easily detected (lane 5, long) while a small amount of oligoadenylated transcript is also visible (lane 4, short). A very strong signal was detected in the unbound fraction (lane 3, unbound). This figure illustrates the problems in detecting long poly(A) tails in a mixed population with the RNase H method. Compare the U and L lanes with the actB northern panel in Figure 4E, where an equally strong signal is visible in the long fraction, and with the RL-PAT for actB, where most of the signal appears in the oligoadenylated bottom band. In addition, the RNase H cleavage experiment confirms that actin mRNA has large percentage of mRNA with a short poly(A) tail.
A summary of a functional analysis for the top 10% of deadenylated mRNAs is shown in Table 2 and in more detail in Supplementary Table 3. Of the 148 non-cross-hybridizing probes in the top 10% of deadenylated mRNAs, 34 encoded proteins involved in RNA-mediated processes, of which six were translation initiation factors and five mitochondrial ribosomal proteins. Cytoplasmic ribosomal proteins are underrepresented in the top of this ranking. Other well-represented categories included cell adhesion (13), cell cycle (10, including cyclin D2 and D3), chromatin components (19, including the histones), proteins involved in ubiquitination and proteasome-mediated degradation (11) and components of the endoplasmatic reticulum and related membrane structures (14).
DISCUSSION
We describe a versatile method which allows for detailed analysis of the length of the poly(A) tail. It detects changes in poly(A) tail length and allows for separation of the RNAs into multiple fractions based on the length of the poly(A) tail of the RNA. This facilitates the analysis of each fraction separately. The fractions with the short poly(A) tails are quite homogeneous while the fractions with the longer poly(A) tails contain a wider variety of poly(A) tail lengths. Small changes in poly(A) tail length are easily detected because a small change in poly(A) tail length results in the shift from one fraction to a neighbouring fraction, even though the capture of the shorter poly(A) tails is not complete (Figure 2). In addition, differences of poly(A) tail lengths between 80 nt (c-mos mRNA in mature oocytes) and 200 nt (cyclin B1 mRNA in mature oocytes) are also apparent, indicating the method works for comparisons between mRNAs as well. Our careful characterization of the unbound fraction allows us to state that A25 tails are 70% depleted from this fraction and any longer tails are depleted more efficiently, while tails of 15 nt or less are only marginally depleted. Therefore any mRNA with a significant amount of signal in the unbound fraction will have tails of 30 nt or less.
When transcripts have a wide variety of poly(A) tail lengths it can be very difficult to detect and quantify them from PAT assays or RNase H treated Northerns because the longer tails are represented by a smear, resulting in a worse signal to background ratio, as can be seen in Figure 4B and C. The poly(A) fractionation protocol combined with northern blotting and analysis of the recovery percentages as in Figure 2C can give a quantitative estimate of oligoadenylated+deadenylated and polyadenylated transcripts from a particular gene, without underestimating the poly(A) fraction, because this method does not rely on the detection of smears.
Until now, the length and regulation of the poly(A) tails of only a few transcripts from higher eukaryotes have been studied. This has led to the assumption that mRNAs with short tails are usually degraded (3). The most well-characterized exception are the histone mRNAs, which are generated by an alternative 3′ processing pathway and stabilized in the cytoplasm by a 3′ hairpin that binds a specific protein complex (45). Histone mRNAs do not normally carry a poly(A) tail, except when targeted for degradation in the nucleus (46), which is probably what we are detecting in our microarray experiment.
We have shown that RNA obtained by RNA fractionation can be used for microarray analysis and that the results obtained from this type of experiment predict the levels of oligoadenylated transcripts for each mRNA. The success of our analysis is probably due to the lack of degraded products in the A25 fraction, as opposed to the unbound fraction. Surprisingly, large numbers of genes, possibly as much as 25% of all expressed mRNAs, are predicted to have significant levels of full-length transcripts with poly(A) tails of less than 30, as we have shown for the beta actin and Akt2 mRNAs. Beta actin is known to be a stable mRNA and the fact that 25% of this mRNA has a very short of no poly(A) tail indicates that this represents a stable deadenylated mRNA (47). In addition, for a full-length oligoadenylated mRNA to accumulate to similar levels as the polyadenylated transcript, the rate of deadenylation has to significantly exceed the rate of mRNA degradation, indicating that deadenylation and mRNA degradation are not as tightly linked as previously thought for many of these mRNAs.
Possibly some of the unbound mRNAs may not be polyadenylated at all in a similar fashion to the histone mRNAs. Alternatively, they may only obtain very short oligoadenylate tails in the nucleus, as has been shown for the mRNA-encoding Xenopus serum albumin, which is efficiently translated despite its homogeneous 17 nt oligoadenylate tail conferred by a poly(A) limiting element (PLE) in its 3′ UTR (48). The cadherin 11 mRNA could conceivably contain a PLE, as no polyadenylated mRNA was detected (Figure 4E). However, all the other mRNAs we have studied have some fraction of polyadenylated mRNA. It appears therefore more likely these mRNAs are exported with a long poly(A) tail and are deadenylated in the cytoplasm, but their decapping and degradation is slow. A recent study of the stability of Hsp70 mRNA in insect cells found that it was removed by just such a type of degradation (49). Our data therefore suggest that this kind of mRNA decay is common in NIH 3T3 cells.
Surprisingly, there is very little overlap between our work and a microarray screen comparing oligo(dT) bound mRNA with mRNA purified with a high-affinity mutant of cap-binding initiation factor eIF4E (50). Notably, the authors found that the ribosomal protein mRNAs were better captured by eIF4E than by oligo(dT), while our data indicate that these mRNAs have predominantly long tails. This is possibly due to the differences in origin of the RNA (adult human liver vs proliferating mouse NIH3T3 fibroblasts).
As we were preparing this manuscript, a microarray study on poly(A) tail length in Saccharomyces cerevisiae was published (51). The authors successfully employed the previously described thermal elution of poly(U) sepharose affinity chromatography to separate mRNA into fractions with different poly(A) tail lengths. Their data do not directly show the efficiency of capture of short oligoadenylate tails, but the early elutions appear to contain significant amounts of A10-20, which end up predominantly in the unbound fractions in our method, confirming that poly(U) sepharose is superior in binding very short oligoadenylate tails. It is unclear, however, whether the fractionation would discriminate effectively between unadenylated RNAs with A rich regions and terminal A10 tracts. Although thermal elution from poly(U) sepharose should be readily applicable to other RNA samples, fraction elution based on temperature is difficult to perform reproducibly without specialist equipment, while the poly(A) fractionation method described here can be performed in any molecular biology laboratory. Strikingly, the functional groups identified by Beilharz and Preiss (51) as being enriched in mRNAs with short poly(A) tails overlap to a high extent with the ones identified in our screen: mRNAs-encoding RNP proteins (but not cytoplasmic ribosomal proteins), cytoskeletal proteins and cell cycle proteins all appear to be overrepresented in the short poly(A) fractions in both yeast and NIH 3T3 cells.
The poly(A) fractionation microarray method is ideally suited to study changes in poly(A) tail length in tissue culture cells such as the cell cycle, apoptosis and amino acid starvation as well as less easily manipulated samples such as brain tissue. In addition, the poly(A) fractionation protocol only requires a small sample size. A few micrograms of total RNA would still allow for separation and subsequent RT-PCR analysis and 40 μg is sufficient for microarray analysis. This can be extremely useful if the amount material is limiting which is often the case for clinical samples.
We have developed a novel and highly reproducible method that significantly improves the analysis of poly(A) tail length of endogenous mRNAs and enables global analysis of poly(A) tail length changes in a large variety of systems using standard laboratory equipment. Poly(A) fractionation should facilitate the study of poly(A) tail metabolism and determine its role in a variety of processes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was funded by Wellcome Trust (076179); Biotechnology and Biological Sciences Research Council (42/G14670). Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Piccioni F, Zappavigna V, Verrotti AC. Translational regulation during oogenesis and early development: the cap-poly(A) tail relationship. Crit. Rev. Biol. 2005;328:863–881. doi: 10.1016/j.crvi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn U, Wahle E. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit. Rev. Biochem. Mol. Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 4.Wilusz CJ, Wormington M, Peltz SW. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 5.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van DS, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 6.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorgoni B, Gray NK. The roles of cytoplasmic poly(A)-binding proteins in regulating gene expression: a developmental perspective. Brief. Funct. Genomic. Proteomic. 2004;3:125–141. doi: 10.1093/bfgp/3.2.125. [DOI] [PubMed] [Google Scholar]
- 8.De Moor CH, Meijer HA, Lissenden S. Mechanisms of translational control by the 3′ UTR in development and differentiation. Semin. Cell Dev. Biol. 2005;16:49–58. doi: 10.1016/j.semcdb.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 9.Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- 10.Verrotti AC, Thompson SR, Wreden C, Strickland S, Wickens M. Evolutionary conservation of sequence elements controlling cytoplasmic polyadenylylation. Proc. Natl Acad. Sci. USA. 1996;93:9027–9032. doi: 10.1073/pnas.93.17.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kashiwabara S, Noguchi J, Zhuang T, Ohmura K, Honda A, Sugiura S, Miyamoto K, Takahashi S, Inoue K, et al. Regulation of spermatogenesis by testis-specific, cytoplasmic poly(A) polymerase TPAP. Science. 2002;298:1999–2002. doi: 10.1126/science.1074632. [DOI] [PubMed] [Google Scholar]
- 12.Luitjens C, Gallegos M, Kraemer B, Kimble J, Wickens M. CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 2000;14:2596–2609. doi: 10.1101/gad.831700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin CY, Kundel M, Wells DG. Rapid, activity-induced increase in tissue plasminogen activator is mediated by metabotropic glutamate receptor-dependent mRNA translation. J. Neurosci. 2004;24:9425–9433. doi: 10.1523/JNEUROSCI.2457-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in aplysia. Cell. 2003;115:893–904. doi: 10.1016/s0092-8674(03)01021-3. [DOI] [PubMed] [Google Scholar]
- 15.Richter JD. Think globally, translate locally: what mitotic spindles and neuronal synapses have in common. Proc. Natl Acad. Sci. USA. 2001;98:7069–7071. doi: 10.1073/pnas.111146498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev. Biol. 2006;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 17.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 18.Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J, Di PF, Lin SC, Gram H, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120:623–634. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 19.Sasayama T, Marumoto T, Kunitoku N, Zhang D, Tamaki N, Kohmura E, Saya H, Hirota T. Over-expression of Aurora-A targets cytoplasmic polyadenylation element binding protein and promotes mRNA polyadenylation of Cdk1 and cyclin B1. Genes Cells. 2005;10:627–638. doi: 10.1111/j.1365-2443.2005.00870.x. [DOI] [PubMed] [Google Scholar]
- 20.Thomadaki H, Tsiapalis CM, Scorilas A. Polyadenylate polymerase modulations in human epithelioid cervix and breast cancer cell lines, treated with etoposide or cordycepin, follow cell cycle rather than apoptosis induction. Biol. Chem. 2005;386:471–480. doi: 10.1515/BC.2005.056. [DOI] [PubMed] [Google Scholar]
- 21.Carrick DM, Lai WS, Blackshear PJ. The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res. Ther. 2004;6:248–264. doi: 10.1186/ar1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scorilas A. Polyadenylate polymerase (PAP) and 3′ end pre-mRNA processing: function, assays, and association with disease. Crit. Rev. Clin. Lab. Sci. 2002;39:193–224. doi: 10.1080/10408360290795510. [DOI] [PubMed] [Google Scholar]
- 23.Groisman I, Jung MY, Sarkissian M, Cao Q, Richter JD. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/s0092-8674(02)00733-x. [DOI] [PubMed] [Google Scholar]
- 24.Zangar RC, Hernandez M, Kocarek TA, Novak RF. Determination of the poly(A) tail lengths of a single mRNA species in total hepatic RNA. BioTechniques. 1995;18:465–469. [PubMed] [Google Scholar]
- 25.Brogna S. Nonsense mutations in the alcohol dehydrogenase gene of Drosophila melanogaster correlate with an abnormal 3′ end processing of the corresponding pre-mRNA. RNA. 1999;5:562–573. doi: 10.1017/s1355838299981359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- 27.Salles FJ, Richards WG, Strickland S. Assaying the polyadenylation state of mRNAs. Methods. 1999;17:38–45. doi: 10.1006/meth.1998.0705. [DOI] [PubMed] [Google Scholar]
- 28.Rassa JC, Wilson GM, Brewer GA, Parks GD. Spacing constraints on reinitiation of paramyxovirus transcription: the gene end U tract acts as a spacer to separate gene end from gene start sites. Virology. 2000;274:438–449. doi: 10.1006/viro.2000.0494. [DOI] [PubMed] [Google Scholar]
- 29.Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T. Messenger RNA deadenylylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA. 1997;94:5628–5633. doi: 10.1073/pnas.94.11.5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pique M, Lopez JM, Mendez R. Cytoplasmic mRNA polyadenylation and translation assays. Methods Mol. Biol. 2006;322:183–198. doi: 10.1007/978-1-59745-000-3_13. [DOI] [PubMed] [Google Scholar]
- 31.Shyu AB, Belasco JG, Greenberg ME. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 32.Graindorge A, Thuret R, Pollet N, Osborne HB, Audic Y. Identification of post-transcriptionally regulated Xenopus tropicalis maternal mRNAs by microarray. Nucleic Acids Res. 2006;34:986–995. doi: 10.1093/nar/gkj492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du L, Richter JD. Activity-dependent polyadenylation in neurons. RNA. 2005;11:1340–1347. doi: 10.1261/rna.2870505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meijer HA, Radford HE, Wilson LS, Lissenden S, De Moor CH. Translational control of maskin mRNA by its 3′ untranslated region. Biol. Cell. 2007;99:239–250. doi: 10.1042/BC20060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 36.Minvielle-Sebastia L, Winsor B, Bonneaud N, Lacroute F. Mutations in the yeast RNA14 and RNA15 genes result in an abnormal mRNA decay rate; sequence analysis reveals an RNA-binding domain in the RNA15 protein. Mol. Cell. Biol. 1991;11:3075–3087. doi: 10.1128/mcb.11.6.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burke P. PolyATract mRNA isolation systems. Promega Notes Magazine. 1996;56:27–29. [Google Scholar]
- 38.Charlesworth A, Cox LL, MacNicol AM. Cytoplasmic polyadenylation element (CPE)- and CPE-binding protein (CPEB)-independent mechanisms regulate early class maternal mRNA translational activation in Xenopus oocytes. J. Biol. Chem. 2004;279:17650–17659. doi: 10.1074/jbc.M313837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bushell M, Stoneley M, Kong Y, Hamilton TL, Spriggs KA, Dobbyn HC, Qin X, Sarnow P, Willis AE. Polypyrimidine tract binding protein regulates IRES-mediated gene expression during apopotosis. Mol. Cell. 2006;23:401–412. doi: 10.1016/j.molcel.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 40.Stebbins-Boaz B, Hake LE, Richter JD. CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c-mos mRNAs and is necessary for oocyte maturation in Xenopus. EMBO J. 1996;15:2582–2592. [PMC free article] [PubMed] [Google Scholar]
- 41.Wollerton MC, Gooding C, Robinson F, Brown EC, Jackson RJ, Smith CW. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB) RNA. 2001;7:819–832. doi: 10.1017/s1355838201010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lane MJ, Paner T, Kashin I, Faldasz BD, Li B, Gallo FJ, Benight AS. The thermodynamic advantage of DNA oligonucleotide ‘stacking hybridization’ reactions: energetics of a DNA nick. Nucleic Acids Res. 1997;25:611–617. doi: 10.1093/nar/25.3.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paris J, Philippe M. Poly(A) metabolism and polysomal recruitment of maternal mRNAs during early Xenopus development. Dev. Biol. 1990;140:221–224. doi: 10.1016/0012-1606(90)90070-y. [DOI] [PubMed] [Google Scholar]
- 44.Paris J, Richter JD. Maturation-specific polyadenylation and translational control: diversity of cytoplasmic polyadenylation elements, influence of poly(A) tail size, and formation of stable polyadenylation complexes. Mol. Cell. Biol. 1990;10:5634–5645. doi: 10.1128/mcb.10.11.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene. 2007;396:373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reis CC, Campbell JL. Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics. 2007;175:993–1010. doi: 10.1534/genetics.106.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dormoy-Raclet V, Menard I, Clair E, Kurban G, Mazroui R, Di MS, von RC, Pause A, Gallouzi IE. The RNA-binding protein HuR promotes cell migration and cell invasion by stabilizing the beta-actin mRNA in a U-rich-element-dependent manner. Mol. Cell. Biol. 2007;27:5365–5380. doi: 10.1128/MCB.00113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng J, Schoenberg DR. mRNA with a <20-nt poly(A) tail imparted by the poly(A)-limiting element is translated as efficiently in vivo as long poly(A) mRNA. RNA. 2005;11:1131–1140. doi: 10.1261/rna.2470905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bonisch C, Temme C, Moritz B, Wahle E. Degradation of the hsp70 and other mRNAs in Drosophila via the 5′-3′ pathway and its regulation by heat shock. J. Biol. Chem. 2007;282:21818–21828. doi: 10.1074/jbc.M702998200. [DOI] [PubMed] [Google Scholar]
- 50.Choi YH, Hagedorn CH. Purifying mRNAs with a high-affinity eIF4E mutant identifies the short 3′ poly(A) end phenotype. Proc. Natl Acad. Sci. USA. 2003;100:7033–7038. doi: 10.1073/pnas.1232347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beilharz TH, Preiss T. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA. 2007;13:982–997. doi: 10.1261/rna.569407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.