Abstract
Homologous recombination provides a means for the in vivo construction of recombinant DNA molecules that may be problematic to assemble in vitro. We have investigated the efficiency of recombination within the Caenorhabditis elegans germ line as a function of the length of homology between recombining molecules. Our findings indicate that recombination can occur between molecules that share only 10 bp of terminal homology, and that 25 bp is sufficient to mediate relatively high levels of recombination. Recombination occurs with lower efficiency when the location of the homologous segment is subterminal or internal. As in yeast, recombination can also be mediated by either single- or double-stranded bridging oligonucleotides. We find that ligation between cohesive ends is highly efficient and does not require that the ends be phosphorylated; furthermore, precise intermolecular ligation between injected molecules that have blunt ends can also occur within the germ line.
INTRODUCTION
In Saccharomyces cerevisiae, it has been shown that relatively short regions of terminal homology (as little as 20 bp) are sufficient to mediate recombination between linear DNA molecules that are simultaneously introduced during transformation (1). The relatively high fidelity and efficiency of recombination in yeast has been exploited for the construction of recombinant molecules that are difficult to generate using strictly in vitro methods (2). Similar technology has been developed in Escherichia coli through use of the lambda Red system (3,4). In the case of Caenorhabditis elegans, it has been known for some time that homologous DNA molecules will undergo recombination to form large extrachromosomal arrays when they are coinjected into the germ line syncytium (5). The efficiency of this recombination is sufficiently high that it is practical for the in vivo construction of recombinant molecules (6). The minimum length of homology necessary for efficient recombination in C. elegans has not been reported, although Mello et al. (5) observed efficient recombination between linear molecules that shared 295 bp of terminal homology at one end. We reasoned that if the minimum length of homology were sufficiently short, then it would be feasible and economical to include a recombination targeting sequence within the 5′portion of a PCR primer; this could permit the rapid in vivo construction of recombinant molecules, such as GFP-tagged rescuing constructs, that may be time-consuming or difficult to generate through other cloning methods. This procedure could also provide a rapid method for preliminary testing of particular recombinant configurations to determine whether it is likely to be worthwhile to undertake a multistep construction.
MATERIALS AND METHODS
Culture of nematodes
Caenorhabditis elegans stocks were maintained as described by Brenner (7), using NGM-lite media (8) and E. coli strain AMA1004 (9). All experiments were done using the N2 wild-type strain of C. elegans (7).
Generation and analysis of transgenics
Worm stocks were maintained at 15°C prior to injection. Microinjection of DNA into the germ line syncytium was performed as described by Mello et al. (5). Injected worms were transferred to seeded plates and allowed to produce progeny at 19.5°C. F1 roller progeny were typically transferred to a new plate and scored for the pharyngeal fluorescence phenotype using an Olympus SZX12 dissecting microscope equipped with epifluorescence and GFP filters or (for scoring weaker fluorescence) mounted on agarose pads and examined using a Nikon Microphot SA compound microscope. For β-galactosidase staining, worms were washed in M9 buffer, frozen at −80°C, then thawed and incubated at 30°C for at least 4 h in staining solution (10), omitting DAPI and kanamycin. The frequency of stable lines was estimated by transferring F1 rollers in small groups (typically five) to individual plates, then examining the F2 progeny for the presence of Rol animals and/or GFP positive animals via dissecting microscope. This results in a mild underestimate of the actual frequency of stable lines, since each plate with F2 Rol and/or GFP(+) animals is counted as representing only a single line.
In all cases where we examined animals using the compound microscope, we found that many of the transgenic progeny exhibited weak to moderate GFP expression within a subset of neurons in the head, plus a single ventral preanal neuron (probably PVPL or PVPR). The only segment that was included in all of our assays was the GFP region, which includes the synthetic introns characteristic of this series of GFP constructs. Therefore, this segment probably contains an enhancer element that drives expression within this neuronal subset.
DNA manipulation
Standard molecular biology methods were used for cloning and PCR amplification of DNA. DNA concentrations were estimated by ethidium bromide staining of agarose gels and comparison between samples and standard markers. Phusion polymerase, restriction enzymes, calf intestinal phosphatase (CIP) and DNA ligase were obtained from New England Biolabs and used according to the manufacturer's instructions.
Construction of plasmids
pEL60: pPD118.33 was digested with XhoI to remove the internal XhoI fragment, then ligated to recircularize. pEL61: pPD118.33 was digested with ApaI and BsaAI, the ApaI end was blunted with Klenow and the plasmid then ligated to remove the 3′ half of GFP. pEL62: pPD118.33 was cut with SmaI and BsaAI and religated to remove the 5′ half of GFP. pEL64: pPD118.33 was digested with NcoI and ligated to an adapter created by annealing oligonucleotides o1304 and o1305. This results in a 47 bp NcoI fragment insertion that generates a stop codon after W57 of the GFP-coding sequence.
Generation of PCR products for use in recombination assays
For initial experiments involving regeneration of GFP (Figures 1 and 2), we used as a template pPD118.33 that had been cleaved with EcoRI and HindIII to eliminate the possibility of amplifying full-length GFP and diluted to 0.1 μg/ml. For experiments in which we scored for the presence of weak GFP signal via compound microscope, we used pEL61 (cut with PvuI) and pEL62 (cut with XhoI) as templates, since neither of these plasmids carries the full-length GFP-coding sequence. For experiments with lacZ, we used as a template pPD87.50 (gift of Andrew Fire, Stanford University) that had been linearized with BamHI and HindIII. All PCR products were purified by chromatography (Qiagen or EpochBiolabs) prior to use in injection mixes. PCR products diagrammed in Figures 3, 4 and 6 were generated using Phusion polymerase (New England Biolabs), which creates blunt-ended products.
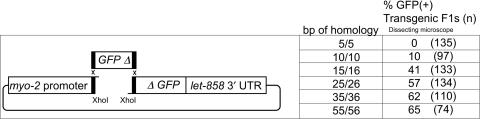
Figure 1.
Recombination between linear molecules to regenerate GFP. pEL60 that has been linearized with XhoI has a 4 nt single-stranded 5′ extension. The amount of homology between this extension, plus the flanking double-stranded DNA at the left/right ends of the recombining PCR product is indicated as ‘bp of homology’. The frequency of GFP(+) F1s is significantly different between each of the first four rows [P < 0.01; z test, (17)]; however, the 25/26, 35/36 and 55/56 categories are not significantly different from each other (P > 0.05) The sizes of the various regions are not drawn to scale. Regions of homology are indicated by black rectangles. GFP was scored by dissecting microscope. The approximate lengths of the indicated regions are as follows: myo-2 promoter, 1.1 kb; GFPΔ, 350 bp plus varying homology; ΔGFP, 780 bp; let-858 3′UTR, 440 bp; plasmid backbone, 2.3 kb. Primer combinations used to generate the recombining fragments were as follows: 5/5, o995 and o996; 10/10, o1009 and o1010; 15/16, o1001 and o1002; 25/26, o999 and o1000; 35/36, o993 and o994; 55/56, o991 and o992.
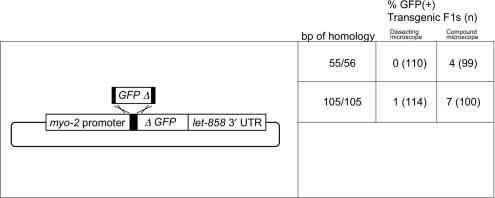
Figure 2.
Recombination between linear and circular molecules to regenerate GFP. Uncut pEL60 was injected in combination with linear PCR products with varying lengths of homology. Regions are as in Figure 1 legend. GFP fluorescence of F1 Rol animals was scored by dissecting microscope in one experiment and by compound microscope in a subsequent set of injections, as indicated. Primer combinations: 55/56, o991 and o992; 105/105, o989 and o990.
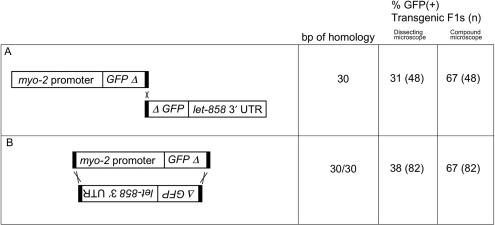
Figure 3.
Comparison between single and double terminal homology. Regions and statistics are as in Figure 1 legend. GFP was scored by both dissecting microscope and compound microscope. The frequency of GFP(+) F1s is not significantly different (P > 0.2) between A and B. Templates and primer combinations: (A) pEL61 cleaved with PvuI, amplified with o1170 and o1240. pEL62 cut with XhoI, amplified with o1171 and o1241. (B) pEL61 cleaved with PvuI, amplified with o1170 and o1240. pEL62 cut with XhoI, amplified with o1241 and o1299.
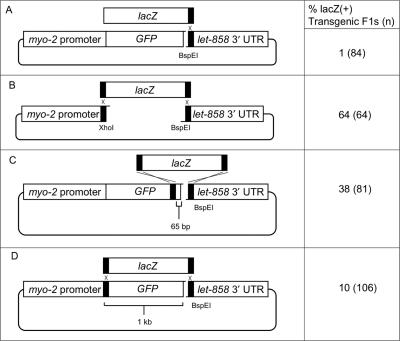
Figure 4.
Recombination assays using lacZ. Regions and statistics are as in Figure 1 legend. The size of the lacZ segment is ∼3.3 kb. GFP was scored in representative worms for cases A, C and D; in each case the lacZ(+) animals were also GFP(+), which is consistent with the efficiency of intramolecular ligation (discussed in text relating to Figure 5) and multiple copy number of transgenes present within extrachromosomal arrays. The depicted locations of homologous recombination events are inferred based on the positions of homologous regions and the fact that homology is almost absolutely necessary to achieve lacZ expression. The frequency of lacZ(+) F1s is significantly different (P < 0.01) in all pairwise comparisons. β-Galactosidase staining was performed as described in Materials and Methods section. The following primer combinations were used to amplify lacZ: A, o1042 and o1047; B, o1041 and o1042; C, o1042 and o1048; D, o1041 and o1042.
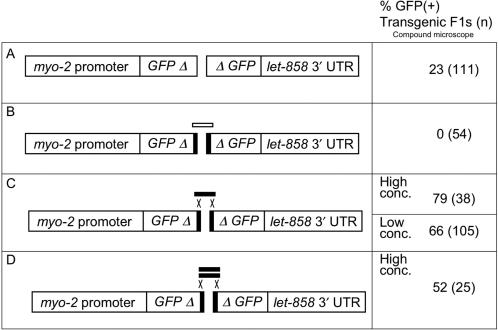
Figure 6.
Use of bridging oligonucleotides to mediate recombination. Regions and statistics are as in Figure 1 legend. The frequencies of GFP(+) F1s in A and B are significantly different from each other and from all other rows (P < 0.01). The values for high and low concentration of olignucleotide in panel C are not significantly different (P > 0.05). The value for low concentration in panel C is not significantly different from the value in panel D. The value for high concentration in panel C is significantly different from the value in panel D (0.05 > P > 0.01). Templates and primers: All panels, pEL61 cut with PvuI and amplified with o1170 and o1236, pEL62 cut with XhoI and amplified with o1171 and o1235. o1237 ss, o1237 + o1238. (A) No added oligonucleotide. (B) Non-homologous oligonucleotide o944. (C) Single-stranded homologous 60-mer, o1237. (D) Annealed oligonucleotides o1237 and o1238.
Annealing of oligonucleotides
Oligonucleotides dissolved in TE were mixed, heated to 95°C, then allowed to cool slowly to room temperature. Annealing was confirmed by running an aliquot on a 4% agarose gel and comparing to the non-annealed oligos.
Formulation of injection mixes
Unless otherwise indicated, all injection mixes contained a standard concentration of 100 μg/ml pRF4 (5). Each recombining fragment was included at a concentration of ∼40 μg/ml. Therefore, in the case of the assays depicted in Figure 1, the small GFPΔ segment is present at ∼10-fold molar excess, whereas, the lacZ segment shown in Figure 3 is present at approximately the same concentration as the plasmid. Plasmid restriction enzyme digests were heat inactivated, then used directly in injection mixes.
Oligonucleotides
All oligonucleotides were obtained lyophilized from Invitrogen and dissolved in TE.
The following oligonucleotides were used:
o944
gatcAtacctgacgctttttatcgcaactctctactgtGAAGGA GAGGATCCgcggccgcggg
o989 GGCCAAAGGACCCAAAGGTATGTT
o990 TGTAGTTCCCGTCATCTTTGAAAAA
o991 CATTTTCAGGAGGACCCTTGGCTAG
o992 TCGGGCATGGCACTCTTGAAAAAGTC
o993 GCTAGCGTCGACGGTACCGGTCAG
o994 AAAGTCATGCTGTTTCATATGATC
o995 CTCGAGGAGCTCCCGAGATCCTATC
o996 CTCGAGAAGCATTGAACACCATAACAG
o999 ACGGTACCGGTCAGATATCACTCG
o1000 TGTTTCATATGATCTGGGTATCTCG
o1001 TCAGATATCACTCGAGGAGCTCCC
o1002 GATCTGGGTATCTCGAGAAGCATTG
o1009 TATCACTCGAGGAGCTCCCGAGAT
o1010 GGTATCTCGAGAAGCATTG
o1015 ACGGTACCGGTCAGATATCACTCGAGG AGCTCCCGAGATCCTATCG
o1016 TGTTATGGTGTTCAATGCTTCTCGAGAT ACCCAGATCATATGAAAC
o1029 CGATAGGATCTCGGGAGCTCCTCGAGT GATATCTGACCGGTACCGT
o1030 GTTTCATATGATCTGGGTATCTCGAGAA GCATGAACACCATAAGA
o1041 CGTCGACGGTACCGGTCAGATATCACTC GAGGGTACCGAGCTCAGAAAAAATG
o1042 AATTCAACGACGTTGGCGTCGATCATCC GGGGGATGTTGAAGAGTAATTGGACT
o1047 GGGTACCGAGCTCAGAAAAAATG
o1048 AGTTTGTAACAGCTGCTGGGATTACACA TGGCACTGCTCCAAAGAAGAAGCGTAAG
o1170 ATGGATACGCTAACAACTTGG
o1171 GCCCAAGCGAGGACAATTC
o1235 GTGCTGAAGTCAAGTTTGAA
o1236 GTGTCTTGTAGTTCCCGTCA
o1237 TTTTCAAAGATGACGGGAACTACAAGAC ACGTGCTGAAGTCAAGTTT GAAGGTGATACCC
o1238 GGGTATCACCTTCAAACTTGACTTCAGC ACGTGTCTTGTAGTTCCC GTCATCTTTGAAAA
o1240 CTTCAAACTTGACTTCAGCACGTGTCTT GTAGTTCCCGTC
o1241 TACAAGACACGTGCTGAAGTCAAGTTTG AA
o1299 TCATTTCCAAGTTGTTAGCGTATCCATG CCCAAGCGAGGACAATTC
o1304 CATGGTGACAATGTACCCATACGATGTT CCAGATTACGCTGGATCCC
o1305 CATGGGGATCCAGCGTAATCTGGAACA TCGTATGGGTACATTGTCAC
RESULTS
Short segments of terminal homology will mediate recombination
In order to develop a sensitive measure of recombination, we began with plasmid pPD118.33 (gift of A. Fire), in which GFP expression is driven within pharyngeal muscle cells by the myo-2 promoter. We created a modified version of this plasmid (pEL60) by deleting a 350 bp XhoI fragment that includes the coding region for amino acids 1–71 of GFP. Next, we used PCR to generate a nested series of DNA molecules, each of which carries the missing portion of GFP, flanked by varying lengths of homology. Coinjection of pEL60 along with the PCR product will regenerate a functional GFP clone if homologous recombination occurs. We included the rol-6(su1006)-containing plasmid pRF4 (5) in supercoiled form, at a standard concentration of 100 μg/ml in all of our injection mixes. The backbone of pRF4 has extensive homology to pPD118.33, so recombination will promote the formation of extrachromosomal arrays that carry both molecules. The rol-6(su1006) gene causes a dominant Roller (Rol) phenotype that makes it possible to unambiguously identify transgenic F1 progeny.
Our results suggest that homologous recombination can occur even when the extent of terminal homology is only 10 bp (Figure 1). The frequency of GFP(+) F1 Rol animals increases with longer stretches of homology, but appears to plateau at ∼65% above 25 bp. In order to provide a rough estimate of the percentage of molecules that underwent successful recombination, we tested three dilutions of intact pPD118.33. At 40, 4 and 0.4 μg/ml, we found that the percentage of GFP positive F1 transgenic animals detectable by dissecting microscope was, respectively, 99% (n = 90), 66% (n = 137) and 1% (n = 184). Although we did not quantitate the intensity level of GFP fluorescence, it was obvious that the brightness level diminished with decreasing levels of pPD118.33. Therefore, these assays established an approximate limit for detection level via dissecting microscope. Since our injection mixes contained ∼40 μg/ml of pEL60, this suggests that ∼10% of the injected molecules underwent successful recombination. It is likely that we would have observed a higher percentage of GFP(+) animals in all of these assays if we had examined the F1 Rol(+) animals by compound microscope, which is a more sensitive means of detecting weak GFP fluorescence (see below).
Targeted recombination between linear and circular molecules does occur, but is not efficient
Since restriction sites are not always present at the desired site of recombination, we tested whether a linear PCR fragment could successfully recombine with a circular plasmid. We found that with 55 or 105 bp of terminal homology, very few or none of the F1 transgenics were GFP(+) when examined by dissecting microscope (Figure 2). Therefore, efficient recombination did not occur in these assays. In order to determine whether low-level recombination does occur in a higher fraction of the F1s, we repeated these experiments and scored the GFP phenotype of F1 Rol animals using a compound microscope to increase the sensitivity of detection. We did detect more GFP(+) animals using this method; however the intensity of GFP was quite weak. Furthermore, none of the F1 rollers in this experiment produced GFP(+) F2 progeny.
Terminal homology at a single end is sufficient to mediate efficient recombination
Since it is has been established that linear DNA molecules can be used for transformation of C. elegans (5,11), we tested whether a single short segment of terminal homology would mediate efficient recombination between two linear segments. As expected based on our previous results, we found that 30 bp of terminal homology was sufficient to permit recombination at a high level (Figure 3A). Adding 30 bp of homology to the other terminus did not further increase the percentage of GFP+ F1s (Figure 3B). In both cases, we were able to obtain stable GFP(+) lines at a useful frequency (3/48 F1s, single homology; 7/76 F1s, double homology). Notably, although all of the lines were derived from Rol F1s, in each case the majority of the GFP(+) lines that we obtained were non-Rol (2/3 and 5/7 for single and double homology, respectively). One likely explanation for this result is that the F1 animals might have carried more than one extrachromosomal array—one with GFP and a separate one with rol-6. This is not unexpected, since the GFP-containing segment does not have homology to the pRF4 plasmid and this is expected to decrease the efficiency of their incorporation into the same extrachromosomal array (5). A non-exclusive possibility is that in some cases the GFP and rol-6-containing molecules did undergo non-homologous recombination and were thus present on the same extrachromosomal array; however, the Rol phenotype might have been absent among the F2s due to partial silencing of transgene expression [it has been established that rol-6(su1006) phenotypic expression is dosage sensitive (5)].
Terminal homology mediates precise molecular joining
The successful regeneration of GFP depicted in Figures 1 and 3 requires precise intermolecular recombination. Imprecise events would cause either insertions or deletions that would prevent GFP from functioning. However, as an independent means of characterizing the fidelity of the recombination junction, we performed PCR to amplify the appropriate region of GFP from two individual F1 GFP(+) animals (generated using the mixture depicted in Figure 3A), then directly sequenced the resulting PCR products. Each animal is expected to contain one or more extrachromosomal arrays, each of which is expected to carry multiple copies of the coinjected molecules. If a high proportion of direct ligations or imprecise recombination events took place, then we would expect the sequence to become difficult to read beginning at the region of overlapping terminal homology. Notably, in both cases, we observed the expected recombination junction and the quality of the sequence remained high throughout the region of homologous overlap. This confirms that precise recombination events are the predominant joining mechanism.
Subterminal homology can mediate moderate levels of recombination
As an alternative method for targeting recombination to locations that do not contain restriction sites, we tested whether linearization adjacent to the desired site of recombination could permit efficient targeting of a recombination event. For these experiments, we performed assays in which recombination was targeted between GFP and a heterologous gene, lacZ (10). We began by assessing the level of background pharyngeal expression of lacZ arising from non-homologous recombination during extrachromosomal array formation (Figure 4A); in this assay, the 3′ end of the lacZ-containing segment can recombine with the myo-2::GFP-containing pPD118.3 plasmid, but no homology exists at the 5′ end. Our results suggest that non-homologous recombination at the 5′ end of lacZ probably does occur, but at a very low level (Figure 4A). As an additional control, we tested whether the presence of short stretches of terminal homology at each end (33 bp/30 bp) would efficiently target the insertion of lacZ (Figure 4B). Consistent with our previous observations, this amount of homology was sufficient to target efficient recombination. Next, we tested the efficiency of recombination when the region of homology at the amino-terminal end of lacZ was internal to the site of linearization of pPD118.33. We found that recombination was moderately efficient when homology was 65 bp internal to the cleavage site (Figure 4C), and occurred at low efficiency when the site of recombination was 1 kb internal (Figure 4D).
Using in vivo recombination to test promoter/enhancer activity
We examined the practicality of using in vivo recombination to assay PCR fragments for promoter function. For these assays, we used plasmid pPD95.77, which carries GFP fused to the unc-54 3′UTR, but lacking a C. elegans promoter (gift of A. Fire, Stanford University). This plasmid was cut with XbaI and HindIII, which cleave immediately upstream of GFP, then mixed with pRF4 plus PCR-amplified promoter segments containing ∼40 bp (43 and 36 bp) of terminal homology at each end. We tested upstream regions from two genes whose expression patterns have been previously described, pes-1 (12) and ehn-3 (13). Since each of these genes is expressed during embyrogenesis or early L1, it was not possible to determine the percentage of F1 Rol animals in which GFP expression was driven by the promoter segment being assayed (although GFP(+) F1 embryos were visible on the plate). Therefore, it was necessary to establish stable Rol lines and then examine the progeny of these animals. In each case, we obtained stable Rol lines at typical frequencies, 10/114 F1s for pes-1 and 6/111 F1s for ehn-3. For ehn-3, 3/6 lines exhibited GFP expression, while all of the pes-1 lines were GFP(+). In each case, the GFP(+) animals exhibited the expression pattern expected for the corresponding gene.
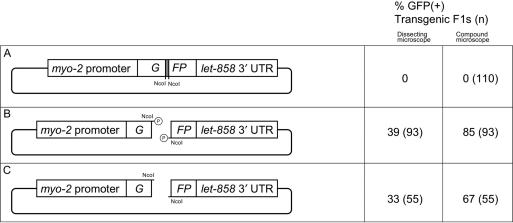
Ligation of complementary cohesive ends occurs efficiently in vivo
In preliminary experiments, we found that linearization of pPD118.33 by cleaving within the GFP-coding sequence did not result in an appreciable reduction in the frequency of GFP(+) F1s (data not shown). This suggested that ligation occurs efficiently in vivo. In order to test this more rigorously, we constructed plasmid pEL64, which carries a 47 bp NcoI fragment inserted into a unique NcoI site within GFP. Animals injected with pEL64 did not produce GFP(+) progeny (Figure 5A). However, when pEL64 that had been cut with NcoI was injected, a large fraction of the transgenic F1s expressed GFP that was bright enough to be visualized by dissecting microscope (Figure 5B). Furthermore, treatment of the digested plasmid with CIP only modestly reduced the percentage of GFP(+) F1 transgenics (Figure 5C). The effectiveness of the CIP treatment was verified by attempting to ligate the treated plasmid in vitro and finding that it was unable to recircularize. Therefore, ligation between complementary cohesive ends occurs efficiently in vivo and does not require that the injected molecules be phosphorylated. As in vitro, intramolecular ligation is likely to be the predominant mechanism responsible for regenerating GFP in these assays; however, in other experiments, we have found that intermolecular ligation of cohesive ends also occurs with reasonably high efficiency (data not shown).
Figure 5.
Intramolecular ligation experiments. Regions and statistics are as in Figure 1 legend. (A) Uncut pEL64 has a 47 bp NcoI fragment (filled rectangle) inserted into GFP. (B) pEL64 cut with NcoI and gel purified to separate it from the NcoI insert. (C) As in B, but treated with phosphatase. GFP was scored by both dissecting microscope and compound microscope. The frequency of GFP(+) F1s scored by compound microscope in B and C is significantly different (P < 0.01).
Oligonucleotide bridges mediate recombination between separate DNA segments
In yeast, it has been found that separate DNA fragments can be joined through homologous recombination by using oligonucleotides that overlap/bridge the desired junction(s) (2). We set out to test whether either single- or double-stranded bridges would stimulate recombination between separate halves of GFP. In order to establish conditions for this assay, we first tested whether the PCR fragments could undergo blunt-end ligation either in the absence of a bridging oligo or in the presence of a non-homologous oligo. For maximal detection of GFP(+), all F1 transgenics were scored on the compound microscope. In the simple blunt-end ligation assay, a significant proportion of the F1s were GFP(+); however, the fluorescence level of most of these was very low, even under the compound microscope (Figure 6A; data not shown). Therefore, in vivo ligation of unphosphorylated blunt ends does occur, but at a very low level. When a non-homologous 63 nt oligonucleotide was included in this mixture, none of the F1 transgenics were GFP(+) (Figure 6B). Possibly, at the relatively high concentration tested (200 μg/ml), the olignucleotide interferes with in vivo ligation by titrating DNA repair enzymes. Regardless of the basis for this inhibition, these results established that the background level of ligation/recombination was sufficiently low that we could test whether addition of homologous oligos would stimulate recombination.
We began by testing injection mixes that included relatively high concentrations (200 μg/ml) of either a single- or double-stranded bridging molecule that has 30 bp of homology to each half of GFP. In the corresponding injection mixes, the bridges are present at ∼100-fold molar excess compared to the halves of GFP. Consistent with the results of Mello et al. (5), we found that with this concentration of oligos, we obtained relatively few F1 transgenics compared to other injection mixes (Figure 6C and D). However, the frequency of GFP(+) F1s was substantially higher than in the control, blunt-end ligation experiment. Furthermore, in both cases, the majority of the GFP(+) F1s animals were easily detectable on the compound microscope (but not on the dissecting microscope). We also found that we were able to obtain stable GFP(+) lines in each case (3/37 for single-stranded; 1/25 for double-stranded). To reduce the inhibitory effect of the oligonucleotides on transformation efficiency, we also tested the single-stranded bridging molecule at a lower concentration (10 μg/ml). Under these conditions, we were able to obtain transgenic F1s more readily, but the percentage of GFP(+) animals was somewhat lower (Figure 6D). Again, we were able to isolate stable GFP(+) lines at a reasonable frequency (4/105). Note that in these experiments (as in those depicted in Figure 3), we identified transgenic F1s based on the Rol phenotype and that the GFP-containing segments do not have homology to the pRF4 rol-6 plasmid.
DISCUSSION
Multiple methods have been developed that enable the generation of recombinant DNA molecules for use in C. elegans. These include PCR-based recombination (14), recombination in yeast (15), recombination in E. coli, in vitro recombination (16) and in vivo recombination within C. elegans (6). Note that in this article we use recombination to refer to any mechanism that joins one segment of DNA to another, regardless of the exact enzymatic sequence of events. However, most of the events that we have assayed are likely to result from homologous recombination, rather than simple joining reactions. Most importantly for practical considerations, our results indicate that certain types of constructs can rapidly and reliably be generated by in vivo recombination that is mediated by relatively short segments of terminal homology. The relative ease and lack of set-up involved in this method would make it useful in cases where a restriction enzyme cleavage site is available near the intended site of recombination and a rapid assay is desirable. For example, this method could be used to rapidly assay the efficacy of fusing GFP to various potential locations within a given cloned gene, or to fuse GFP to a gene contained within a large cosmid clone. Furthermore, this recombination-based methodology enables combinatorial in vivo construction of recombinant molecules at junctions that do not include recognition sites for site-specific recombinases.
We found that intramolecular ligation/circularization of injected molecules occurs efficiently in vivo and this reaction does not require phosphorylation of the 5′ termini. Since we found that circular molecules are not highly recombinogenic, such events could potentially interfere with homologous recombination. However, the recombination efficiencies that we observed are relatively high and recombination efficiency was not obviously elevated when complementary cohesive ends were not present. Therefore competition with intramolecular ligation/circularization does not drastically interfere with the recombination reaction.
We found that either single- or double-stranded bridges are capable of mediating recombination between separate DNA molecules. Although the efficiency of this recombination was relatively low compared to simple binary recombination between molecules that have terminal overlap, such bridges could be used to mediate recombination in cases where both of the DNA fragments to be joined are refractory to PCR amplification.
ACKNOWLEDGEMENTS
We thank Robert Q. Shanks and Barbara Conradt for helpful discussions and Andy Fire for plasmids. This work was supported by NIH RO1GM49785. Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hua SB, Qiu M, Chan E, Zhu L, Luo Y. Minimum length of sequence homology required for in vivo cloning by homologous recombination in yeast. Plasmid. 1997;38:91–96. doi: 10.1006/plas.1997.1305. [DOI] [PubMed] [Google Scholar]
- 2.Raymond CK, Sims EH, Olson MV. Linker-mediated recombinational subcloning of large DNA fragments using yeast. Genome Res. 2002;12:190–197. doi: 10.1101/gr.205201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat. Rev. Genet. 2001;2:769–779. doi: 10.1038/35093556. [DOI] [PubMed] [Google Scholar]
- 4.Dolphin CT, Hope IA. Caenorhabditis elegans reporter fusion genes generated by seamless modification of large genomic DNA clones. Nucleic Acids Res. 2006;34:e72. doi: 10.1093/nar/gkl352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boulin T, Etchberg JF, Hobert O. Ambros V, editor. Reporter gene fusions. WormBook. 2006. http://www.wormbook.org. doi/10.1895/wormbook.1.106.1. [DOI] [PMC free article] [PubMed]
- 7.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun AY, Lambie EJ. gon-2, a gene required for gonadogenesis in Caenorhabditis elegans. Genetics. 1997;147:1077–1089. doi: 10.1093/genetics/147.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casadaban MJ, Martinez-Arias A, Shapira SK, Chaou J. β−Galactosidase gene fusions for analyzing gene expression in Escherichia coli and yeast. Methods Enzymol. 1993;100:293–299. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- 10.Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 11.Stinchcomb DT, Shaw JE, Carr SH, Hirsh D. Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol. Cell. Biol. 1985;5:3484–3496. doi: 10.1128/mcb.5.12.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hope IA. PES-1 is expressed during early embryogenesis in Caenorhabditis elegans and has homology to the fork head family of transcription factors. Development. 1994;120:505–514. doi: 10.1242/dev.120.3.505. [DOI] [PubMed] [Google Scholar]
- 13.Mathies LD, Henderson ST, Kimble J. The C. elegans hand gene controls embryogenesis and early gonadogenesis. Development. 2003;130:2881–2892. doi: 10.1242/dev.00483. [DOI] [PubMed] [Google Scholar]
- 14.Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- 15.Sassi HE, Renihan S, Spence AM, Cooperstock RL. Gene CATCHR—gene cloning and tagging for Caenorhabditis elegans using yeast homologous recombination: a novel approach for the analysis of gene expression. Nucleic Acids Res. 2005;33:e163. doi: 10.1093/nar/gni164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hope IA, Stevens J, Garner A, Hayes J, Cheo DL, Brasch MA, Vidal M. Feasibility of genome-scale construction of promoter:reporter gene fusions for expression in Caenorhabditis elegans using a multisite gateway recombination system. Genome Res. 2004;14:2070–2075. doi: 10.1101/gr.2463804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore DS, McCabe GP. Introduction to the Practice of Statistics. New York, USA: W.H. Freeman; 1998. [Google Scholar]