Abstract
Many nucleic acid enzymes and aptamers have modular architectures that allow them to retain their functions when combined with other nucleotide sequences. This modular function facilitates the engineering of RNAs and DNAs that have more complex functions. We sought to create new DNA aptamers that bind cellulose to provide a module for immobilizing DNAs. Cellulose has been used in a variety of applications ranging from coatings and films to pharmaceutical preparations, and therefore DNA aptamers that bind cellulose might enable new applications. We used in vitro selection to isolate aptamers from a pool of random-sequence DNAs and subjected two distinct clones to additional rounds of mutagenesis and selection. One aptamer (CELAPT 14) was chosen for sequence minimization and more detailed biochemical analysis. CELAPT 14 aptamer variants exhibit robust binding both to cellulose powder and paper. Also, an allosteric aptamer construct was engineered that exhibits ATP-mediated cellulose binding during paper chromatography.
INTRODUCTION
Single-stranded DNA (ssDNA) oligonucleotides are capable of forming intricate structures that give rise to complex chemical functions (1–3). For example, DNA aptamers are engineered molecules capable of binding to specific ligands with high affinity (4,5). Aptamers are typically generated using in vitro selection methods (6) that enrich a population of random-sequence RNAs or DNAs for sequences that can bind to target ligands. However, recent findings have revealed that metabolite-binding RNA aptamers exist naturally as components of gene control elements called riboswitches (7). A diverse assortment of riboswitches regulates the expression of metabolic genes in many eubacteria (8–10) and in some eukaryotes (11–13).
Engineered and naturally occurring aptamers can show exquisite specificity and high affinity for their target ligands. However, riboswitch aptamers are usually larger, use more complex architectures and exhibit greater affinity and specificity compared to their engineered counterparts (14). These disparities presumably could be overcome when creating new aptamers by applying more stringent in vitro selection conditions with larger nucleic acid constructs. Moreover, engineered aptamers do not necessarily require riboswitch-like affinities and specificities to find utility in various biotechnology applications. Engineered aptamers have been generated for a variety of targets, ranging from metabolites such as arginine (15) and ATP (16), drugs such as theophylline (17) and cocaine (18), to macromolecules such as proteins (19–23). Aptamers have been readily adapted to be useful in many affinity-based separation or detection processes. In several cases it is possible to substitute an aptamer for an existing antibody or other receptor with relatively few modifications to the application protocol (24–31). Additionally, several allosteric aptamer systems have been engineered to function as biosensors that convert ligand binding into a physical or chemical readout (32–38).
DNA aptamers are particularly well suited for biotechnology applications because of their chemical stability, ease of production via enzymatic or solid-phase chemical synthesis and compatibility with a great diversity of molecular biology methods. If engineered DNA aptamers can be made using simple methods to perform with affinities and specificities like riboswitch aptamers, the applications for engineered aptamers would be greatly expanded. To facilitate the isolation of aptamers by in vitro selection, we sought to create a small DNA aptamer that can selectively bind to a solid support that is inexpensive and compatible with various chemical separation technologies. Such an aptamer could then be used in allosteric selection strategies (37,38) that permit the isolation of new aptamers.
Cellulose was chosen as a target for aptamer development because it is an inexpensive and readily available biopolymer that is used for coatings, laminates, optical films and sorption media, as well as additives in pharmaceuticals, foodstuffs and cosmetics (39). Additionally, cellulose papers are often used in a variety of experiments for blotting and filtration. These characteristics provide many opportunities for the development of applications with aptamers that can be selectively immobilized to cellulose.
Unfortunately, DNAs can bind to cellulose surfaces non-specifically under typical aqueous conditions, and this might interfere with some molecular biology applications. In this study, we used in vitro selection to isolate cellulose-binding DNA aptamers that are functional in conditions chosen to reduce non-specific interactions. Two representative DNA aptamers were subjected to additional rounds of mutagenesis and in vitro selection, and the secondary structure model for the best-performing aptamer was examined by mutational analysis and dimethyl sulfate (DMS) probing. This further-optimized aptamer also was engineered to produce a two-component allosteric aptamer that preferentially binds cellulose when an adjoining aptamer binds ATP. These results demonstrate that allosteric aptamers can be integrated with cellulose-based chromatography matrices under conditions that reduce non-specific interactions.
MATERIALS AND METHODS
In vitro selection
Cellulose powder for column chromatography was purchased from Sigma-Aldrich. DNA oligonucleotides were purchased from Sigma-Genosys. The starting DNA library was synthesized with the sequence 5′-CGACGTCGCTCGAATGC-N70-CGCCGAGCTAGAGGTCCTTC where N represents an equal mixture of A, G, C and T. Primer 1 (5′-CGACGTCGCTCGAATGC) and primer 2 (5′-AAAAAAAATAATACGACTCACTATAGGAAGGACCTCTAGCTCGGCG) were used for amplification of the DNA library ensemble and individual members.
In vitro selection was initiated by applying 300 pmol of the original random-sequence DNA pool (generation zero or ‘G0’) to ∼15 mg of cellulose powder that was pre-equilibrated with binding buffer (see Results and Discussion section) in a 2 ml Spin-X Centrifuge Tube Filter (Corning). This step and subsequent chromatography steps were conducted at 23°C, and washes and elutions were processed by centrifugation for 1 min at 9000g. The DNA–cellulose mixture was washed using four 200 µl aliquots of binding buffer, and then the bound fraction was eluted by incubating for 10 min with 200 µl elution buffer (see Results and Discussion section). The recovered DNA was precipitated by the addition of 2.5 volumes of cold ethanol in the presence of 40 pmol each of PCR primers 1 and 2. After centrifugation, the DNA pellet was suspended in PCR buffer [1.5 mM MgCl2, 20 mM NaCl, 10 mM Tris–HCl (pH 8.3), 0.01% gelatin] with 2.5 mM each of the four dNTPs and 1 U/µl Taq DNA polymerase. G1 DNAs were produced by thermal-cycling the mixture to yield near full amplification of the selected DNA templates. A portion (6 pmol) of the amplified G1 DNA was then used as template in an additional single-stranded PCR containing 40 pmol of only primer 1 and using the PCR buffer described above. Ten PCR cycles were used to generate the single-stranded population, which was then applied to a freshly prepared column as described above. Limiting the number of PCR cycles to 10 and increasing the amount of double-stranded template helped reduce the amount of alternative products formed during the single-primer PCR. Additional steps for each round of selection were conducted as described for the first round.
After 15 rounds of in vitro selection, individual DNA aptamers were cloned using a TOPO TA cloning kit with the TOPO 2.1 vector (Invitrogen) following the protocol supplied by the manufacturer. Colonies were chosen and grown in rich media overnight, plasmid DNAs were recovered using a QIAprep kit (QIAGEN) and the cloned inserts were analyzed by sequencing. Sequencing was performed by the DNA Analysis Facility on Science Hill at Yale University using an Applied Biosystems 3730 DNA analyzer.
Aptamer reselection
Parental DNA clones CELAPT 3 (5′-CGACGTCGCTCGAATGCTGATAGTAGTTCTCCTTTAAGGAGTTTGGGTGGTG GGTGGGTGTTAGATGTTGAGATCGTTAGCATGTTTCGCCGAGCTA) and CELAPT 14 (5′-CGACGTCGCTCGAATGCCGGGCTCGCGTTGCCGAGGGGGTGGGTTTGGGTCACCA CTGGCGTAGGAAGCCAAGGGTGTGGTGTGCAGCGCCGAGCTA) were synthesized with a degeneracy (40) of 0.12 at each underlined position while the remainder of the positions were held constant. This yields molecules that have an average of seven mutations relative to the parental sequence. Moreover, all possible molecules with up to seven mutations have a high probability of being represented in the reselection population at least once. To avoid contamination by other DNAs from the initial selection, primer 2 analogs selective for CELAPT 3 or CELAPT 14 were established by extending the 3′ end 8 nt into the portion of the DNAs that were originally randomized. This yielded primer 3 (5′-TAGCTCGGCGAAACATGC) for the CELAPT 3 reselection and primer 4 (5′-TAGCTCGGCGCTGCACAC) for CELAPT 14 reselection. The mutagenized populations were subjected to additional rounds of in vitro selection using the protocol that was used to generate the original aptamers. The populations were cloned and sequenced using the protocol described above.
Column-based cellulose affinity experiments
Radiolabeled ssDNA was prepared by PCR in the presence of [α-32P]dGTP (GE Healthcare) using primer 1 and using primers 2, 3 and 4 with a 3′ terminal ribonucleotide. This produces a double-stranded population where the antisense strand is cleavable via alkali-mediated digestion. The ribose linkage was digested using 0.2 N NaOH at 90°C for 15 min and the two strands were separated by denaturing 6% polyacrylamide gel electrophoresis (PAGE). The full-length single-stranded sense DNA was visualized by autoradiography, excised and eluted from the gel by crushing the gel and soaking in 200 mM NaCl, 10 mM Tris–HCl (pH 7.5) and 1 mM ethylenediaminetetraacetic acid (EDTA) for 15 min with shaking. The ssDNA was precipitated with ethanol and recovered as described above.
Approximately 3 pmol of ssDNA was applied to a 2 ml Spin-X Centrifuge Tube Filter (spin column) containing ∼20 mg of cellulose powder that was pre-equilibrated with binding buffer. The column was washed and eluted as described for the selection. Fractions were collected at each step and were quantified using a Tricarb 2900TR liquid scintillation counter (Packard).
Alternatively, pressure flow columns were constructed using 1000 µl pipette tips with column supports made of rolled cellulose blotting paper and packed with ∼100 mg of cellulose powder. These columns were used for additional binding analyses where the ssDNA was applied to the top of the column and positive pressure was applied using a pipette to push the buffer through. This protocol was also used to screen alternate polysaccharides for DNA binding by replacing the cellulose column packing with starch, sephadex, sepharose or sephacryl, which were all purchased from Sigma-Aldrich.
Paper chromatography
5′ 32P-labeled ssDNA (0.5 μl; ∼3 pmol) was manually applied in spots on 100% cotton fiber blotting paper #703 (VWR Scientific). The blotting paper was placed on edge with the DNA spots arranged horizontally (above the buffer level) in a buffer reservoir, and the binding buffer was allowed to travel ∼6–8 cm. The resulting paper chromatogram was imaged using a Storm PhosphorImager (GE Healthcare). In the absence of 0.01% (w/v) sodium dodecyl sulfate (SDS) in the binding buffer, non-specific interactions between DNA and cellulose cause all DNA to remain at the origin.
Aptamer truncations
PCR primers unique to either CELAPT 3 or CELAPT 14 were used to generate a series of truncated DNAs that represent successive 10-nt deletions from their 3′ ends. The truncated PCR products were used as templates for additional PCRs in which the reverse primers contained a 3′ terminal ribonucleotide allowing for alkaline digestion and isolation of the appropriate ssDNA strand. The ssDNA products were purified and recovered as described above, and were screened for cellulose-binding function by paper chromatography.
After defining the 3′ boundary of each aptamer, a series of truncations were made on the 5′ end by chemically synthesizing the constructs of interest. Synthetic DNA constructs were 5′ 32P-labeled using T4 polynucleotide kinase and [γ-32P]ATP (GE Healthcare) following the protocol supplied by the enzyme manufacturer (New England Biolabs).
Purification of chemically synthesized aptamers
Some depurination (41) and other defects can occur during the chemical synthesis of DNA oligonucleotides. The DNA sequences examined in this study are purine rich and, as expected, we detected significant amounts of depurination as revealed by cleavage of internucleotide linkages at abasic sites by β elimination in the presence of alkali (42) at elevated temperature (data not shown). When synthetic DNA was used in binding assays, damaged and non-functional DNA was discarded by first passing the DNA through a cellulose column. The functional DNA was recovered by eluting the bound fraction using the selection methods described above. This cleanup step significantly increased the percentage of synthetic DNAs that bind to cellulose. However, some non-functional molecules in the population are likely caused by misfolding and not chemical damage, and subsequently some fraction of molecules will misfold after denaturing and refolding.
DNA structural probing
DMS was used to methylate the N7 position of guanosine residues (43). DNA was methylated by adding 0.5 μl of DMS to 1 pmol of 5′ 32P-labeled DNA in a 20 μl reaction and incubating for 1 min at 23°C. The reaction was quenched with 80 μl of quench buffer (1.5 M sodium acetate, 20 mM 2-mercaptoethanol). The methylated DNA was recovered by precipitation as described above. The DNA was resuspended in binding buffer and applied to a cellulose column as described for the cellulose column-binding assays. DNAs bound to the column were recovered by incubation in elution buffer and then precipitated as described above. The recovered DNA was resuspended in 0.2 N NaOH and incubated at 70°C for 30 min to cleave abasic sites caused by purine methylation. Cleavage fragments produced by β elimination (42) were analyzed by denaturing PAGE and imaged using a PhosphorImager.
RESULTS AND DISCUSSION
In vitro selection of cellulose-binding DNA aptamers
An initial population of ∼1014 random-sequence DNA oligonucleotides was subjected to 15 rounds of in vitro selection to favor molecules that bind cellulose. The DNA population was applied to a cellulose column pre-equilibrated with binding buffer containing 5 mM MgCl2, 20 mM Tris–HCl (pH 7.5 at 23°C), 100 mM NaCl and 0.01% (w/v) SDS. Although most ssDNA binds non-specifically to cellulose using simple buffered solutions, the addition of 0.01% SDS was found to reduce binding to near undetectable levels. After loading the DNA pool, the column was washed with binding buffer, and DNAs that remained on the column were isolated by the addition of denaturing elution buffer consisting of 7 M urea and 300 mM EDTA. This elution buffer composition was used to elute cellulose-binding DNAs in a previous effort to isolate DNA aptamers for cellulose (44).
DNAs eluted from the cellulose affinity column during each round were amplified by PCR to yield double-stranded DNA products. Progress of the selection process was monitored by using non-denaturing agarose gel electrophoresis and ethidium bromide staining to detect the PCR products. The number of PCR cycles needed to fully amplify the population was reduced as the selection advanced (data not shown), indicating that a progressively larger portion of the population was eluting from the column as successive rounds of selection were completed.
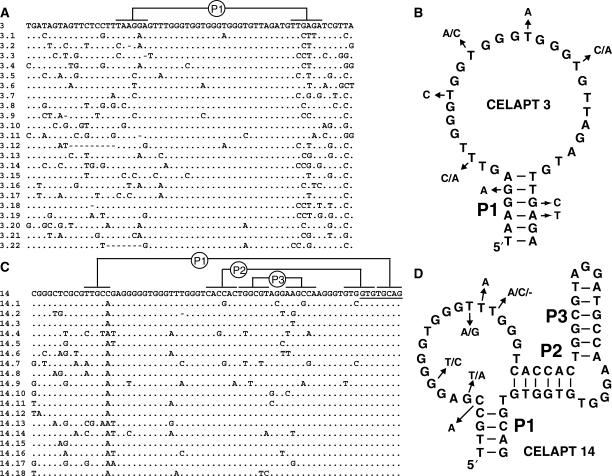
Of 20 clones from G15 that were sequenced, only clones ‘CELAPT’ 1 and 2, and clones CELAPT 3, 16 and 17 were similar or identical in sequence (Figure 1A). All other sequences appeared to be distinct, indicating that a diversity of DNA structures can bind to cellulose under the selection conditions used. Nine clones were randomly chosen for cellulose-binding tests using a column-based affinity assay (see Materials and Methods section). The fraction of the total DNA applied to the column that bound was established to aid in selecting the best-performing clones for further analysis. Clones CELAPT 3 and CELAPT 16 are nearly identical in sequence, and expectedly both exhibited near identical levels of binding. CELAPT 14 performed the best in this initial screening, with almost 35% of the DNA loaded onto the column remaining bound after extensive washing (Figure 1B).
Figure 1.
Sequences and functions of G15 DNAs selected to bind cellulose. (A) Sequences of individual clones recovered from G15 of the initial in vitro selection lineage. Sequences of the primer-binding sites are not shown. Clones 8, 12 and 19 failed to yield readable sequences. (B) Cellulose-binding characteristics for several clones from the G15 population. The fraction of radiolabeled DNA bound to cellulose was determined by eluting bound material and using a scintillation counter to quantify the amount of eluted material compared to the total amount of DNA applied to the column (see Materials and Methods section).
CELAPT 3 and CELAPT 14 were chosen for further optimization by in vitro selection. Although CELAPT 3 and CELAPT 18 exhibited similar levels of binding, the former was chosen for reselection because the CELAPT 3 aptamer class was represented by 3 of the 20 clones examined from the G15 population (Figure 1A). Both parental CELAPT 3 and CELAPT 14 sequences were mutagenized during chemical synthesis to generate the starting DNA pools for reselection (see Materials and Methods section). The original random-sequence domains of both clones were mutagenized to yield a population wherein all possible sequences carrying seven mutations or less are represented. However, eight nucleotides on the 3′ end of the random regions were not permitted to mutate, which provides invariant primer-binding sites that are unique for each construct. This reduces the possibility of amplifying contaminating DNAs.
Secondary structure models for two cellulose aptamer classes
After 11 and 7 rounds of reselection, respectively, the CELAPT 3 and CELAPT 14 populations were cloned and ∼20 were sequenced from each lineage. Alignment of the sequence variants revealed several segments of interest within the mutagenized region of the parental molecules (Figure 2). For example, CELAPT 3 variants (Figure 2A) tolerate mutations on both the 3′ and 5′ ends of the mutagenized region. In contrast, a relatively invariant segment was observed near the center of this region. This pattern of mutation acquisition indicates that the central portion of the mutagenized region is required for aptamer function, whereas the flanking segments are less important.
Figure 2.
Sequence alignments and structural models of variant cellulose aptamers representing two structural classes. (A) Sequence alignment of CELAPT 3 variants. The invariant 3′ region bound by the extended primer 3 binding site is not shown. Deleted residues are indicated with dashes, dots indicate the same base identity as the parent sequence (top line), and variations from the parent sequence are indicated by the nucleotides depicted. Lines identify possible base-paired nucleotides. (B) Predicted secondary structure model for CELAPT 3. Arrows indicate sites of commonly acquired mutations among the variants. (C) Sequence alignment of CELAPT 14 variants. The invariant binding site for primer 4 is underlined. Other details are as described in A. (D) Secondary structure model of CELAPT 14. Details are as described in B.
Some evidence of covariation also was observed between two segments (Figure 2A). Mutations in one segment correlate with nucleotide identities or mutations in the other in a manner that creates or retains base-pairing potential. This pattern strongly suggests that these two segments form a base-paired stem whose stability is important for cellulose-binding activity. Furthermore, clone CELAPT 3.22 has sustained a large deletion of the first base-pairing segment, yet retains the potential to form a similar stem using nucleotides located further upstream. These findings are consistent with a secondary structure model for CELAPT 3 variants involving a single base-paired stem (P1) wherein the large hairpin loop carries all of the most highly conserved nucleotides (Figure 2B).
A notable feature of the conserved loop sequences of CELAPT 3 is the presence of four invariant blocks of G residues. This arrangement is characteristic of DNAs that form G-quadruplex structures, wherein four strand segments are arranged in a quadruplex that is stabilized by hydrogen bonding and base-stacking interactions between guanine bases (45,46). Although G-rich sequences have been observed in other structured DNAs (47,48) that are likely to form G-quadruplexes, in one instance (16) X-ray crystallography revealed that the DNA forms a different 3D fold (49).
The pattern of mutation acquisition in the CELAPT 14 reselection lineage (Figure 2C) was more complex than that observed for CELAPT 3, suggesting that CELAPT 14 variants fold into a larger and more complex structure. Three regions of base-pairing potential (P1, P2 and P3) with evidence of covariation were identified, and these regions are interspersed with segments of the mutagenized domain that remain highly conserved. Furthermore, CELAPT 14 also carries four blocks of invariant G residues that reside in bulges formed between the three stems (Figure 2D). Thus, CELAPT 14 might also form a G-quadruplex, although the overall architecture for this aptamer is substantially different than that predicted for CELAPT 3.
The high G content of the selected DNAs is similar to that observed in the previous selection for DNAs that bind to cellulose or cellobiose (44). Many of the DNAs recovered, in the previous study carry clusters of G residues that are mainly separated by T residues. These characteristics are indicative of G-quadruplex structures, though notably no significant affinity for cellulose was observed with G-quadruplex-forming DNAs (44). Despite the presence of G residue clusters, there is no notable sequence or structural similarities between the DNAs isolated previously and those isolated in the current study.
Paper chromatography with cellulose aptamers
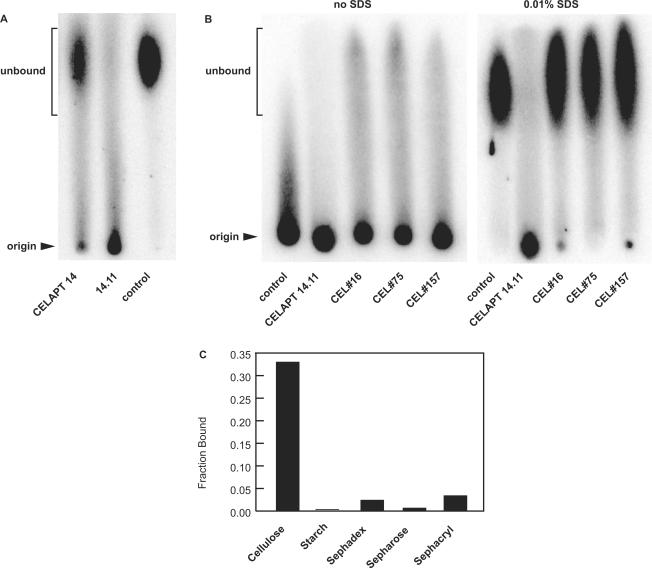
In addition to conducting assays using cellulose column chromatography, we expected that cellulose aptamers would also migrate differently than other DNAs on cellulose-based paper chromatograms. This hypothesis was tested by spotting radiolabeled CELAPT 14, CELAPT 14.11 and random-sequence G0 (control) DNAs on cotton fiber blotting paper and developing the chromatogram using binding buffer as the solvent (Figure 3A). Although much of the CELAPT 14 DNA migrates near the solvent front (unbound), a substantial portion remains at the origin. This result is consistent with that observed by cellulose column chromatography, where ∼35% of CELAPT 14 binds to the cellulose matrix. Interestingly, the variant CELAPT 14.11 is retained at the origin of the paper chromatogram to a far greater extent, as expected if reselection favored DNA aptamers with more optimal cellulose-binding characteristics. In contrast, almost all of the control G0 DNA migrates near the solvent front, which confirms that little non-specific binding of DNA to cellulose occurs with the binding buffer used for in vitro selection and for paper chromatography.
Figure 3.
Paper chromatography of cellulose aptamers. (A) Paper chromatogram of parental CELAPT 14 aptamer, CELAPT 14.11 and G0 random-sequence DNA (control). (B) Paper chromatograms of several DNA samples conducted in the absence (left) or presence of SDS (right). The sequences of the cellulose aptamers CEL#16, CEL#75 and CEL#157 are reported elsewhere (44). The addition of 0.01% SDS to the binding buffer disrupted non-specific interactions, but also aptamer binding for the aptamers from the literature. (C) Specificity of the truncated CELAPT MINI DNA for cellulose matrix versus other polysaccharide matrices (see Materials and Methods section for experimental details).
The characteristics of CELAPT 14.11 on paper chromatograms were compared with those for several cellulose- or cellobiose-binding DNA aptamers published previously (44). Cellobiose is the disaccharide component of cellulose, and therefore we expected these DNAs to remain bound near the origin upon development of the paper chromatogram. Radiolabeled G0 random-sequence control and CELAPT 14.11 DNAs were spotted on blotting paper adjacent to radiolabeled CEL#16, CEL#75 and CEL#157 aptamers, and paper chromatograms were developed with binding buffer without or with 0.01% SDS as indicated (Figure 3B). If SDS was not included in the buffer, all aptamers and the control DNA remained at the origin. In contrast, only CELAPT 14.11 remained at the origin when 0.01% SDS was included in the buffer. These results indicate that the cellulose-binding DNAs reported previously (44) might rely on molecular contacts that are blocked when SDS adheres to the cellulose matrix. In contrast, CELAPT 14.11 was selected to remain functional in the presence of SDS, and therefore its function is not disrupted by the addition of the detergent. Furthermore, 0.01% (v/v) Tween 20 also prevents non-specific binding, and CELAPT 14 class aptamers retain cellulose-binding activity when this detergent replaces SDS in the binding buffer. This indicates that the aptamers are not targeting an SDS–cellulose matrix when binding.
A truncated DNA aptamer based on CELAPT 14.3, called CELAPT MINI (see below) was tested for binding to alternative common polysaccharide matrices (Figure 3C). No substantive binding was observed to starch, sephadex, sephacryl or sepharose matrices. Less than 5% of the input DNA bound in all cases, compared to >30% for cellulose. While cellulose is comprised of β(1–4) linked glucose subunits, starch, sephadex and sephacryl are all comprised of primarily α(1–4) linkages. Additionally, glucose and cellobiose (the disaccharide subunit of cellulose) both were unable to compete for DNA binding to the bulk cellulose matrix (data not shown). CELAPT MINI most likely recognizes some structural unit larger than a disaccharide, wherein the structure of the polysaccharide is important for aptamer recognition.
Minimization and analysis of cellulose aptamer structures
A succession of 3′ and 5′ truncations of 10 nt each were made using PCR with variant CELAPT 3 and CELAPT 14 constructs. Each construct was screened for cellulose binding by column chromatography to determine the minimal length required for function. A series of synthetic constructs were made for CELAPT 3.1 to determine the minimal aptamer structure (data not shown). The loss of either side of the proposed stem caused a significant loss in binding, although it did not completely disrupt function (∼5% bound). Additionally, the CELAPT 3.1 constructs continuously leak off of the column during washing, indicating that the interaction with cellulose might be weaker than CELAPT 14 variants, which exhibit relatively low leakage. These characteristics of CELAPT 3 constructs combined with the robust performance of CELAPT 14 variants prompted us to focus on CELAPT 14 for the remainder of the analysis and engineering efforts.
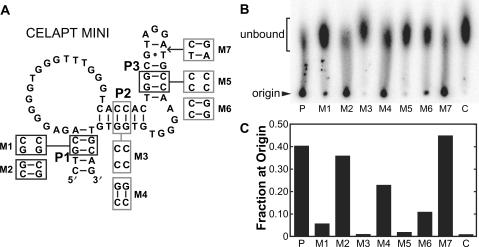
The architecture of CELAPT 14 variants makes them more attractive components of an allosteric aptamer system than CELAPT 3 variants because either P1 or P3 conceivably could serve as attachment points for additional DNA motifs. The CELAPT 14.3 variant was chosen for further analysis because it exhibited the most robust cellulose-binding activity among several variants tested in a preliminary screen (data not shown). The minimal aptamer sequence (CELAPT MINI) was defined (Figure 4A) based on the truncation results (data not shown) and the phylogeny produced by reselection (Figure 2C). The C-to-A mutation that was observed in all CELAPT 14 variants examined is predicted to permit a base pair to form that extends the P1 stem into the bulge between P1 and P2 (Figure 2D). Since the 3′ side of P1 was not permitted to mutate during reselection, only the 5′ side of P1 could acquire mutations that stabilize this predicted base-paired element.
Figure 4.
Mutational analysis of CELAPT MINI. (A) Mutations resulting in the disruption or subsequent restoration of base pairing in stems P1, P2 and P3 are depicted. M7 carried two additional base pairs in P3. (B) Paper chromatogram used to reveal the effects of the mutations described in A. P and C designate lanes spotted with parental CELAPT MINI DNA and G0 random-sequence DNA, respectively. See Materials and Methods section for experimental details. (C) Plot depicting the fraction of DNA remaining at the origin versus the total DNA spotted per lane for the paper chromatogram shown in B.
The secondary structure model for CELAPT MINI was further tested by creating a series of synthetic DNAs that disrupt or restore base pairing in stems P1, P2 and P3 (Figure 4). For example, mutant M1 carries two nucleotide changes in P1 that disrupt base pairing, while mutant M2 carries an additional 2 nt changes that restore pairing (Figure 4A). Constructs M1 and M2 exhibit a loss of cellulose binding and subsequent restoration of cellulose binding, respectively, as observed using both paper chromatography (Figure 4B) and cellulose column chromatography (Figure 4C). Thus, the P1 structure is important for aptamer function, but sequence forming this structure is not. Likewise, P2 and P3 base pairing was assessed and similar results were observed. These results are consistent with the secondary structure model proposed for the CELAPT 14 class of aptamers.
Evidence for G-quadruplex formation by CELAPT 14 class aptamers
Three spans of conserved G residues in the bulge separating stems P1 and P2, along with the span of conserved G residues in the bulge separating P2 and P3, could comprise the nucleotides needed to form a G-quadruplex (45,46). In a G-quadruplex, one plane of four nucleotides is stabilized in part by four hydrogen bonds between the exocyclic amine at the 2 position of each guanine base with the N7 of each adjacent guanine base. Therefore, chemical alteration of these functional groups will destabilize G-quartet formation and also should disrupt the function of DNAs that rely on this substructure (e.g. Ref. 47).
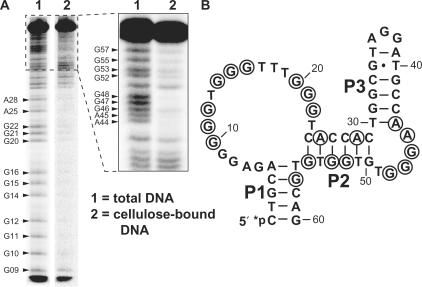
DMS is a reagent that methylates predominantly the N7 positions of purine nucleotides in DNA and RNA (43,50) and thus disrupts interactions that are required for stable formation of G-quadruplexes and other structures. The N-glycosidic linkages of the methylated purines are destabilized and susceptible to depurination and subsequent alkaline-mediated β elimination (42). The locations of purines whose N7 positions were essential for cellulose binding by CELAPT MINI were determined by incompletely methylating the DNA with DMS, separating the resulting modified DNAs into two samples using cellulose affinity purification and cleaving the DNAs in the two isolates by depurination and β elimination (Figure 5A). This DMS probing assay revealed that the N7 positions of four sets of triplet deoxyguanosine residues (Figure 5B, nucleotides 10–12, 14–16, 20–22 and 46–48) are critical for cellulose binding. Modification of these residues prevents binding of the DNA to a cellulose column, which is consistent with the possibility that CELAPT 14 class aptamers form a three-tiered G-quadruplex. Eight additional guanine or adenine residues also exhibit sensitivity to DMS modification. However, methylation at some of these N7 sites is not as disruptive when compared to methylation of sites residing in the triple deoxyguanosine residues described above (Figure 5). The presence of these additional methylation-sensitive sites indicates that the DNA folds into a complex shape that involves numerous other tertiary contacts to the N7 positions of purines. Also, some of these sites might not be important for tertiary structure formation, but these functional groups might be directly involved in making contacts to the cellulose matrix.
Figure 5.
Identification of sites of purine N7 methylation that disrupt cellulose binding by a CELAPT 14 class aptamer. (A) PAGE separation of cleavage products produced by alkali treatment of DMS-modified 5′ 32P-labeled DNAs. Lane 1 contains DMS-treated DNAs that were subjected to cleavage by β elimination to reveal an unbiased distribution of purine methylation sites. Lane 2 contains DMS-treated DNAs that were subsequently enriched for those that retain aptamer function by using a cellulose affinity column prior to cleavage by β elimination. Loss of a band in lane 2 indicates disruption of function by DMS methylation at the N7 of the purine residue designated (arrows). Inset represents a second PAGE analysis of the reaction that more clearly resolved the larger cleavage products. (B) Mapping of DMS-sensitive nucleotides on the secondary structure model of CELAPT MINI. Encircled bases indicate positions that are strongly affected by N7 methylation.
If a G-quadruplex structure is being formed, there is the possibility that this structure might be forming between two or more DNA strands, which would add complexity above that expected if only one DNA strand is needed to form a cellulose-binding pocket. However, we do not observe any increase in cellulose-binding activity with preparations of individual DNA aptamers when concentrations of DNA used are increased (data not shown). This indicates that intermolecular DNA complexes that might be favored at higher DNA concentrations are unlikely to be active. Furthermore, we observed that some DNA aptamer preparations lost cellulose-binding activity after long-term storage at −20°C. Although the samples remained inactive after simple precipitation with ethanol, alkaline-mediated denaturation of the DNA was found to restore cellulose-binding activity (data not shown). This suggests that the formation of stable alternate structures are detrimental to aptamer formation, and perhaps these structures include G-quadruplexes that form between two or more DNA molecules.
Allosteric aptamer design
An allosterically regulated aptamer was constructed to determine if CELAPT MINI was amenable to additional modifications, which might make it useful as a sensor system using paper chromatography or as a tool for allosteric aptamer selection. One strategy proven useful for creating allosteric ribozymes and deoxyribozymes involves the integration of an aptamer in the loop of a hairpin that must be base paired to permit robust catalytic activity (e.g. 32–35,51). The P3 stem of the CELAPT MINI aptamer does not appear to require a specific nucleotide sequence, although its structure is important for cellulose binding (Figure 4). Furthermore, the length of this stem can be expanded (Figure 4, M7) without disrupting activity, suggesting that this stem is unlikely to be in a sterically confined pocket. These characteristics make P3 a desirable location for the attachment of other DNA motifs by employing modular rational design strategies.
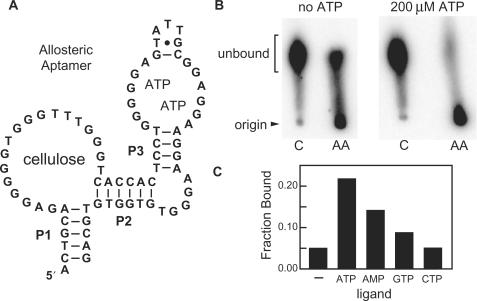
A series of conjoined aptamer designs were made by fusing a DNA aptamer that binds adenosine (or its 5′ phosphorylated derivatives such as ATP) to the P3 stem of CELAPT MINI. The ATP-binding aptamer (16) has been used previously to create ATP-sensing DNA enzymes (34,52), and an atomic-resolution model for the aptamer bound to two ATP molecules has been proposed based on X-ray diffraction data (49). We synthesized the dual aptamer construct or allosteric aptamer (Figure 6A) where P3 of the cellulose aptamer was replaced by the sequence for the ATP-binding DNA aptamer reported previously (16,49). This engineered construct was tested for cellulose-binding activity using both paper chromatography and cellulose column chromatography.
Figure 6.
Allosteric aptamer fusion between CELAPT MINI and an ATP-binding aptamer. (A) P3 of CELLAPT MINI (Figure 4) was replaced with the ATP-binding DNA aptamer sequence reported previously (16,49). (B) Paper chromatograms of 5′ 32P-labeled G0 random-sequence DNAs (C) and allosteric aptamer DNAs (AA) developed in the absence of ATP (left) or in the presence of 200 µM ATP. (C) Fraction of DNA bound to a cellulose affinity column in binding buffer only (−), or in 200 µM each of the ligands indicated.
Although the allosteric aptamer (AA, Figure 6B) exhibits substantial binding to cellulose in the absence of ATP, the addition of 200 µM ATP induces the majority of the DNAs to bind cellulose. Similarly, the allosteric aptamer exhibits the highest fraction of DNA bound to a cellulose-packed column when 200 µM ATP or AMP are present, while GTP and CTP cause little or no change in the cellulose-binding activity of the DNA (Figure 6C). These results reveal that variant DNAs based on the CELAPT 14 class of aptamers are amenable to allosteric control. Furthermore, the demonstration of ATP-dependent changes in mobility of the allosteric aptamer serves as a simple prototype of a biosensor, wherein ligand-triggered changes in DNA migration could be used to report the presence or absence of analytes that are targeted by engineered allosteric aptamers. Molecules with such properties could be used as inexpensive biosensors or as tools for the allosteric selection of new aptamers.
CONCLUSIONS
Our results demonstrate that a diversity of DNA aptamers for cellulose can be created even when buffer conditions are adjusted to eliminate the high level of non-specific binding that typically occurs. Both of the aptamer classes examined in greater detail use base-pairing interactions to form structural components required for function. Interestingly, both carry blocks of conserved G residues that could be involved in forming G-quadruplex structures. If true, these structures can be very stable and might form a core around which other essential aptamer structures are presented.
Of the two classes of cellulose aptamers examined in detail, the CELAPT 14 class appears to be particularly amenable to the construction of allosteric aptamer systems whose cellulose-binding function can be modulated by the binding of a ligand to a second aptamer. The general architecture of the allosteric construct presented here could be expanded to create other constructs that are triggered by almost limitless numbers of target ligands either by using modular rational design or by using in vitro selection. In particular, the latter approach could be applied to create variants of the conjoined construct described in this study (Figure 6) that are likely to exhibit greater dependencies on ATP for cellulose-binding activity. Such refinements, or alterations in ligand-binding specificity, would be necessary to create allosteric DNAs that have characteristics suitable for practical application.
Currently, the use of modular rational design to create allosteric DNAs is limited severely by the fact that very few DNA aptamers for desirable targets exist. However, the cellulose aptamers created in this study might be useful for accelerating the in vitro selection of novel aptamer classes. Previously (37), a two-step allosteric aptamer selection strategy was used to isolate conjoined aptamers that function allosterically. First, two populations of aptamers were created by in vitro selection that bond to two different ligands. Second, the two populations were fused and in vitro selection was applied to enrich for molecules that were able to bind each ligand in a mutually exclusive manner. An alternative approach to allosteric aptamer selection could harness a cellulose aptamer like those reported herein to immobilize a nucleic acid population (aptamer coupled to a random-sequence domain) to a cellulose surface. Members of the population that form a second aptamer and either induce or inhibit function of the adjoining cellulose aptamer could be selectively released from the surface, amplified and subjected to further rounds of selective amplification.
This strategy is similar to that utilized recently to generate structure-switching aptamers (38) by tethering a random-sequence population to beads using a biotinylated oligonucleotide. The advantage of such selection strategies is that immobilization of the target ligands is not require, thus eliminating one barrier to the rapid generation of new aptamers. This technique also allows the full surface of the ligand to be available for binding interactions since no linker chemistry is required which may obscure functional groups. The evolving aptamers can entirely engulf the ligand, which is observed for some natural aptamers that exhibit high affinity and selectivity for their metabolite ligands (e.g. 53–59). Additionally, the use of cellulose aptamers in this strategy would take advantage of a low-cost and readily available medium and would permit the resulting aptamers to be integrated with cellulose-based separation technologies that have been used in scientific research for decades.
ACKNOWLEDGEMENTS
We thank members of the Breaker Laboratory for helpful discussion. This work was supported by NIH grants DK070270 and HV28186. Research in the Breaker laboratory is also supported by the Howard Hughes Medical Institute. Funding to pay the Open Access publication charges for this article was provided by the Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Breaker RR. DNA enzymes. Nat. Biotechnol. 1997;15:427–431. doi: 10.1038/nbt0597-427. [DOI] [PubMed] [Google Scholar]
- 2.Emilsson GM, Breaker RR. Deoxyribozymes: new activities and new applications. Cell Mol. Life Sci. 2002;59:596–607. doi: 10.1007/s00018-002-8452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breaker RR. Natural and engineered nucleic acids as tools to explore biology. Nature. 2004;432:838–845. doi: 10.1038/nature03195. [DOI] [PubMed] [Google Scholar]
- 4.Gold L, Poliski B, Uhlenbeck O, Yarus M. Diversity of oligonucleotide functions. Annu. Rev. Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 5.Osborne SE, Ellington AD. Nucleic acid selection and the challenge of combinatorial chemistry. Chem. Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 6.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 7.Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- 8.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 9.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 10.Schwalbe H, Buck J, Furtig B, Noeske J, Wohnert J. Structures of RNA switches: insight into molecular recognition and tertiary structure. Angew. Chem. Int. Ed. Engl. 2007;46:1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- 11.Sudarsan N, Barrick JE, Breaker RR. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubodera T, Watanabe M, Yoshiuchi K, Yamashita N, Nishimura A, Nakai S, Gomi K, Hanamoto H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the introns containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 13.Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- 14.Breaker RR. Riboswitches and the RNA World. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3rd. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2006. [Google Scholar]
- 15.Harada K, Frankel AD. Identification of two novel arginine binding DNAs. EMBO J. 1995;14:5798–5811. doi: 10.1002/j.1460-2075.1995.tb00268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huizenga DE, Szostak JW. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 17.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 18.Stojanovic MN, de Prada P, Landry DW. Aptamer-based folding fluorescent sensor for cocaine. J. Am. Chem. Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 19.Jellinek D, Green LS, Bell C, Janjic N. Inhibition of receptor-binding by high-affinity RNA ligands to vascular endothelial growth-factor. Biochemistry. 1994;33:10450–10456. doi: 10.1021/bi00200a028. [DOI] [PubMed] [Google Scholar]
- 20.Hornung V, Hofmann HP, Sprinzl M. In vitro selected RNA molecules that bind to elongation factor Tu. Biochemistry. 1998;37:7260–7267. doi: 10.1021/bi972969e. [DOI] [PubMed] [Google Scholar]
- 21.Boiziau C, Dausse E, Yurchenko L, Toulme JJ. DNA aptamers selected against the HIV-1 trans-activation-responsive RNA element form RNA-DNA kissing complexes. J. Biol. Chem. 1999;274:12730–12737. doi: 10.1074/jbc.274.18.12730. [DOI] [PubMed] [Google Scholar]
- 22.Hesselberth JR, Miller D, Robertus J, Ellington AD. In vitro selection of RNA molecules that inhibit the activity of ricin A-chain. J. Biol. Chem. 2000;275:4937–4942. doi: 10.1074/jbc.275.7.4937. [DOI] [PubMed] [Google Scholar]
- 23.Andreola ML, Pileur F, Calmels C, Ventura M, Tarrago-Litvak L, Toulme JJ, Litvak S. DNA aptamers selected against the HIV-1 RNase H display in vitro antiviral activity. Biochemistry. 2001;40:10087–10094. doi: 10.1021/bi0108599. [DOI] [PubMed] [Google Scholar]
- 24.Drolet DW, Moon-McDermott L, Romig TS. An enzyme-linked oligonucleotide assay. Nat. Biotechnol. 1996;14:1021–1025. doi: 10.1038/nbt0896-1021. [DOI] [PubMed] [Google Scholar]
- 25.Potyrailo RA, Conrad RC, Ellington AD, Hieftje GM. Adapting selected nucleic acid ligands (aptamers) to biosensors. Anal. Chem. 1998;70:3419–3425. doi: 10.1021/ac9802325. [DOI] [PubMed] [Google Scholar]
- 26.Davis KA, Lin Y, Abrams B, Jayasena SD. Staining of cell surface human CD4 with 2′-F-pyrimidine-containing RNA aptamers for flow cytometry. Nucleic Acids Res. 1998;26:3915–3924. doi: 10.1093/nar/26.17.3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinjung F, Klussmann S, Erdmann VA, Scheller FW, Furste JP, Bier FF. High-affinity RNA as a recognition element in a biosensor. Anal. Chem. 1998;70:328–331. [Google Scholar]
- 28.Jayasena SD. Aptamers: an emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999;45:1628–1650. [PubMed] [Google Scholar]
- 29.Lee M, Walt DR. A fiber-optic microarray biosensor using aptamers as receptors. Anal. Biochem. 2000;282:142–146. doi: 10.1006/abio.2000.4595. [DOI] [PubMed] [Google Scholar]
- 30.Liss M, Petersen B, Wolf H, Prohaska E. An aptamer-based quartz crystal protein biosensor. Anal. Chem. 2002;74:4488–4495. doi: 10.1021/ac011294p. [DOI] [PubMed] [Google Scholar]
- 31.Liu JW, Mazumdar D, Lu Y. A simple and sensitive “dipstick” test in serum based on lateral flow separation of aptamer-linked nanostructures. Angew. Chem. Int. Ed. Engl. 2006;45:7955–7959. doi: 10.1002/anie.200603106. [DOI] [PubMed] [Google Scholar]
- 32.Tang J, Breaker RR. Rational design of allosteric ribozymes. Chem. Biol. 1997;4:453–459. doi: 10.1016/s1074-5521(97)90197-6. [DOI] [PubMed] [Google Scholar]
- 33.Seetharaman S, Zivarts M, Sudarsan N, Breaker RR. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nat. Biotechnol. 2001;19:336–341. doi: 10.1038/86723. [DOI] [PubMed] [Google Scholar]
- 34.Levy M, Ellington AD. ATP-dependent allosteric DNA enzymes. Chem. Biol. 2002;9:417–426. doi: 10.1016/s1074-5521(02)00123-0. [DOI] [PubMed] [Google Scholar]
- 35.Hesselberth JR, Robertson MP, Knudsen SM, Ellington AD. Simultaneous detection of diverse analytes with an aptazyme ligase array. Anal. Biochem. 2003;312:106–112. doi: 10.1016/s0003-2697(02)00441-4. [DOI] [PubMed] [Google Scholar]
- 36.Stojanovic MN, Kolpashchikov DM. Modular aptameric sensors. J. Am. Chem. Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 37.Wu LH, Curran JF. An allosteric synthetic DNA. Nucleic Acids Res. 1999;27:1512–1516. doi: 10.1093/nar/27.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nutiu R, Li YF. In vitro selection of structure-switching signaling aptamers. Angew. Chem. Int. Ed. Engl. 2005;44:1061–1065. doi: 10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
- 39.Klemm D, Heublein B, Fink HP, Bohn A. Cellulose: fascinating biopolymer and sustainable raw material. Angew. Chem. Int. Ed. Engl. 2005;44:3358–3393. doi: 10.1002/anie.200460587. [DOI] [PubMed] [Google Scholar]
- 40.Breaker RR, Joyce GF. Inventing and improving ribozyme function—rational design versus iterative selection methods. Trends Biotechnol. 1994;12:268–275. doi: 10.1016/0167-7799(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 41.Septak M. Kinetic studies on depurination and detritylation of CPG-bound intermediates during oligonucleotide synthesis. Nucleic Acids Res. 1996;24:3053–3058. doi: 10.1093/nar/24.15.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Küpfer PA, Leumann CJ. The chemical stability of abasic RNA compared to abasic DNA. Nucleic Acids Res. 2007;35:58–68. doi: 10.1093/nar/gkl948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lawley PD, Brookes P. Further studies on alkylation of nucleic acids and their constituent nucleotides. Biochem. J. 1963;89:127–138. doi: 10.1042/bj0890127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Q, Goldstein IJ, Mei HY, Engelke DR. DNA ligands that bind tightly and selectively to cellobiose. Proc. Natl Acad. Sci. USA. 1998;95:5462–5467. doi: 10.1073/pnas.95.10.5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis JT. G-quartets 40 years later: from 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 46.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y, Liu Y, Breaker RR. Capping DNA with DNA. Biochemistry. 2000;39:3106–3114. doi: 10.1021/bi992710r. [DOI] [PubMed] [Google Scholar]
- 48.Breaker RR. DNA aptamers and DNA enzymes. Curr. Opin. Chem. Biol. 1997;1:26–31. doi: 10.1016/s1367-5931(97)80105-6. [DOI] [PubMed] [Google Scholar]
- 49.Lin CH, Patel DJ. Structural basis of DNA folding and recognition in an AMP-DNA aptamer complex: distinct architectures but common recognition motifs for DNA and RNA aptamers complexed to AMP. Chem. Biol. 1997;4:817–832. doi: 10.1016/s1074-5521(97)90115-0. [DOI] [PubMed] [Google Scholar]
- 50.Ehresmann C, Baudin F, Mogel M, Romby P, Ebel J.-P, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Soukup GA, Breaker RR. Engineering precision RNA molecular switches. Proc. Natl Acad. Sci. USA. 1999;96:3584–3589. doi: 10.1073/pnas.96.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nutiu R, Mei S, Liu ZJ, Li YF. Engineering DNA aptamers and DNA enzymes with fluorescence-signaling properties. Pure Appl. Chem. 2004;76:1547–1561. [Google Scholar]
- 53.Mandal M, Boese B, Barrick JE, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 54.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 55.Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 57.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 58.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edwards TE, Ferreé-D’Amaré A. Crystal structures of the Thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]