Abstract
Concerted, stochastic and sequential mechanisms of action have been proposed for different hexameric AAA+ molecular motors. Here we report the crystal structure of the E1 helicase from bovine papillomavirus, where asymmetric assembly is for the first time observed in the absence of nucleotide cofactors and DNA. Surprisingly, the ATP-binding sites adopt specific conformations linked to positional changes in the DNA-binding hairpins, which follow a wave-like trajectory, as observed previously in the E1/DNA/ADP complex. The protein's assembly thus maintains such an asymmetric state in the absence of DNA and nucleotide cofactors, allowing consideration of the E1 helicase action as the propagation of a conformational wave around the protein ring. The data imply that the wave's propagation within the AAA+ domains is not necessarily coupled with a strictly sequential hydrolysis of ATP. Since a single ATP hydrolysis event would affect the whole hexamer, such events may simply serve to rectify the direction of the wave's motion.
Accession No. PDB2V9P
INTRODUCTION
Papillomaviral E1 protein is a member of the helicase superfamily III (SFIII) of AAA+ ATPases (1–4). Its N-terminal half (residues 1–300) consists of a regulatory domain and a sequence-specific DNA-binding domain (DBD) involved in the recognition of the replication origin, while the C-terminal half can function alone as a helicase (5). The helicase domain (HD) of the protein can be subdivided into two portions: the oligomerization domain (residues ∼300–378) and AAA+ domain (residues 378–605) containing functional sites involved in ATP hydrolysis and ssDNA translocation (6). The X-ray structure of bovine papillomavirus (BPV-1) E1 helicase (residues 308–577) in complex with a single-stranded DNA molecule and ADP (E1/ADP/DNA) (6) revealed that the surface of the most constricted part of the central tunnel of the hexamer is lined by a DNA-binding hairpin. This hairpin (also known as the ‘pre-sensor 1 β-hairpin’) has been shown to be essential for the function of E1 (5) and SV40 Large Tumor Antigen (LTag) helicase (7). These two helicases belong to the same SFIII clade (8) and have very similar 3D structures (6,9) in spite of the little overall amino acid sequence similarity.
Different ‘motor’ models of DNA translocation through the central tunnel were proposed following X-ray structure determination for several hexameric helicases (1,4). The X-ray structures of LTag (residues 251–627) in the apo-form and in complexes with ATP and its analogs showed symmetrical hexamers with the nucleotide-binding sites of all subunits being in identical conformations. Structural data pointed to a concerted ‘all or none’ movement of the six symmetrically arranged DNA-binding hairpins along the central tunnel in response to the state of nucleotide triphosphate hydrolysis (9). A different picture has emerged from the recent X-ray structure of the E1/ADP/DNA complex determined at 3.15 Å resolution (6), where the hairpins form direct contacts with the ssDNA. In this structure, the vertical positions of six adjacent DNA-binding hairpins (along the central tunnel) follow a wave-like trajectory. This asymmetric arrangement [called a ‘staircase’ in (6)] correlated with a sequential variation of the state of the six ATP-binding sites. These sites were classified on the basis of atomic details of nucleotide cofactor coordination as ATP-type, ADP-type and apo-type.
In the previous account, the reasons for the asymmetry in the E1 hexamer were unclear. The supposition from the structure of E1 in complex with DNA and ADP was that either one or both of these cofactors conferred asymmetry to the hexamer. At the same time, structural similarity with the LTag suggested that the asymmetric arrangement in E1 is induced solely by bound DNA. Another uncertainty concerned the two different states of E1 observed in the two hexamers present in the asymmetric unit (6). These two states were attributed to two different intermediate steps in DNA translocation and their observation raised questions about the degree of structural variability and the number of possible stable states of the E1 hexamer. It was also not clear whether any of the two DNA-bound hexamers were in the ground (energy minima) state preferred by the ligand-free protein assembly. The structure of the E1 protein in the absence of DNA and nucleotide cofactors, reported here, permits these issues to be addressed.
MATERIALS AND METHODS
Purification, crystallization and X-ray data collection
The BPV-1 El helicase domain (E1HD) was expressed and purified from Escherichia coli as described before (13). The protein was concentrated using a 10 K ultrafiltration membrane (Filtron) to 20 mg/ml in buffer containing 10 mM HEPES pH 7.5, 0.2 M NaCl, 5 mM MgCl2, 5 mM DTT. Crystallization by hanging drop vapor diffusion, by mixing 1 μl of protein with 1 μl of reservoir (0.32 M K-phosphate pH 7.5, 0.2 M NaCl, 10–12% PEG3350 and 0.1 M MES pH 7.5), yielded flat single crystals which grew to a size of 0.4 × 0.4 × 0.1 mm3 at +18°C. Diffraction in cryo-solutions and at room temperature was limited by 9–10 Å resolution. However, we noticed that the diffraction quality of some crystals improved to 4.5 Å after 5–6 h of slow drying achieved by equilibrating hanging drops over a solution containing 1.5 M NaCl and 30% PEG 3350. Further tests of ∼150 different crystals led to the identification of a single crystal that diffracted significantly better. Exposing different areas of this crystal at BM14 beamline, ESRF, showed that one of the four corners of the plate-like crystal had significantly lower mosaicity and produced stronger diffraction; this allowed collection of a complete X-ray data set to ∼3.0 Å resolution. The data were processed using DENZO and SCALEPACK, Table 1 (14).
Table 1.
Data collection and structure refinement statistics
| Data collection | |
|---|---|
| Space group | P212121 |
| Unit cell dimensions, (Å) | a = 135.1, b=180.6, c = 187.5 |
| Wavelength (Å) | 0.976 |
| Resolution, Å (outer shell) | 25–3.0(3.11–3.0) |
| Unique reflections | 86,130(6572) |
| Redundancya | 4.8(3.5) |
| Completeness (%) | 94.0(72.6) |
| <I/σ(I)> | 13.5(2.0) |
| Wilson B-factor (Å2) | 67 |
| Rmergeb (%) | 10.8(51.3) |
| Structure refinement | |
| Resolution range (Å) | 3.0–25 |
| Number of reflections in refinement | 85,189 |
| R-factorc (%) | 21.3 |
| Number of reflections used for Rfree | 853 |
| Free R-factorc (%) | 27.2 |
| Number of protein atoms | 26,332 |
| Number of water molecules | 38 |
| Average B-factor (Å2) | 77 |
| R.m.s.d. | |
| Bond lengthsd (Å) | 0.01 (0.02) |
| Bond anglesd (degree) | 1.2 (2.0) |
| Ramachandran statistics | |
| Most favored regions (%) | 90.5 |
| Allowed regions (%) | 9.5 |
| Disallowed regions(%) | 0 |
aThe average number of observations of the same reflection.
bThe value of the merging R factor between equivalent measurements of the same reflection, RI = ∑ |I − <I>| / ∑ I.
cCrystallographic R-factor, R(free) = ∑ ||Fo| − |Fc|| / ∑ |Fo|.
dR.m.s.d. from the standard values are given with target values in parentheses.
Structure determination and refinement
The DNA–nucleotide-free E1 structure was determined by molecular replacement with MOLREP (15), using the E1 hexamer A–F of the E1/ADP/DNA complex (6) as the search model. After rigid body refinement using REFMAC5 (16,17), a model was built and completed using COOT (18) and QUANTA (Accelrys Inc.). During the refinement (16,17), non-crystallographic symmetry restraints were applied separately to four groups of subunits (BCDHIJ, EK, FL and AG), to residues 314–375, 378–492, 517–545 and 559–572. The electron density for the ATP-binding sites was modeled by phosphate and magnesium ions, solvent atoms were added manually into the peaks in the difference electron density maps using 4σ cutoff level. The final refinement cycles utilized translation-libration-screw-motion (TLS) parameters. The side chains of Lys425 residues of subunits A,B,C,D,G,H,I,J,L were not defined in the electron density maps and their side-chain atoms were removed from the model. Residues 306–309, 550–555 and 580–605 were not modeled due to the lack of a clear electron density.
RESULTS
Hexameric assembly of DNA–nucleotide-free E1
The X-ray structure determination of the DNA–nucleotide-free E1 (residues 299–605) at 3.0 Å resolution became possible after significant improvement of the diffraction quality of crystals by partial dehydration (Table 1). For most subunits, the final refined model includes residues 300–579 (Figure 1). Like in the crystals of the E1/ADP/DNA complex [residues 308–577 (6)], the asymmetric unit contains two hexamers (formed by subunits A–F and G–L) but their packing is completely different: ‘head to tail’ in the current structure and ‘head to head’ in the E1/ADP/DNA complex. Unlike in the E1/ADP/DNA complex, the two DNA–nucleotide-free E1 hexamers are essentially identical and could be superimposed with the Cα r.m.s.d. of 0.35 Å; we thus refer throughout to a single hexamer.
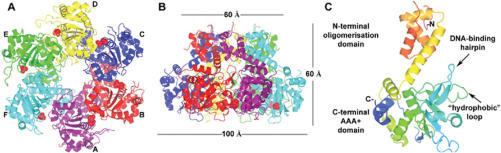
Figure 1.
Structure of the DNA–nucleotide-free E1. (A and B) Ribbon diagrams of the E1 hexamer viewed along and perpendicular to the 6-fold axis. Phosphate and magnesium ions are shown as van der Waals models. (C) E1 monomer in the same orientation as in (B) rainbow-colored with N-terminus in red and C-terminus in blue. All Figures were prepared using CCP4MG (19).
The width of the central tunnel of the DNA–nucleotide-free E1 hexamer varies along its length, with the van der Waals diameter of its narrow parts being ∼13 and ∼11 Å, for the oligomerization and the AAA+ domains, respectively (Figure 1). The C-terminal domain of BPV1 E1 could be readily superposed with the AAA+ domains of SV40 LTag helicase (Cα r.m.s.d. of 2.5 Å for 236 aligned residues) and adeno-associated virus (AAV) Rep40 (10) (Cα r.m.s.d. of 3.2 Å for 195 aligned residues). In common with LTag and Rep40 helicases, the oligomerization domain of E1 (residues 300–378) is composed of α-helices but their three-dimensional arrangement is different.
Like in the E1/ADP/DNA complex (6), the interface between subunits A and F of the DNA–nucleotide-free structure is more open than other interfaces, with only 1450 Å2 (9%) of subunit surfaces buried in the contact area, compared with ∼1800 Å2 (13%) buried at all other interfaces. However, a significant gap observed between the A and F pair of subunits in the E1/ADP/DNA complex, is somewhat reduced in the DNA–nucleotide-free E1 hexamer. Inspection of the current structure shows that like in the E1/ADP/DNA complex, the DNA-binding hairpins are arranged asymmetrically, their positions follow a wave-like trajectory (Figure 2). Analysis shows that two factors contribute to this asymmetry: (i) different orientation of AAA+ domains and (ii) flexibility of the DNA-binding hairpin (Figure 3).
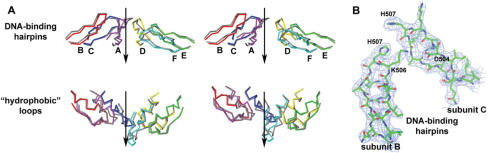
Figure 2.
Wave-like arrangement of DNA-binding hairpins (residues 500–514) and ‘hydrophobic loops’ (residues 453–471) in the DNA–nucleotide-free E1. (A) Cα traces are shown in stereo with the reported structure in color and the E1/ADP/DNA complex in gray. The direction of DNA translocation is indicated by arrow. For clarity, the loops and hairpins are separated. (B) Electron density maps corresponding to two adjacent DNA-binding hairpins of DNA–nucleotide-free E1 calculated with maximum likelihood weighted coefficients 2|Fo|−|Fc| and contoured at 1.25σ.
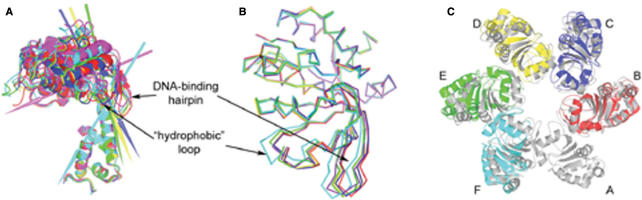
Figure 3.
Conformational variability. (A) Six subunits overlapped by their oligomerization domains and shown as ribbon diagrams. Each rotational axis relates a particular subunit with subunit A and is colored accordingly. (B) Cα diagrams of six superposed C-terminal domains. (C) Subunit A (gray) superposed with all other subunits (color) by its oligomerization domain. For clarity only core regions of C-terminal domains are shown.
Asymmetry of the AAA+ domains
While the oligomerization domains of the E1 hexamer form a symmetric collar, the AAA+ domains are arranged asymmetrically. AAA+ domains of different subunits are related by rotations of up to 14° around different axes (Figure 3A) resulting in a positional deviation of the AAA+ domains by up to ∼7.5 Å when the subunits are superposed by their oligomerization domains (Figure 3A and C). Principal component analysis revealed two dominating rotational modes in the experimentally observed mobility of the AAA+ domains (Supplementary Data).
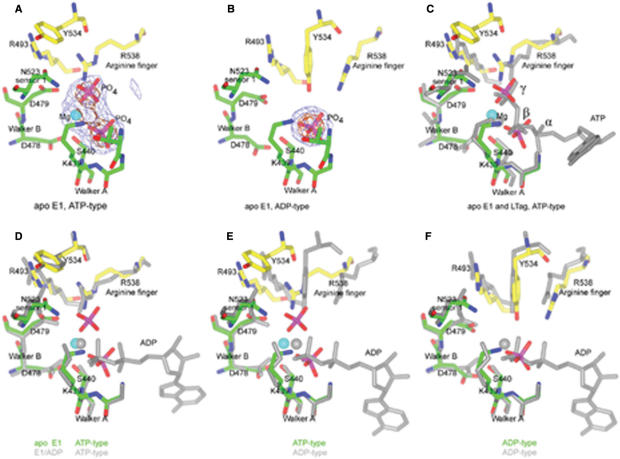
ATP-binding sites are located at the interfaces of adjacent AAA+ domains. One subunit contributes Walker A (GPPNTGKS), Walker B (AALVDD) and sensor-1 (VTSNI) motifs, which coordinate the phosphates of the nucleotide whereas adjacent subunit presents an arginine finger (Arg538) essential for ATP hydrolysis, sensor-2 (Lys425) and several other residues participating in the active site formation. Two types of electron density are observed for the ATP-binding sites (Figure 4A, B). The first type is observed for the three sites formed by subunits A/B, B/C and C/D and corresponds to two phosphate ions and one magnesium ion. These three E1 active sites were superimposed with those of the SV40 LTag helicase, for which the X-ray structure in complex with ATP is available (Figure 4C). Superposition shows that the phosphate ions in the DNA–nucleotide-free E1 structure are mimicking γ- and β-phosphates of the ATP molecule. The phosphate mimicking the γ-phopshate of ATP which was absent in the E1/ADP/DNA complex (6), is coordinated by the arginine finger and residues of Walker A,B and sensor-1 motifs. A different picture is observed for the ATP-binding sites formed by subunits D/E, E/F and F/A. The electron density was modeled by a single phosphate ion (Figure 4B). This phosphate corresponds to the β-phosphate of ATP in the LTag/ATP complex; it is coordinated exclusively by amino acid residues of the Walker A motif.
Figure 4.
ATP-binding sites of the DNA–nucleotide-free E1. E1 molecule is labeled ‘apo E1’ to reflect absence of DNA and nucleotide cofactors. E1/ADP/DNA complex is labeled ‘E1/ADP’. Residues belonging to adjacent subunits are shown in green and yellow, with oxygen atoms colored in red and nitrogen atoms in blue. (A, B) Difference electron density maps for an ATP-type and ADP-type coordination site, respectively, calculated after omission of phosphate and magnesium ions from the model. Maps are contoured at 3σ (blue) and 6.5σ (brown). (C) ATP-type site of the current structure superposed with the ATP-binding site of the SV40 LTag complexed with ATP. (D–F) Superpositions of different ATP-binding sites of DNA–nucleotide-free E1 with ATP-binding sites of E1/ADP/DNA complex (gray).
Analysis of the six ATP-binding sites of the DNA–nucleotide-free E1 hexamer shows they exist in three different states. Three types of nucleotide coordination by the active site residues were also observed for the complex of E1 with DNA and ADP: ATP-type, ADP-type and apo-type. Comparison of all ATP-binding sites of the DNA–nucleotide-free E1 with those of the E1/ADP/DNA complex (6) shows that the configuration of three active sites formed by subunits A/B, B/C and C/D, with two bound phosphates and a magnesium ion, corresponds to the ATP-type of nucleotide coordination (Figure 4D). Active sites formed by subunits D/E and E/F with one bound phosphate ion assume the ADP-type coordination, where Y534 is sandwiched between R493 and R538 (Figure 4E and F). The active site formed by the F–A interface is the only site that superposes poorly with all ATP-binding sites of the E1/ADP/DNA complex but could be classified as an apo-type. This type of coordination is a derivative of the ADP-type but with shifted active site residues. This shift is about ∼3.5 Å in the case of the E1/ADP/DNA apo site (6) and ∼2 Å in the corresponding site of the DNA–nucleotide-free E1.
Organization of DNA-binding segments
Flexibility of the DNA-binding hairpins relative to the AAA+ domains results in Cα deviations of up to 4.5 Å (Figure 3B). This flexibility and the asymmetric arrangement of subunits contribute to the wave-like arrangement of the DNA-binding hairpins, which is essentially the same as in the hexamer A–F of the E1/ADP/DNA complex (6) (Figure 2). In this assembly, the DNA-binding hairpins of subunits B and F occupy the top and bottom positions, respectively, in the orientation shown on Figure 1B. The electron density in the area of the DNA-binding hairpins is clear (Figure 2B) except for the extended end of the hairpin in subunit A (residues 505–509). This hairpin in the structure of the E1/ADP/DNA complex is the only one which does not make contacts with the DNA and is partially disordered. Interactions between adjacent hairpins observed in the E1/ADP/DNA complex (including the salt bridge K506-D504 and hydrogen bonding interactions between K506 and the main chain carbonyl oxygens of R505 and K508) are preserved in the absence of DNA and ADP. It is therefore possible that these interactions stabilize the asymmetric arrangement of the hairpins and of the whole hexamer.
Depending on a particular subunit of the E1/ADP/DNA complex (6), the conformation of H507 corresponds, to either αL or to an adjacent disallowed area of the Ramachandran's plot. This conformation is stabilized by contacts with the phosphates of the DNA backbone. In the structure of the DNA–nucleotide-free E1 reported here, H507 of all six subunits were modeled in the αR conformation. Such conformation is not possible in the case of the E1/ADP/DNA complex since the main chain carbonyl oxygen of H507 would clash with the DNA backbone phosphate. In the E1/ADP/DNA complex, the main chain amide proton forms a hydrogen bond with the DNA backbone phosphate stabilizing the unfavorable conformation. The tension created by the disallowed conformation of H507 could have biological meaning since the potential transition into the αR conformation correlates with the proposed direction of DNA translocation through the complex.
In the structure of the E1/ADP/DNA complex (6), additional van der Walls contacts with the sugar moiety of DNA are made by the conserved F464 residue from an adjacent ‘hydrophobic loop’. This residue was shown to be important for ssDNA binding (5). In the structure of DNA–nucleotide-free E1 the hydrophobic loops (residues 457–467) are in van der Waals interactions with the DNA-binding hairpins of corresponding subunits; the six such loops follow the asymmetric arrangement seen in the hairpins.
DISCUSSION
We determined the structure of the DNA–nucleotide-free E1 helicase using crystals obtained at neutral pH and in the presence of phosphate and magnesium. Unexpectedly, in the absence of bound DNA and ATP analogs, E1 subunits are positioned asymmetrically within the hexamer (Figure 3C). As in the structure of the E1/ADP/DNA complex (6), the positions of the DNA-binding hairpins along the tunnel follow a wave-like trajectory, while the ATP-binding sites are present in the three different states of ATP coordination (ATP-, ADP- and apo-type). We argue that the asymmetric state observed in the DNA–nucleotide-free E1 is not induced by bound magnesium and phosphate ions since chemically identical protein chains are equally exposed to the same excessive molar concentrations of each ion present during crystallization. Thus, we consider that differences in the number of phosphate and magnesium ions bound to different sites and differences in their coordination are best explained as an intrinsic feature of the E1 assembly. However, it is of course likely that subunit–subunit interactions within the hexameric assembly and conformations of individual residues coordinating phosphates and magnesium are stabilized by bound ions. The overall conformation of the DNA–nucleotide-free E1 hexamer resembles the conformation of the A-F hexamer observed in complex with DNA and nucleotide cofactor (6).
Several conclusions could be drawn from this unexpected observation. First, it appears that an asymmetric arrangement of subunits is an intrinsic feature of the E1 oligomer, which is not induced by binding of DNA or nucleotide cofactors. Second, independent experimental observations of essentially the same conformational state in both hexamers of the DNA–nucleotide-free E1 and in one of the two hexamers observed in the structure of the E1/ADP/DNA complex (6) suggest that this is the ground energy minima state of the system. Such a state is observed repeatedly notwithstanding the absence or presence of ADP/DNA and in spite of the differences in the crystallization conditions and in the protein fragments used (306–577 for the ADP/DNA complex and 299–605 for the DNA–nucleotide-free E1). The repeated observation of the same state points against significant structural variability of the E1 hexamer suggesting that the number of its favored states is limited. Third, the observed assembly of chemically equivalent E1 protein chains in the asymmetric hexamer suggests that such a hexamer is an integral interrelated system which would be maintained irrespective of whether nucleotide cofactors occupy all consecutive sites observed in the E1/ADP/DNA complex (6). In such system any event in a single subunit or at a single subunit–subunit interface, like ATP binding or hydrolysis, will affect the whole hexamer.
The essence of the ‘coordinated escort’ mechanism of DNA translocation, derived from the structure of the E1/ADP/DNA complex (6), is that each DNA-binding hairpin captures one nucleotide of ssDNA and migrates through the tunnel using the energy of ATP hydrolysis, thus translocating the DNA. In this mechanism, ATP is hydrolyzed sequentially from one subunit to the next (6). Our data support the proposed mechanism of conformational changes, but indicate that DNA translocation is not necessarily coupled with a strictly sequential hydrolysis of ATP around the hexamer. Indeed, we consider it more appropriate to discuss the mechanism in terms of the states of the whole hexamer. Accordingly, the working cycle of the E1 system could be viewed as transitions of the hexamer between a limited set of such discrete states; two of them have been observed experimentally (6). We note that because E1 subunits maintain the asymmetric state in the absence of nucleotide cofactors and DNA, the conformational wave could migrate across the six subunits due to thermal motion (especially at limiting ATP concentrations) and ATP binding or hydrolysis would only rectify the direction of the wave's movement. Equally, since a single hydrolysis event would affect the whole hexamer, a firing event at a single site may result in the wave's propagation across more than one AAA+ domain.
How exactly the hexameric helicases work has been the subject of much debate. Based on the structural data, different mechanisms were put forward, in spite of the similarities in the fold and oligomer architecture. A sequential mechanism of DNA translocation has been proposed for the T7 gp4 helicase (11) and for a related RecA-like φ12 RNA packaging ATPase P4 (12). A sequential mechanism has been also proposed for the BPV-1 E1 helicase, on the basis of the asymmetric arrangement of subunits in the E1/ADP/DNA complex (6). In contrast, structural studies on LTag (9), the closest homolog of E1 (28% identity for 130 aligned residues), suggested a concerted mechanism of action following observation of the symmetric arrangement of the subunits in complexes with different nucleotide cofactors (but in the absence of DNA).
The data reported here allow a comparison of the hexameric assemblies of E1 and LTag in the absence of DNA. This comparison shows significant differences in the interactions between the flexible AAA+ domains and the oligomerization domains forming the rigid collar. The data suggest that a reduced interaction surface area results in weaker interactions in E1 allowing asymmetric assembly of AAA+ domains in spite of the highly symmetric arrangement of the oligomerization domains. In contrast, additional interactions between AAA+ and oligomerization domains in LTag stabilize the symmetric assembly of AAA+ domains. It remains to be seen whether these inter-domain interactions are disrupted in a functional complex of LTag with DNA, facilitating an asymmetric arrangement of AAA+ domains.
In conclusion, detailed understanding of the underlying molecular principles linking chemical events of ATP hydrolysis with the mechanical events of DNA translocation requires kinetic, mutagenesis and single molecule studies of the E1 system. The concept of the integral, interrelated system introduced here on the basis of the DNA–nucleotide-free structure of E1 dictates that single ATP hydrolysis events induce conformational changes in the whole system. Equally, this concept allows consideration of the E1 helicase action as the propagation of a conformational wave around the protein ring. It remains to be seen if the DNA translocation requires sequential hydrolysis of ATP at all consecutive sites or whether such events are probabilistic. In the latter case, the conformational wave migrates across the six subunits due to Brownian motion until an ATP-binding (and hydrolysis) event in one of the six sites takes place, thus rectifying the direction of the wave's motion.
Protein Data Bank accession code
Coordinates and structure factors of the DNA–nucleotide-free E1 have been deposited with the Protein Data Bank (PDB), accession code 2v9p.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by a Wellcome Trust Fellowship (067416 to A.A.A.) and Yorkshire Cancer Research (S284 and personal support to C.M.S.). We thank David Burgin for help during protein production and Martin Walsh for help during the data collection at the UK beamline BM14 (ESRF, Grenoble), which is supported by the UK BBSRC, EPSRC and MRC Research Councils. Funding to pay the Open Access publication charges for this article was provided by the Wellcome Trust.
Conflict of interest statement. None declared.
REFERENCES
- 1.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu. Rev. Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 2.Soultanas P, Wigley DB. DNA helicases: 'inching forward'. Curr. Opin. Struct. Biol. 2000;10:124–128. doi: 10.1016/s0959-440x(99)00059-7. [DOI] [PubMed] [Google Scholar]
- 3.Hickman AB, Dyda F. Binding and unwinding: SF3 viral helicases. Curr. Opin. Struct. Biol. 2005;15:77–85. doi: 10.1016/j.sbi.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 5.Castella S, Bingham G, Sanders CM. Common determinants in DNA melting and helicase-catalysed DNA unwinding by papillomavirus replication protein E1. Nucleic Acids Res. 2006;34:3008–3019. doi: 10.1093/nar/gkl384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 7.Shen J, Gai D, Patrick A, Greenleaf WB, Chen XS. The roles of the residues on the channel beta-hairpin and loop structures of simian virus 40 hexameric helicase. Proc. Natl Acad. Sci. USA. 2005;102:11248–11253. doi: 10.1073/pnas.0409646102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119:47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 10.James JA, Escalante CR, Yoon-Robarts M, Edwards TA, Linden RM, Aggarwal AK. Crystal structure of the SF3 helicase from adeno-associated virus type 2. Structure. 2003;11:1025–1035. doi: 10.1016/s0969-2126(03)00152-7. [DOI] [PubMed] [Google Scholar]
- 11.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 12.Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–755. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 13.Castella S, Burgin D, Sanders CM. Role of ATP hydrolysis in the DNA translocase activity of the bovine papillomavirus (BPV-1) E1 helicase. Nucleic Acids Res. 2006;34:3731–3741. doi: 10.1093/nar/gkl554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. New York: Academic Press; 1997. [DOI] [PubMed] [Google Scholar]
- 15.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 16.CCP4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 17.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 18.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 19.Potterton E, McNicholas S, Krissinel E, Cowtan K, Noble M. The CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1955–1957. doi: 10.1107/s0907444902015391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.