Abstract
Saccharomyces cerevisiae DNA polymerase δ (Pol δ) and DNA polymerase ε (Pol ε) are replicative DNA polymerases at the replication fork. Both enzymes are stimulated by PCNA, although to different levels. To understand why and to explore the interaction with PCNA, we compared Pol δ and Pol ε in physical interactions with PCNA and nucleic acids (with or without RPA), and in functional assays measuring activity and processivity. Using surface plasmon resonance technique, we show that Pol ε has a high affinity for DNA, but a low affinity for PCNA. In contrast, Pol δ has a low affinity for DNA and a high affinity for PCNA. The true processivity of Pol δ and Pol ε was measured for the first time in the presence of RPA, PCNA and RFC on single-stranded DNA. Remarkably, in the presence of PCNA, the processivity of Pol δ and Pol ε on RPA-coated DNA is comparable. Finally, more PCNA molecules were found on the template after it was replicated by Pol ε when compared to Pol δ. We conclude that Pol ε and Pol δ exhibit comparable processivity, but are loaded on the primer-end via different mechanisms.
INTRODUCTION
At least three DNA polymerases are required for eukaryotic genome replication: DNA polymerase alpha (Pol α), DNA polymerase delta (Pol δ) and DNA polymerase epsilon (Pol ε) (1). Pol α initiates DNA synthesis on both the leading and lagging strands by synthesizing a RNA/DNA hybrid primer. The replicative DNA polymerases, Pol δ and Pol ε, then extend the DNA synthesis from the primer. In addition to the three DNA polymerases, DNA replication requires additional replication factors: the single-stranded DNA-binding protein (RPA), the clamp loader (RF-C) and the clamp (PCNA) (2). At first RPA may appear to have a relatively simple task in interacting with and stabilizing single-stranded DNA (ssDNA). However, the ability to interact with ssDNA places RPA in the middle of several processes that involve DNA replication and repair (3).
The precise role of Pol ε during DNA replication has been difficult to define. However, genetic analysis and chromatin immunoprecipitation assays have shown that Pol ε participates at the origins of DNA replication during the establishment of replication forks (4,5). Furthermore, chromatin immunoprecipitation assays have demonstrated that Pol ε remains associated with the fork as it progress from the origin (6,7). This supports a model in which Pol ε is responsible for the replication of one strand at the replication fork. Other results supporting this one polymerase-one strand model include the ability of both Pol δ and Pol ε to be purified as monomers with regard to the catalytic subunit (8,9). In vitro DNA replication with cell-free Xenopus extracts is dependent on Pol ε, and immunodepletion experiments suggest that Pol ε and Pol δ have unique functions (10,11). Genetic experiments in yeast demonstrated that Pol δ and Pol ε proofread opposite strands, in agreement with the observed strand asymmetry in replication fidelity (12–14). Together with the unique property of Pol δ that allows idling to maintain a ligatable nick, genetic experiments firmly support a model by which Pol δ is solely responsible for the synthesis of the lagging strand (15–17). Recently, the first direct evidence for Pol ε to function as the leading strand DNA polymerase in Saccharomyces cerevisiae was identified using an altered error signature from a pol2 allele with a mutation in the active site (18).
There are still unresolved questions regarding the function of Pol ε. While replication fidelity studies in yeast suggest that Pol ε generally replicates the leading strand (18), studies with POL2 partial deletion mutants indicate that under conditions of Pol ε dysfunction, Pol δ can replicate the leading strand (19). Pol ε has also been suggested to play an important role at late firing origins (20), and in this model Pol ε would replicate certain regions of the genome. Pol ε has been shown to promote epigenetic silencing in S. cerevisiae (21,22). In addition to these functions, Pol ε acts as a sensor for the S-phase checkpoint in S. cerevisiae (23). Pol ε interacts with ssDNA, and is inhibited by ssDNA when RPA is excluded from the polymerase assay (24). This has been proposed to be one mechanism by which Pol ε may sense DNA damage during S-phase (25). However, ssDNA is not considered to be long-lived in vivo as RPA efficiently coats ssDNA. Early in vitro assays suggested that Pol ε may be the leading strand polymerase based on the measured processivity (26). However, in vitro studies where Pol ε replicated single-stranded circular DNA templates in the presence of PCNA showed that Pol ε was much less efficient than Pol δ, potentially due to the presence of ssDNA in the reaction mix. The high affinity for ssDNA was thought to inhibit the processivity of Pol ε.
The purpose of this study was to clarify differences between the leading- and lagging-strand DNA polymerases by challenging the S cerevisiae polymerases in various assays. As all previous studies have been under conditions where the loading efficiency at the primer-end has masked the true processivity, we first assessed the PCNA-dependent processivity of Pol δ and Pol ε. Second, we assessed the influence of RPA on the processivity of Pol ε and Pol δ. Finally, we determined whether Pol ε interacts with PCNA, independent of PCNA binding to DNA. The ability to interact with PCNA off DNA is typical for proteins that depend on a PCNA interaction motif to be efficiently loaded onto the primer termini. Here we show that Pol ε and Pol δ are loaded onto the primer termini via distinct mechanisms. Remarkably, we find that Pol ε and Pol δ are similarly processive, with surprisingly short replication products resulting from a single encounter with the template.
MATERIALS AND METHODS
Proteins and DNA templates
Pol ε, Pol δ, RPA, RF-C and PCNA were purified as described previously (9,27–30). Oligonucleotides used in the surface plasmon resonance analysis and replication assays were purchased from DNA Technologies Inc. and Sigma-Proligo. Single-stranded M13mp18 DNA was primed with a synthetic 60-mer (complementary to position 6355–6295), and single-stranded pBluescript II SK(+) was primed with a synthetic 50-mer (complementary to position 678–628).
Holoenzyme assays
The standard 30 μl reaction contained 40 mM Tris–HCl, pH 7.8, 0.2 mg/ml BSA, 1 mM DTT, 100 µM each of dGTP, dATP and dTTP, and 25 μM of dCTP, [α-32P]dCTP (1 μCi, Amersham Bioscience), 8 mM MgAc2, 1 mM ATP, 125 mM NaAc, 100 fmol RFC, 1 pmol PCNA, 40 fmol of single-primed M13mp18 template or 100 fmol of single-primed pBluescript II SK(+), 100 fmol of Pol ε or Pol δ and RPA as indicated in the figures. The reactions were incubated at 30°C for indicated times. The reactions were stopped by adding 6 μl of stop solution containing 60 mM EDTA, 1% SDS, 0.1% bromophenol blue and 0.1% xylene blue, and were subsequently loaded onto a 1% alkaline agarose gel, containing 30 mM NaOH and 2 mM EDTA. The gels were run at 25 V for 16 h, neutralized with Tris pH 7.5, dried, placed on a phosphoimager screen (Fujifilm) and scanned with a Typhoon 9400 phosphoimager (GE Healthcare).
For measurements of processivity, [γ-32P]ATP labeled 50-mer primer was annealed to pBluescript SKII+, and [α-32P]dCTP was omitted from the reaction mixture. In these reactions, 2.4 fmol of Pol δ or Pol ε was added to meet the required single-hit conditions (2.4 fmol polymerase to 80 fmol primer–template). The resulting products were separated on denaturing 8% polyacrylamide gels and 2% alkaline agarose gels. Sequencing ladder was prepared using Thermo Sequenase sequencing kit (USB) according to the manufacturer's instructions. Results were quantified using ImageQuant TL software (GE Healthcare).
The loading of PCNA was measured with [γ-32P]ATP as described previously (31). The standard reaction was scaled up to 500 fmol of single-primed pBluescript II SK(+), 500 fmol RFC, 13 pmol PCNA, 60 pmol of RPA and 500 fmol of either Pol δ or Pol ε. The reactions were incubated for 7.5 min at 30°C and stopped by the addition of EDTA to a final concentration of 50 mM. The reactions were immediately loaded onto a 2 ml BiogelA-5m column, equilibrated in 10 mM Tris–HCl pH 7.5, 1 mM EDTA, 5% glycerol, 50 mM NaCl, 1 mM β-mercaptoethanol, 8 mM MgAc2 and 50 µg/ml BSA. Two drops per fraction were collected, and the amount of PCNA in each fraction was quantified in a scintillations counter and then plotted.
Preparation of DNA for biomolecular interaction analysis
A 60-mer oligonucleotide with biotin at the 5′-end (BIA1) was used as a template strand and as a ssDNA substrate (Table 1). A 40-mer complementary to the 3′ end of the template oligonucleotide (BIA2) was annealed to BIA1 to create a primed substrate. The double-stranded substrate was created by annealing a complementary 60-mer oligonucleotide (BIA3) to BIA1. The complementary oligonucleotides were mixed in 100 mM sodium chloride, incubated at 75°C for 5 min and cooled to room temperature. Annealed templates were gel purified by non-denaturing 10% PAGE.
Table 1.
Oligonucleotides used in surface plasmon resonance experiments
| BIA1 | 5′-Biotin-TGTGGAATTGTGAGCGGAACACCAAACACATA TAACCCCCATCATCACGAATTCACTGG-3′ |
| BIA2 | 5′-CCAGTGAATTCGTGATGATGGGGGTTATATGTGTTT GGTG-3′ |
| BIA3 | 5′-CCAGTGAATTCGTGATGATGGGGGTTATATGTGTTTG GTGTTCCGCTCACAATTCCACA-3′ |
Surface plasmon resonance
Interactions of the polymerases with DNA and PCNA were monitored using a BIAcore 3000 surface plasmon resonance biosensor instrument. Streptavidin was immobilized on a CM5 chip according to the manufacturer's instructions. Biotinylated primed DNA, dsDNA and ssDNA were individually immobilized onto first, third and fourth streptavidin surfaces, respectively, of the streptavidin-coated chip. Approximately 175 RU of primed and dsDNA templates and 95 RU of ssDNA template were immobilized, reflecting equal molar amounts of substrate. The second surface was left underivatized to correct for refractive index changes, non-specific binding and instrument drift. For the PCNA chip, ∼500RU of PCNA were covalently immobilized on the surface of the dextran chip (CM5) by the amine method, according to the manufacturer's instructions.
All interactions were monitored at 20°C. Polymerases were injected at indicated concentrations in running buffer of 25 mM HEPES pH 7.6, 10% glycerol, 200 mM sodium acetate pH 7.8, 8 mM magnesium acetate, 1 mM dithiotreitol, 50 μM dGTP, 50 μM dTTP, 0.005% P-20 and 0.2 mg/ml BSA, at a flow rate of 50 μl/min for 60 s with a 60 s long dissociation phase. The running buffer was supplemented with 500 pM RPA when the DNA on the chip surface was coated with RPA.
RESULTS
Analysis of products formed by Pol ε and Pol δ in a holoenzyme assay
Several previous studies have measured the activity of Pol δ and Pol ε with and without auxiliary factors, such as RPA, PCNA or RFC. However, many of these studies were performed with proteolytic forms of Pol ε (32,33) and direct comparative studies of the two enzymes carried out under identical experimental conditions are lacking. With a currently well-established overproduction system for Pol ε, all the required factors from S. cerevisiae can be over-expressed and purified to homogeneity from a well-defined source.
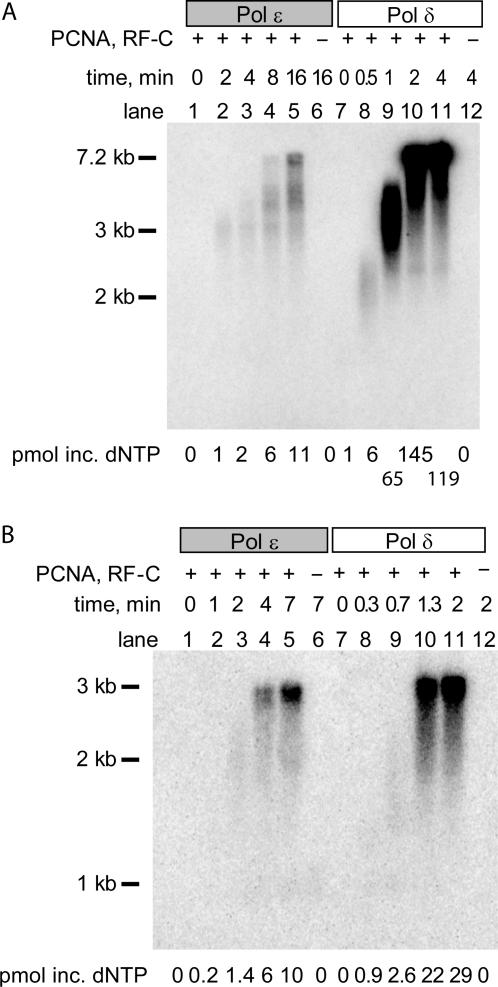
We first investigated whether our over-expressed Pol ε showed biochemical properties similar to previously described in experiments (26,32,32), which were generally carried out on single-primed ssDNA templates from M13mp18. In our initial experiments, we added a sufficiently large molar excess of RPA to coat all ssDNA in the reaction. We had previously found that the stability of Pol ε was significantly improved during purification when chloride ions were replaced with acetate ions in all buffers (9). To maintain the chloride-ion concentration at a low level, we adjusted the salt concentration in the reaction mix with 125 mM NaAc instead of 66 mM NaCl. Under these conditions, both Pol ε and Pol δ are stimulated by the processivity clamp, PCNA, when replicating the template (32,34). Time-course analysis showed that Pol δ replicated the M13 ssDNA in 2 min (Figure 1A). In contrast, Pol ε requires at least 8 min to replicate the entire template (Figure 1A). This was consistent with previous studies of Pol ε and Pol δ, where Pol ε was incubated for 30 min and Pol δ for 2–4 min (32,34,34). The single-stranded M13mp18 template can form many hairpin loops, presenting strong pause sites. For this reason, we also used single-stranded pBluescript II SK(+) as a template, which has a lower tendency to form secondary structures that cause replication pause sites (Figure 1B). Again, Pol δ was approximately four times faster when replicating the template with the help of PCNA. Pol δ needed ∼1 min to replicate the entire template (3 kb), while Pol ε needed ∼4 min.
Figure 1.
Comparison of the replication efficiency of Pol ε and Pol δ when stimulated by PCNA. Polymerase replication was assessed over time on single-primed circular ssDNA templates, (A) M13mp18 DNA and (B) pBluescript II SK(+). PCNA and RFC were included in lanes 1–5 and 7–11. No PCNA or RFC was added in lanes 6 and 12. Products of the reactions were separated by electrophoresis on a 1% alkaline agarose gel.
Influence of RPA on the holoenzyme assay
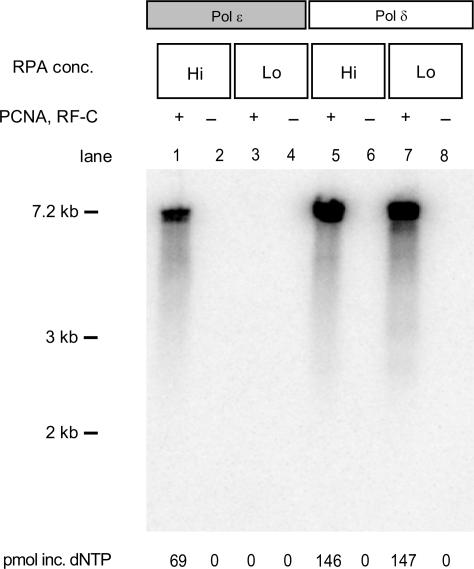
Pol ε has previously been shown to have affinity for ssDNA (25). However, ssDNA is not abundant in vivo, but instead is efficiently coated by RPA. To determine whether Pol δ and Pol ε are sensitive to ssDNA partially coated with RPA, we lowered the RPA concentration in a holoenzyme assay to a level where only ∼20% of the M13mp18 ssDNA was coated with RPA (36) (Figure 2). Based on the previous experiment, we incubated Pol ε for 15 min and Pol δ for 4 min. Under these conditions, the activity of Pol δ was only mildly affected by the presence of naked ssDNA (Figure 2, lane 5 and 7). In contrast, Pol ε was not able to synthesize sufficient amounts of DNA to be detected on the alkaline agarose gel (Figure 2, lane 3). Several explanations are possible as to why Pol ε is inhibited but not Pol δ. One possibility is that ssDNA specifically inhibits the activity of Pol ε. Alternatively, RPA could increase the efficiency of Pol ε loading onto the 3′-terminus of the primer. A third possibility is that Pol ε processivity depends on RPA to melt secondary structures on the template. To distinguish between these hypotheses, we measured the true processivity of Pol ε and Pol δ in the presence of the accessory proteins.
Figure 2.
RPA influences the PCNA-dependent stimulation of Pol ε but not Pol δ. RPA was added at varying concentrations. Reactions in lanes 1 and 2 and 5 and 6 contained 10 pmol RPA, and 2 pmol RPA was added in lanes 3 and 4 and 7 and 8. Reactions with Pol ε (lanes 1–4) were incubated for 15 min; reactions with Pol δ (lanes 5–8) were incubated for 4 min. Products of the reactions were separated by electrophoresis on a 1% alkaline agarose gel.
Processivity of Pol ε and Pol δ
Holoenzyme assays discussed above have often been described as processivity assays. One potential conclusion is that Pol δ is more processive than Pol ε when stimulated by PCNA (Figures 1 and 2). However, in this assay, the DNA polymerase is added in excess or equimolar amounts to template. These conditions do not allow the processivity of the enzyme to be measured due to the occurrence of multiple binding events. To determine if the processivity of Pol ε is affected by RPA and to measure the true processivity of Pol δ and Pol ε, we diluted Pol δ and Pol ε to a concentration where single-hit criteria were met (37). Under these conditions, the DNA polymerase does not encounter an already extended primer terminus when recycled to a new primer terminus. We used similar reaction conditions as described in Figures 1 and 2, but with 30-fold molar excess of end-labeled primer–template over Pol δ and Pol ε to meet the criteria for single-hit conditions. This was confirmed by quantification of the replication products at two different time points, followed by the calculation of the termination probability at a given nucleotide as described previously (38). The termination probability at a given nucleotide at both incubation times was within the variation interval of two independent experiments.
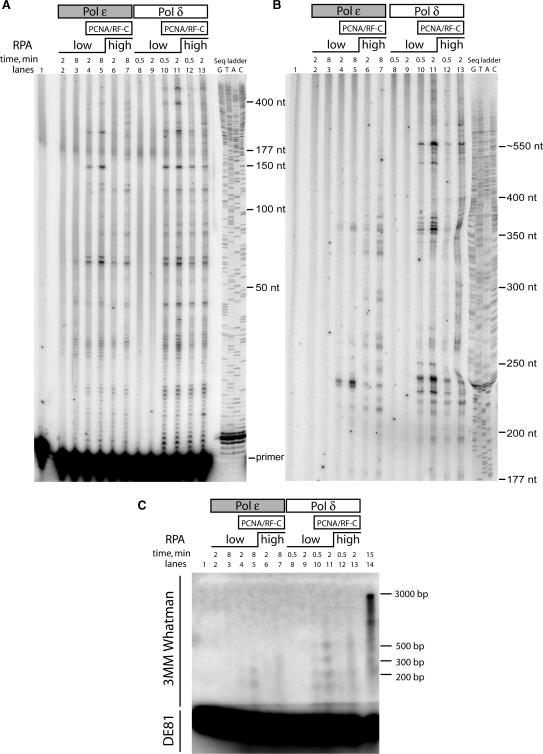
In the first assay, we added 2.7 pmol RPA to the reaction prior to adding Pol ε or Pol δ (lanes 2 and 3 and lanes 8 and 9, Figure 3A–C). On this ssDNA partially coated with RPA and in the absence of PCNA, Pol ε was more processive than Pol δ and was able to synthesize 64-nt long products where a weak pause site was located. In contrast, Pol δ was unable to synthesize products longer than 6 nt. Next, we preloaded PCNA with the help of RFC prior to adding Pol δ and Pol ε, and found that the processivity of both Pol δ and Pol ε was stimulated by PCNA. Pol ε was able to synthesize up to 377-nt long products with the help of PCNA, while Pol δ was able to synthesize products that were longer than 600 nt. We analyzed the products on a 2% alkaline agarose gel, but the amount of products at each length did not allow detection of any products longer than ∼550 nt (Figure 3C). We found that the processivity of Pol ε was stimulated ∼6-fold by PCNA, while, in comparison, PCNA stimulated the processivity of Pol δ by at least 100-fold; these approximations are an underestimation, as we were unable to accurately determine the length of the longest replication products.
Figure 3.
Processivity of Pol ε and Pol δ. The replication products from a single reaction mix were separated on three different gels: (A) 8% polyacrylamide gel separating products in the range of 0–300 nt, (B) 8% polyacrylamide gel separating products in the range of 177–600 nt and (C) a 2% alkaline agarose gel separating products in the range of 100–3000 nt. All gels were dried on a 3MM Whatmann paper, except the 2% alkaline agarose where the section with the end-labeled 50-mer primer was blotted onto a DE81 paper (as indicated on the left side). Reaction times are indicated above each lane. An end-labeled oligonucleotide was annealed to Bluescript II SK(+) ssDNA (lane 1) and 80 fmol primer–template was added in all reactions. RPA (2.7 pmol) was added to the reactions in lanes 2–5 and 8–11. RPA (21 pmol) was added to reactions in lanes 6 and 7 and 12 and 13. Pol ε (2.4 fmol) was added to lanes 2–7 and Pol δ (2.4 fmol) was added to lanes 8–13. RFC and PCNA were added in lanes 4–7 and 10–13. Lane 14 of gel (C) contains 100 fmol of Pol δ to demonstrate where the full-length template is migrating. A sequencing ladder with the identical template was used as a molecular weight marker to the right of gel (A) and (B). A 100 bp molecular weight marker was used on the 2% alkaline agarose gel and stained with ethidium bromide. The migration of each band is indicated to the right of gel (C).
These processivity experiments were carried out in the presence of a low RPA concentration, which has a significant effect on the activity of Pol ε in a regular holoenzyme assay (Figure 2). To analyze the potential effect of saturating levels of RPA on processivity, we assessed activity in the presence of higher levels (21 pmol) of RPA and found that a higher concentration of RPA modestly stimulated the processivity of both Pol δ and Pol ε. Distribution of products synthesized by Pol ε changed more significantly than distribution of products synthesized by Pol δ; for example, the 150 nt products disappeared and several additional pause sites at high molecular mass appeared in lanes 6 and 7. However, the maximal processivity of Pol ε or Pol δ was not significantly affected. For example, the longest detectable products (which are barely visible in Figure 3B, but with a 2-fold intensity over background level when quantified) synthesized by Pol ε increased from 377 to ∼550 nt. No replication products 3 kb in length were detected on the alkaline agarose gel (Figure 3C). From these results we conclude that RPA only has a modest effect on the processivity of Pol ε in the presence of PCNA.
Affinity between the DNA polymerases and DNA
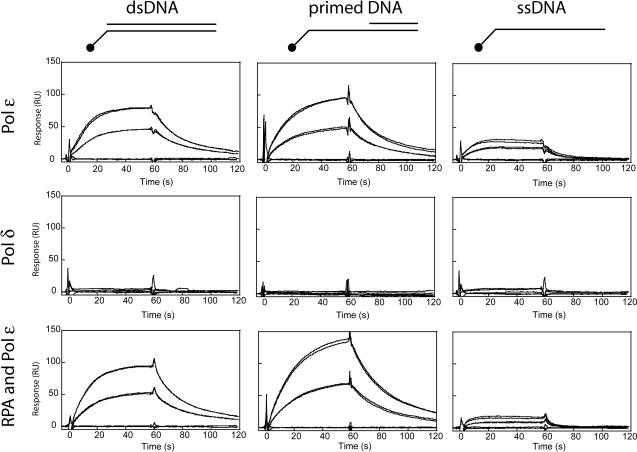
To study the physical interaction between the polymerases and nucleic acids, we used the surface plasmon resonance technique. We captured biotin-labeled ssDNA, double-stranded DNA or primed DNA onto a streptavidin-coated chip in a Biacore 3000. The fourth channel only exposed a streptavidin-coated chip as a reference surface. Pol δ or Pol ε was injected at 10 or 20 nM concentrations and we measured the interactions with the various surfaces (Figure 4). We found that, at these concentrations, Pol δ did not interact with any of the surfaces. In contrast, we detected a high affinity of Pol ε for ssDNA, primed DNA, as well as double-stranded DNA (Figure 4). Pol ε affinity for primed DNA was slightly higher than for dsDNA, while its affinity for ssDNA was about one-third of its affinity for primed DNA. It was not possible to determine the dissociation constant, as the interaction with DNA is complex and the association and dissociation curves cannot be fitted to a 1:1 (Langmuir)-binding model. We next chose to saturate the DNA on the chip with RPA to determine the potential effect of RPA on the interaction between Pol ε and the nucleic acids (Figure 4). Under these conditions, the affinity of Pol ε for ssDNA decreased, the affinity for dsDNA was unchanged, and the affinity for the primed DNA increased. This data suggests that the presence of RPA on ssDNA could inhibit Pol ε from binding non-specifically to ssDNA and promote binding to template–primer junctions.
Figure 4.
Measurement of the affinity between DNA and Pol δ or Pol ε. DNA affinity was analyzed using surface plasmon resonance. Approximately 175 RU of primed and dsDNA templates and 95 RU of ssDNA template were immobilized, reflecting equal molar amounts of substrate. Pol δ or Pol ε was injected at a concentration of 20 and 10 nM. Each injection was repeated twice. After each injection, 0.5 M sodium chloride was injected for 5 s, followed by a buffer blank injection. The top row illustrates the interactions between Pol ε and primed DNA, double-stranded DNA, and single-stranded DNA. The middle row illustrates the interactions between Pol δ and primed DNA, double-stranded DNA and single-stranded DNA. The bottom row illustrates the interactions between Pol ε and primed DNA, double-stranded DNA and single-stranded DNA when the DNA was saturated with RPA.
Affinity between the DNA polymerases and PCNA
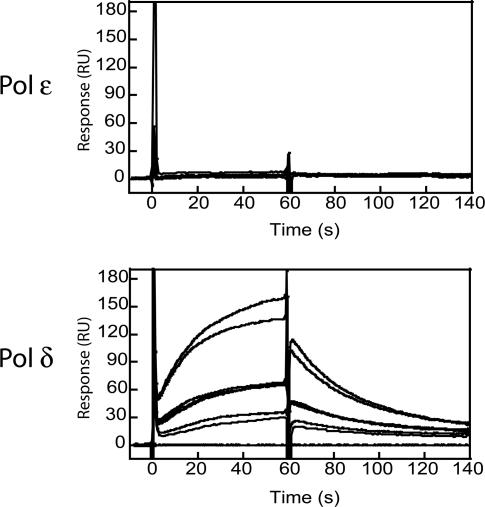
Extensive studies on the interaction between Pol δ and PCNA have shown that Pol δ is dependent on different types of interactions with PCNA. A conserved PCNA interaction motif at the extreme C-terminus of the smallest subunit of Pol δ is essential for the binary interaction between PCNA and Pol δ, i.e. in the absence of template–primer (35). Such a motif has been found in the primary sequence of the catalytic subunit of Pol ε (39), however, the motif was centrally located in the polypeptide and not at the C-terminus or N-terminus as found in all other known PCNA-binding partners. We then used immobilized PCNA on the chip to determine whether Pol ε has a high affinity for PCNA in the absence of a template–primer, and confirmed Pol δ's interaction with PCNA as previously described (35). In contrast, we found that Pol ε was unable to interact with PCNA at the same concentrations (Figure 5), suggesting that the mechanism by which Pol ε functionally interacts with PCNA is distinct to that of Pol δ.
Figure 5.
Interaction between Pol ε and PCNA in solution. Pol δ or Pol ε, at concentrations of 11.5, 23 and 46 nM, were injected onto PCNA immobilized on the surface of the dextran chip (CM5) (∼500 RU). Each injection was repeated twice, and after each injection, 0.5 M sodium chloride was injected for 5 s, followed by a buffer blank injection.
Efficiency by which PCNA is loaded on the primer–template
The RFC clamp-loader binds to the 3′-end of the primer and loads the PCNA clamp. Pol ε has a high affinity for the primer, and the cryo-EM structure of Pol ε suggested that a specific domain of Pol ε binds to the primer, thus possibly blocking the 3′-end of the primer (24,40). This led us to ask whether the efficiency by which PCNA is loaded onto the primer could be suppressed in the presence of excess Pol ε, thus resulting in a lower replication efficiency when compared to Pol δ.
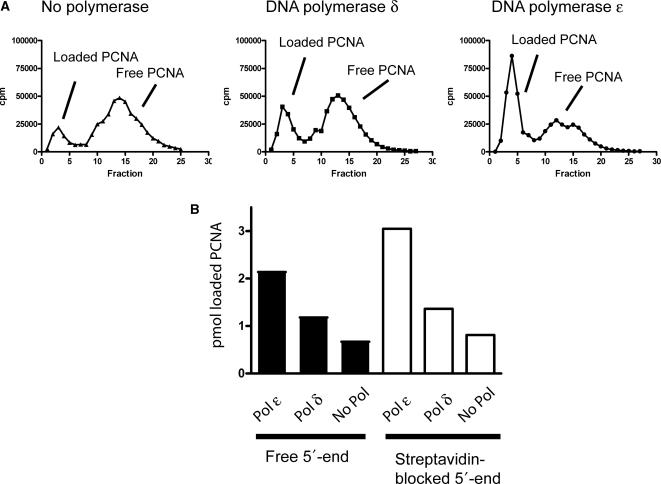
To test this hypothesis, we first measured the amount of PCNA molecules loaded onto a circular ssDNA template when either Pol δ or Pol ε replicated single-stranded pBluescript II SK(+) template (same conditions as in Figure 1B). We generated a tagged version of PCNA and 32P-labeled the purified protein. The labeled PCNA was added to replication reactions and incubated for 7.5 min at 30°C, allowing for full replication of the entire circular template. The reactions were subsequently loaded onto a BiogelA-5 m gel filtration column to separate free PCNA from PCNA loaded on the circular template (3200 bp) (Figure 6A). We found that, on average, ∼4.3 PCNA molecules were loaded onto the template when Pol ε replicated the circular template (Figure 6B). In contrast, only ∼2.4 PCNA molecules were loaded on the template when Pol δ replicated the circular template, and in the absence of polymerase, 1.3 PCNA molecules were loaded on the template. Together, this indicated that more PCNA molecules were loaded onto the template during Pol ε replication compared to Pol δ. We considered the possibility that PCNA could slide off the primers onto the ssDNA. To inhibit this, we repeated the experiment with a primer labeled with a biotin at the 5′-end. The addition of streptavidin blocked the ability of PCNA to slide off the dsDNA onto ssDNA and prevented its dissociation from the circular template. The observed trend under these modified conditions was similar, with 6.1 PCNA molecules per template when Pol ε was added, 2.7 PCNA molecules per template when Pol δ was added, and 1.6 PCNA molecules per template when no polymerase was added (Figure 6B).
Figure 6.
Loading of PCNA on a single-primed circular template. 32P-labeled PCNA was added to a holoenzyme assay with Pol ε, Pol δ or in the absence of polymerase. The reaction with single-primed pBluescript II SK(+) was carried out for 7 min at 30°C, stopped and the PCNA molecules loaded onto the circular template were separated from free PCNA molecules over a BiogelA-5 m column. (A) Elution profiles of reactions where Pol ε or Pol δ was added or the polymerase was omitted. (B) The amount of PCNA loaded onto 0.5 pmol of primed template.
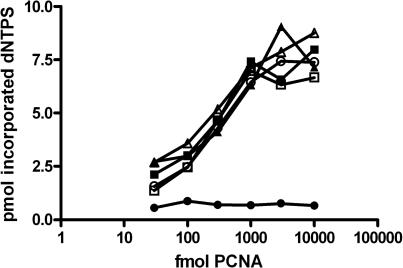
We next determined if the loading of PCNA was the rate-limiting step in replication assays with Pol ε. To address this possibility, we added a 2-fold molar excess of Pol ε over template to the replication assay and assembled a series of reactions using varying concentrations of PCNA, while keeping RFC at a constant level. We found that increasing the amount of PCNA resulted in increased polymerase activity. We next asked if there was a shift in the PCNA response curve when the level of RFC was varied (Figure 7). No changes were detected, indicating that PCNA loading by RFC is not the rate-limiting step in the reaction. Both experiments demonstrated that the tail domain does not interfere with RFC's capability to load PCNA. Instead, it appears that Pol ε has a slow on-rate allowing RFC to load one or several PCNA molecules when Pol ε is disassociated from the primer terminus. Pol δ exhibits a fast on-rate via the PIP-box and the ability to interact with PCNA off DNA, restricting the accessibility for RFC to load multiple PCNA molecules on a single template (Figure 6).
Figure 7.
Assessment of the loading of PCNA as the rate-limiting step in a holoenzyme assay with Pol ε. A series of PCNA-dependent replication assays were carried out, with each reaction having the amount of PCNA indicated on the X-axis. Each series had a constant amount of RFC as follows: filled circle, no RFC added; open circle, 10 fmol RFC; open square, 30 fmol RFC; filled square, 35 fmol RFC; open triangle, 114 fmol RFC and filled triangle, 350 fmol RFC.
DISCUSSION
PCNA interacts with a large number of proteins involved in both DNA repair and DNA replication. In these interactions, PCNA functions as a docking platform to position the enzymes where they should be active, while simultaneously stimulating the activity of the enzymes. PCNA is also a processivity factor for both Pol δ and Pol η in yeast, functioning analogous to clamps in other organisms and viruses (31). However, the role of PCNA as a processivity factor for Pol ε has not been completely clear, due to poor stimulation of Pol ε in holoenzyme assays with single-primed M13mp18 ssDNA, and genetic experiments where mutations in the putative PCNA interaction motif did not result in lethality (33).
The side-by-side comparisons in our replication assays clearly showed that S. cerevisiae Pol ε is not as efficient as Pol δ when replicating M13mp18 ssDNA, and several possibilities could explain the decreased efficiency of Pol ε. One hypothesis is that Pol δ is much more processive than Pol ε when stimulated by PCNA. Alternatively, Pol ε may be inhibited by the presence of ssDNA in the assay, or has difficulty functioning when secondary structures are formed on the template. In this study, we measured the true processivity of Pol δ and Pol ε in the presence of RPA, PCNA and RFC using steady-state kinetics. Our analyses revealed that Pol δ is only slightly more processive than Pol ε. Remarkably, the processivity does not appear to differ >2-fold, and neither polymerase synthesizes significant amounts of replication products longer than 600 nt (Figure 3B and C). From these observations, we can make the following conclusions. First, the processivity of Pol ε, which is comparable to that of Pol δ, cannot explain the relative low overall synthetic capacity of Pol ε (Figures 3 and 1). Second, the addition of RPA does not significantly affect the processivity of the PCNA–Pol ε ternary complex (Figure 3). Thus, allows us to exclude secondary structures of the ssDNA as being a critical factor. There is still a possibility that ssDNA uncoated by RPA could inhibit the enzymatic activity of Pol ε. Our surface plasmon resonance experiments clearly showed that Pol ε has a high affinity for ssDNA and that the affinity decreased when the ssDNA was coated with RPA (Figure 4). In contrast, Pol δ has a much lower affinity for ssDNA. This is consistent with the results from replication assays showing that Pol δ is less sensitive than Pol ε to ssDNA (Figure 2). However, in normal holoenzyme assays, the excess RPA coats all available ssDNA, thus excluding the inhibition of uncoated ssDNA to be a major factor in the decreased efficiency of Pol ε compared to Pol δ (Figure1).
The assembly of a Pol ε–PCNA ternary complex involves the clamp-loader (RFC), the clamp (PCNA) and Pol ε. RFC must first recognize the 3′-terminus of the primer, followed by the loading of PCNA onto the primer, Finally, the polymerase binds both PCNA and the 3′- terminus of the primer. The detected high affinity of Pol ε for dsDNA, coupled with the structural analysis of Pol ε interacting with the dsDNA of the primer, suggested that Pol ε could interfere with RFC's capability to bind to the primer [Figure 4, (24,40)]. This suggests that the rate-limiting step could be the loading of PCNA, not allowing PCNA to be sufficiently loaded. However, we have shown that even adding substoichiometric amounts of RFC in the holoenzyme assay does not alter the ability of PCNA to stimulate Pol ε (Figure 7). In agreement with this, we found that even more PCNA was loaded onto a circular template when Pol ε was replicating the template when compared to Pol δ (Figure 6). Again this suggests that Pol ε is not blocking the 3′-termini for RFC, leading to the conclusion that the rate-limiting step is not the loading of RFC or the processivity of Pol ε, but most likely the mechanism by which Pol ε is loaded onto the PCNA–primer–template junction. Pol δ is much more efficiently loaded and will rapidly replicate long templates in in vitro replication assays, while a slower mechanism of Pol ε loading onto the primer terminus would allow RFC to load multiple PCNA molecules.
PCNA-binding partners are known to carry a PCNA interaction motif, or the PIP-box. Pol δ was previously demonstrated to contain two separate surfaces that interact with PCNA (35). The conserved PCNA interaction motif that is essential for the interaction in solution has been proposed to play a role in localizing the enzyme, while the second interaction region is important in stimulating the enzymatic activity on the DNA. We have shown that Pol ε does not have a high affinity for PCNA in solution. This may be significant for the efficiency by which Pol ε is loaded on the primer terminus, as well as tethering Pol ε to the template, if Pol ε loses contact with the template. In addition, point mutations in the putative PIP-box in the Pol ε catalytic subunit did not affect DNA replication in vivo, although the mutant was somewhat sensitive to DNA damage (33). Whether this motif or additional motifs are responsible for the observed stimulation of Pol ε by PCNA requires further investigation.
Pol ε has two small subunits that interact with dsDNA, and which may be important for the epigenetic silencing of telomeres (21,22,22). The identification of a unique structural dsDNA-interacting domain (40) may explain how Pol ε is localized to sites where it should replicate DNA. At the same time, Pol δ depends on PCNA to be localized to sites where Pol δ replicates DNA, and this allows the two replicative DNA polymerases to function with limited competition at the replication fork. In addition, Pol ε is loaded at the origins of DNA replication prior to Pol δ (4,6). It is possible that the high affinity of Pol ε for dsDNA allows its independent binding to origins, while Pol δ must wait until PCNA is loaded after the first primer is synthesized by Pol α. Pol δ is responsible for the synthesis of Okazaki fragments on the lagging strand, which requires efficient loading of Pol δ, possibly via the PIP-box, by a mechanism which might be shared with other enzymes on the lagging strand, e.g. the flap-endonuclease, FEN-1, and DNA ligase I. Pol ε has been shown to participate in the leading strand synthesis (18). It is possible that the loading of a leading strand polymerase could occur via a PIP-box-independent mechanism, such as with other factors that assist or stabilize Pol ε at the origins allowing sufficient time for loading Pol ε onto the PCNA processivity clamp, including Sld2, Sld3, Dpb11 or the GINS complex (4,41–43)
ACKNOWLEDGEMENTS
This work was supported by the Swedish Research Council (to E.J.), The Swedish Cancer Society (to E.J.), Magnus Bergvalls stiftelse (to E.J.), the Fund for Basic-Science oriented Biotechnology at Umeå University (to E.J.), Wallenberg foundation (to E.J.), Kempe foundation (to O.C.) and by grant GM32431 from the National Institutes of Health (to P.M.B.). Funding to pay the Open Access publication charges for this article was provided by the Swedish Research Council.
Conflict of interest statement. None declared.
REFERENCES
- 1.Garg P, Burgers PM. DNA polymerases that propagate the eukaryotic DNA replication fork. Crit. Rev. Biochem. Mol. Biol. 2005;40:115–128. doi: 10.1080/10409230590935433. [DOI] [PubMed] [Google Scholar]
- 2.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 3.Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- 4.Masumoto H, Sugino A, Araki H. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol. Cell. Biol. 2000;20:2809–2817. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feng W, Rodriguez-Menocal L, Tolun G, D’Urso G. Schizosacchromyces pombe Dpb2 binds to origin DNA early in S phase and is required for chromosomal DNA replication. Mol. Biol. Cell. 2003;14:3427–3436. doi: 10.1091/mbc.E03-02-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aparicio OM, Weinstein DM, Bell SP. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 7.Hiraga S, Hagihara-Hayashi A, Ohya T, Sugino A. DNA polymerases alpha, delta, and epsilon localize and function together at replication forks in Saccharomyces cerevisiae. Genes Cells. 2005;10:297–309. doi: 10.1111/j.1365-2443.2005.00843.x. [DOI] [PubMed] [Google Scholar]
- 8.Johansson E, Majka J, Burgers PM. Structure of DNA polymerase delta from Saccharomyces cerevisiae. J. Biol. Chem. 2001;276:43824–43828. doi: 10.1074/jbc.M108842200. [DOI] [PubMed] [Google Scholar]
- 9.Chilkova O, Jonsson BH, Johansson E. The quaternary structure of DNA polymerase epsilon from Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:14082–14086. doi: 10.1074/jbc.M211818200. [DOI] [PubMed] [Google Scholar]
- 10.Waga S, Masuda T, Takisawa H, Sugino A. DNA polymerase epsilon is required for coordinated and efficient chromosomal DNA replication in Xenopus egg extracts. Proc. Natl Acad. Sci.USA. 2001;98:4978–4983. doi: 10.1073/pnas.081088798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukui T, Yamauchi K, Muroya T, Akiyama M, Maki H, Sugino A, Waga S. Distinct roles of DNA polymerases delta and epsilon at the replication fork in Xenopus egg extracts. Genes Cells. 2004;9:179–191. doi: 10.1111/j.1356-9597.2004.00716.x. [DOI] [PubMed] [Google Scholar]
- 12.Shcherbakova PV, Pavlov YI. 3′–>5′ exonucleases of DNA polymerases epsilon and delta correct base analog induced DNA replication errors on opposite DNA strands in Saccharomyces cerevisiae. Genetics. 1996;142:717–726. doi: 10.1093/genetics/142.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karthikeyan R, Vonarx EJ, Straffon AFL, Simon M, Faye G, Kunz BA. Evidence from mutational specificity studies that yeast DNA polymerases [delta] and [epsi] replicate different DNA strands at an intracellular replication fork. J. Mol. Biol. 2000;299:405–419. doi: 10.1006/jmbi.2000.3744. [DOI] [PubMed] [Google Scholar]
- 14.Pavlov YI, Newlon CS, Kunkel TA. Yeast origins establish a strand bias for replicational mutagenesis. Mol. Cell. 2002;10:207–213. doi: 10.1016/s1097-2765(02)00567-1. [DOI] [PubMed] [Google Scholar]
- 15.Jin YH, Obert R, Burgers PM, Kunkel TA, Resnick MA, Gordenin DA. The 3′–>5′ exonuclease of DNA polymerase delta can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc. Natl Acad. Sci. USA. 2001;98:5122–5127. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin YH, Ayyagari R, Resnick MA, Gordenin DA, Burgers PM. Okazaki fragment maturation in yeast. II. Cooperation between the polymerase and 3′-5′-exonuclease activities of Pol delta in the creation of a ligatable nick. J. Biol. Chem. 2003;278:1626–1633. doi: 10.1074/jbc.M209803200. [DOI] [PubMed] [Google Scholar]
- 17.Garg P, Stith CM, Sabouri N, Johansson E, Burgers PM. Idling by DNA polymerase delta maintains a ligatable nick during lagging-strand DNA replication. Genes Dev. 2004;18:2764–2773. doi: 10.1101/gad.1252304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kesti T, Flick K, Keranen S, Syvaoja JE, Wittenberg C. DNA polymerase epsilon catalytic domains are dispensable for DNA replication, DNA repair, and cell viability. Mol. Cell. 1999;3:679–685. doi: 10.1016/s1097-2765(00)80361-5. [DOI] [PubMed] [Google Scholar]
- 20.Fuss J, Linn S. Human DNA polymerase epsilon colocalizes with proliferating cell nuclear antigen and DNA replication late, but not early, in S phase. J. Biol. Chem. 2002;277:8658–8666. doi: 10.1074/jbc.M110615200. [DOI] [PubMed] [Google Scholar]
- 21.Tsubota T, Tajima R, Ode K, Kubota H, Fukuhara N, Kawabata T, Maki S, Maki H. Double-stranded DNA binding, an unusual property of DNA polymerase epsilon, promotes epigenetic silencing in Saccharomyces cerevisiae. J. Biol. Chem. 2006;281:32898–32908. doi: 10.1074/jbc.M606637200. [DOI] [PubMed] [Google Scholar]
- 22.Iida T, Araki H. Noncompetitive counteractions of DNA polymerase epsilon and ISW2/yCHRAC for epigenetic inheritance of telomere position effect in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004;24:217–227. doi: 10.1128/MCB.24.1.217-227.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Navas TA, Zhou Z, Elledge SJ. DNA polymerase epsilon links the DNA replication machinery to the S phase checkpoint. Cell. 1995;80:29–39. doi: 10.1016/0092-8674(95)90448-4. [DOI] [PubMed] [Google Scholar]
- 24.Tsubota T, Maki S, Kubota H, Sugino A, Maki H. Double-stranded DNA binding properties of Saccharomyces cerevisiae DNA polymerase epsilon and of the Dpb3p-Dpb4p subassembly. Genes Cells. 2003;8:873–888. doi: 10.1046/j.1365-2443.2003.00683.x. [DOI] [PubMed] [Google Scholar]
- 25.Maki S, Hashimoto K, Ohara T, Sugino A. DNA polymerase II (epsilon) of Saccharomyces cerevisiae dissociates from the DNA template by sensing single-stranded DNA. J. Biol. Chem. 1998;273:21332–21341. doi: 10.1074/jbc.273.33.21332. [DOI] [PubMed] [Google Scholar]
- 26.Hamatake RK, Hasegawa H, Clark AB, Bebenek K, Kunkel TA, Sugino A. Purification and characterization of DNA polymerase II from the yeast Saccharomyces cerevisiae. Identification of the catalytic core and a possible holoenzyme form of the enzyme. J. Biol. Chem. 1990;265:4072–4083. [PubMed] [Google Scholar]
- 27.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- 28.Gomes XV, Gary SL, Burgers PM. Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J. Biol. Chem. 2000;275:14541–14549. doi: 10.1074/jbc.275.19.14541. [DOI] [PubMed] [Google Scholar]
- 29.Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Mol. Cell. Biol. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgers PM, Gerik KJ. Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase delta. J. Biol. Chem. 1998;273:19756–19762. doi: 10.1074/jbc.273.31.19756. [DOI] [PubMed] [Google Scholar]
- 31.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc. Natl Acad. Sci. USA. 2005;102:18361–18366. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burgers PM. Saccharomyces cerevisiae replication factor C. II. Formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases delta and epsilon. J.Biol. Chem. 1991;266:22698–22706. [PubMed] [Google Scholar]
- 33.Dua R, Levy DL, Li CM, Snow PM, Campbell JL. In vivo reconstitution of Saccharomyces cerevisiae DNA polymerase epsilon in insect cells. Purification and characterization. J. Biol. Chem. 2002;277:7889–7896. doi: 10.1074/jbc.M108546200. [DOI] [PubMed] [Google Scholar]
- 34.Lee SH, Pan ZQ, Kwong AD, Burgers PM, Hurwitz J. Synthesis of DNA by DNA polymerase epsilon in vitro. J. Biol. Chem. 1991;266:22707–22717. [PubMed] [Google Scholar]
- 35.Johansson E, Garg P, Burgers PM. The Pol32 subunit of DNA polymerase delta contains separable domains for processive replication and proliferating cell nuclear antigen (PCNA) binding. J. Biol. Chem. 2004;279:1907–1915. doi: 10.1074/jbc.M310362200. [DOI] [PubMed] [Google Scholar]
- 36.Kim C, Snyder RO, Wold MS. Binding properties of replication protein A from human and yeast cells. Mol. Cell. Biol. 1992;12:3050–3059. doi: 10.1128/mcb.12.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bambara RA, Fay PJ, Mallaber LM. Methods of analyzing processivity. Methods Enzymol. 1995;262:270–280. doi: 10.1016/0076-6879(95)62023-0. [DOI] [PubMed] [Google Scholar]
- 38.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 39.Dua R, Levy DL, Li CM, Snow PM, Campbell JL. In vivo reconstitution of Saccharomyces cerevisiae DNA polymerase epsilon in insect cells. Purification and characterization. J. Biol. Chem. 2002;277:7889–7896. doi: 10.1074/jbc.M108546200. [DOI] [PubMed] [Google Scholar]
- 40.Asturias FJ, Cheung IK, Sabouri N, Chilkova O, Wepplo D, Johansson E. Structure of Saccharomyces cerevisiae DNA polymerase epsilon by cryo-electron microscopy. Nat. Struct. Mol. Biol. 2006;13:35–43. doi: 10.1038/nsmb1040. [DOI] [PubMed] [Google Scholar]
- 41.Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 42.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 43.Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]