Abstract
Homologous recombination (HR) is the major mechanism used to repair double-strand breaks (DSBs) that result from replication, but a study of repair of DSBs specifically induced during S-phase is lacking. Using an inverted-repeat assay in which a DSB is generated by the encountering of the replication fork with nicks, we can physically detect repair by sister-chromatid recombination (SCR) and intra-chromatid break-induced replication (IC-BIR). As expected, both events depend on Rad52, but, in contrast to previous data, both require Rad59, suggesting a prominent role of Rad59 in repair of replication-born DSBs. In the absence of Rad51, SCR is severely affected while IC-BIR increases, a phenotype that is also observed in the absence of Rad54 but not of its paralog Rdh54/Tid1. These data are consistent with SCR occurring by Rad51-dependent mechanisms assisted by Rad54, and indicate that in the absence of strand exchange-dependent SCR, breaks can be channeled to IC-BIR, which works efficiently in the absence of Rad51. Our study provides molecular evidence for inversions between repeats occurring by BIR followed by single-strand annealing (SSA) in the absence of strand exchange.
INTRODUCTION
Double-strand break (DSB) repair must be highly efficient in all organisms; and indeed, a single unrepaired DSB has been shown to be lethal to Saccharomyces cerevisiae (1). In dividing cells, homologous recombination (HR) is thought to be the main mechanism to repair DNA breaks that arise during replication, as supported by the observations that recombination proteins only form foci during S phase (2) and that in G1 cells HR is practically absent (3,4). HR has also been proposed to play a role in restart of stalled or collapsed replication forks (5,6).
HR needs an intact homologous DNA sequence as a template for repair. Depending on the nature and location of the donor used, the outcome can be different (7). As soon as a sequence is replicated (during S and in G2), any damage in one chromatid can be repaired by sister chromatid recombination (SCR). Since sister chromatids are identical to each other, repair by SCR is accurate and has been shown to be the preferred recombinational DSB repair mechanism (8–10). DSBs can also be repaired using as donor ectopically located homologous sequences (ectopic recombination), or the allelic copy of the homologous chromosome (allelic recombination), in which cases repair can result in loss or reorganization of genetic information.
SCR events involving a reciprocal exchange between chromatids (sister-chromatid exchange, SCE) can be detected cytologically in mammalian cells (11). Nonetheless, cytological studies are insufficient to provide a mechanistic model for SCR. As recombination between sisters occurs between two identical DNA sequences generated by DNA replication, its genetic and molecular analysis is difficult. To overcome this problem, SCR has been generally studied as unequal recombination events occurring between intrachromosomal heteroalleles (9,10,12,13). Given the low frequency of spontaneously occurring HR, many studies have been based on mitotic recombination events induced by DNA damaging agents or by sequence-specific endonucleases. The extensively used endonuclease HO of S. cerevisiae, which cuts at a specific 117-bp sequence (HO site), has become an excellent tool to study DSB-induced recombinational repair (7). Nonetheless, given the high efficiency of cleavage produced by HO, after replication the two duplicated 117-bp HO sites in both chromatids are cleaved, impeding their repair by SCR. To overcome this problem, we have recently reported that HO-induced DSBs made at a 21-bp HO-cleavage site occur with efficiency around 10%. Based on this mini HO-site we have developed new substrates for the specific in vivo analysis of SCR in circular minichromosomes (8). Indeed, we have shown that these DSBs mainly occur during replication by the conversion of single-stranded breaks produced by HO, ensuring that one chromatid remains intact and, therefore, competent as repair template (14).
The steps and proteins involved in HR are well defined in S. cerevisiae (15,16). HR starts with a 5′-resection of the ends of the DSB. Rad52, probably in concert with Rad59, promotes the invasion of the resection-generated 3′-OH single-stranded ends on the homologous duplex DNA. This invasion is facilitated and stabilized by strand exchange catalyzed by the RecA-homolog Rad51, together with Rad54, a member of the SNF-SWI family of ATPases, and the Rad55-Rad57 heterodimer (15,16). Instead, the Rad54 paralog, Rdh54/Tid1, seems to have a more specialized role in allelic recombination (17,18). All these steps are common to most known HR events, regardless of the pathway and repair template used. Nevertheless, some HR mechanisms may also occur in the absence of Rad51-dependent strand exchange. Thus, break-induced replication (BIR), in which the invading end primes extensive DNA synthesis, can occur in the absence of Rad51 (19), although Rad51 participates in wild-type events (20). Also, the single-strand annealing (SSA) mechanism of DSB repair, responsible for most deletions occurring between direct repeats (21,22), is Rad51 independent (23).
The purpose of this study was to establish the genetic and molecular bases of SCR initiated by replication-born DSBs in comparison with other forms of HR repair, such as intrachromatid recombination occurring by break-induced replication (IC-BIR). Using circular minichromosomes in which DSBs arise by replication through single-strand DNA nicks induced by the HO endonuclease at a 21-bp HO site (14), we found that repair of DSBs occurs primarily by SCR, and is dependent on Rad51, Rad59 and Rad54, but not on Tid1/Rdh54. Instead, IC-BIR is a minor event stimulated in rad51Δ and rad54Δ cells, unaffected by tid1Δ/rdh54Δ and decreased in rad59Δ cells. Altogether, our work provides molecular evidence for a role of Rad59 in HR in RAD51 cells and for the BIR followed by SSA model invoked to explain inversions between repeats in the absence of Rad51 (24).
MATERIAL AND METHODS
Strains, plasmids and oligonucleotides
All strains used were isogenic to W303-1A. The wild-type WS (MATa-inc ade2-1 his3-11,15 trp1-1 ura3-1 ade3::GAL-HO leu2Δ::SFA1) and MAWR-4C (MATa-inc ade2-1 his3-11,15 trp1-1 ura3-1 ade3::GAL-HO leu2Δ-k) strains were described previously (8,25). Deletion mutants were obtained by gene replacement of the corresponding ORF by the KanMX4 cassette as described (26). All deletions were confirmed by PCR and Southern analyses (data not shown). Isogenic double mutants were obtained by genetic crosses. Plasmid pRS316-TINV and pCM189-leu2HOr were previously described (8,25).
Analysis of HO cleavage
WS cells carrying a bar1Δ allele (WS-bar1), used as wild-type controls, were grown at 30°C to an OD of 0.4 in synthetic complete medium lacking uracil (SC-ura) with 2% raffinose as carbon source. The bar1Δ alelle was used to enhance the effect of α-factor. The culture was split into two halves, one of which was supplemented with 0.2 µM α-factor to arrest cells at G1. After 4 h, 2% galactose was added to induce HO expression. For α-factor release, cells were washed twice with pre-warmed fresh medium without α-factor, and 50 µg/ml Pronase (SIGMA, USA) was added to remove any trace of α-factor. Yeast samples were taken at different times and DNA was isolated for Southern blot analysis. In the experimental conditions used, HO cleavage is recurrent, as cells are maintained in 2% galactose after α-factor release.
Physical analysis of recombination
Mid-log phase cultures of yeast cells grown in SC-ura 3% glycerol-2% lactate were split. 2% glucose or 2% galactose was added to repress or induce HO expression, respectively. Yeast samples were taken at different times and DNA was isolated for Southern blot analysis.
Genetic analysis of recombination
It was performed as previously described (25) except that HO cleavage was induced for 2 h. The recombination frequencies shown are the average of three median frequencies obtained by independent fluctuation tests, each performed with six independent colonies.
Miscellaneous
DNA isolation was performed as previously described (8). Southern analyses, yeast growth conditions, DNA labeling and quantification with a Fuji FLA3000 were performed according to standard procedures. In each case a representative experiment of at least three performed is shown. Variation was below 20% in all cases.
RESULTS
An in vivo molecular assay for the analysis of the repair of replication-born DSBs via SCR and IC-BIR
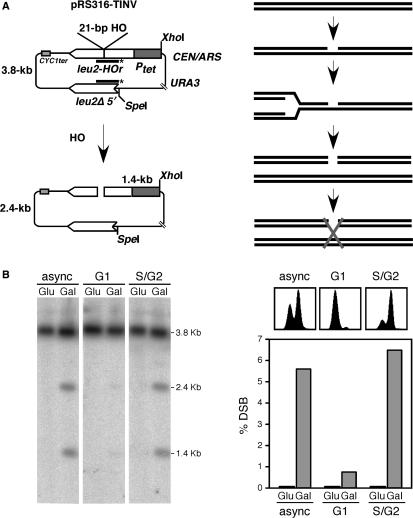
For the molecular detection of SCR we used the circular minichromosome pRS316-TINV (Figure 1A) containing two 1.2-kb leu2 inverted repeats, one of which carried a 21-bp HO site, in yeast strains containing the HO endonuclease gene under the control of the GAL10 promoter. In this plasmid, cleavage at the HO site can be detected by the appearance of 2.4 and 1.4-kb fragments after SpeI-XhoI digestion. In a parallel study we provided evidence that at this mini HO-site, the HO endonuclease produces primarily ssDNA nicks that are converted into DSBs during replication (14). This permits cleavage in only one chromatid, leaving the other intact and competent for repair via SCR. As can be seen in Figure 1B, DSB-containing molecules accumulated at levels above 5% after 2 h of HO induction in cycling cells, whereas such molecules remained below 1% in G1-arrested cells. Nevertheless, when most cells have completed replication after G1-arrest release, DSBs reached levels similar to those observed in asynchronous cultures (Figure 1B, S/G2).
Figure 1.
Analysis of cleavage of a 21-bp HO site located in the circular plasmid pRS316-TINV. (A) Scheme of plasmid pRS316-TINV (left) and the proposed mechanism by which a 21-bp HO-generated nick is converted into a DSB after replication (right). The line marked with an asterisk indicates the 0.7-kb ClaI-EcoRV LEU2 probe used for hybridization experiments. (B) Southern analysis of HO cleavage in asynchronous cultures (async) in cells arrested in G1 (G1) and after α-factor release (S/G2) in the WS-bar1 wild-type control strain. DNA samples were taken 2 h after HO induction (Gal) or from non-induced controls (Glu), and cut with XhoI and SpeI before electrophoresis. FACS pattern of cultures (right top) and quantification data of DSBs (right bottom) are shown.
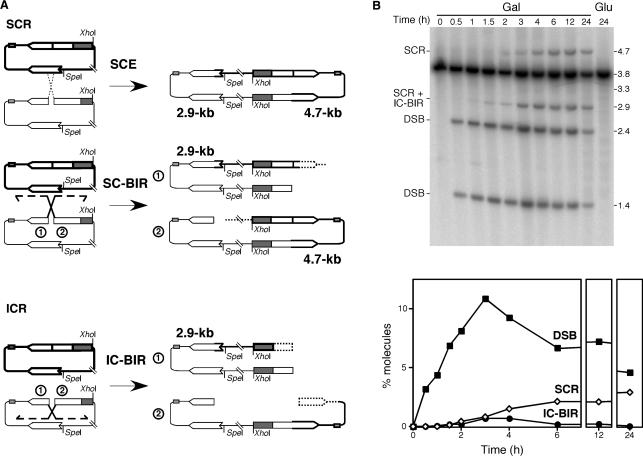
The pRS316-TINV circular minichromosome containing inverted repeats permits to study SCR (8), as well as intrachromatid repeat recombination (ICR). We have previously shown that equal SCR events can be analyzed by the appearance of dimers that are resolved by 2D-gel electrophoresis (8), and that unequal events can be easily scored by the appearance of new restriction fragments as a valid estimate of SCR (8,14). Here we used this latter approach as a measurement of SCR. As can be seen in Figure 2A, repair of the replication-born DSB at the mini-HO site in the pRS316-TINV plasmid, occurring by unequal SCR, either by reciprocal exchange (SCE) or by BIR, leads to 2.9 and 4.7-kb SpeI-XhoI fragments, whereas repair by ICR occurring by BIR only leads to the 2.9-kb band and two type of fragments that can be as large as 1.4 and 3.3-kb if DNA synthesis reaches the end of the invaded template.
Figure 2.
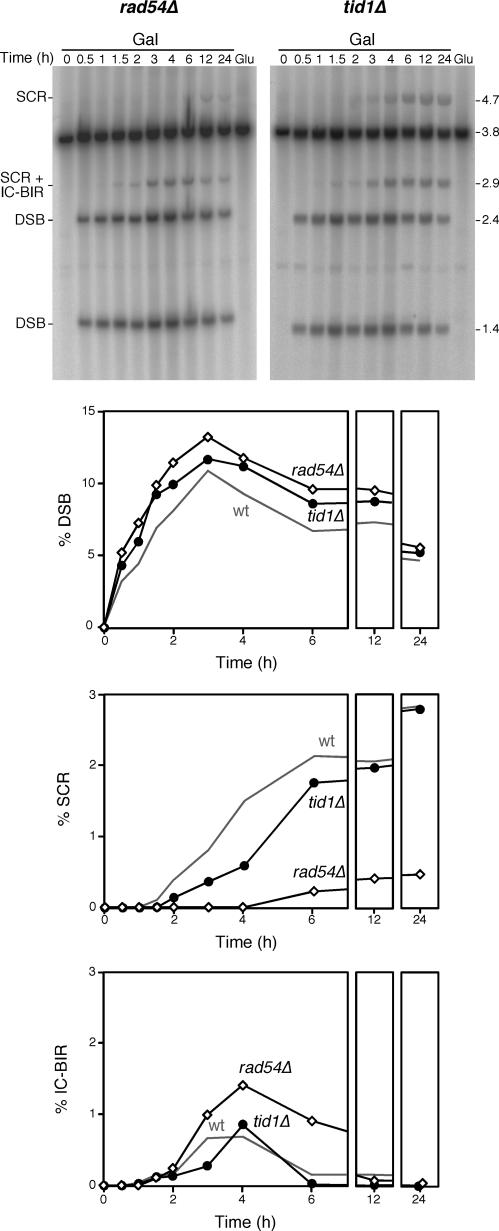
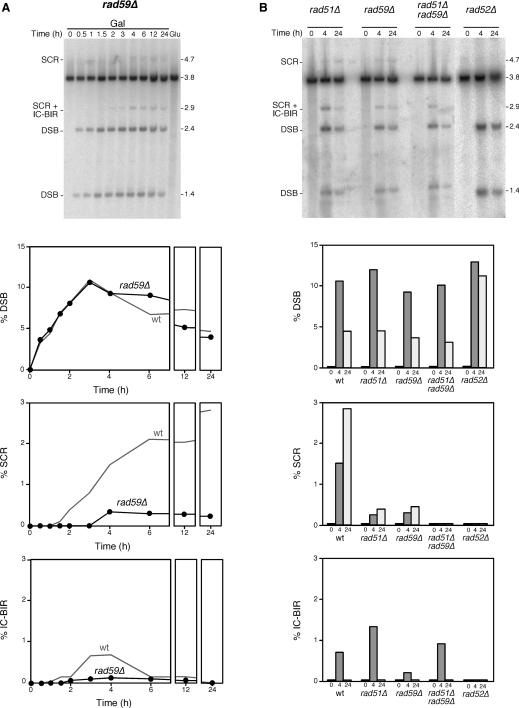
Physical analysis of replication-born HO-induced DSB repair. (A) Scheme of the two distinguishable recombination events initiated by a replication-born HO-induced DSB in plasmid pRS316-TINV carrying two inverted repeats. Fragments generated by HO cleavage and XhoI SpeI digestion, as detected by the LEU2 probe (shown in Figure 1), are indicated with their corresponding size. Sister chromatid recombination (SCR; top) either occurring by reciprocal exchange (SCE) or by BIR (SC-BIR) leads to the formation 2.9 and 4.7-kb fragments. Intra-chromatid recombination (ICR) events occurring by BIR (IC-BIR; bottom) give rise to the 2.9-kb band as well. Note that BIR events (SC-BIR and IC-BIR) are shown as the result of one-ended invasion events for each DSB end (indicated as 1 or 2 for clarification). In addition BIR can result in putative fragments of undefined length (dashed lines) depending on the extent of DNA synthesis. In the case of IC-BIR, since the template is circular, DNA synthesis could be unlimited (rolling circle-like replication) but it would result in 3.8-kb fragments undistinguishable from the parental plasmid. In contrast IC-BIR would be limited by the end of the invaded template, so additional bands of 1.4 kb (undetectable, as it coincides with one of the cleavage products) and 3.3 kb would appear. Intra-chromatid reciprocal exchanges and gene conversions are not detectable in this assay. (B) Kinetics of HO-induced DSB formation and its repair. Yeast DNA was isolated from cells grown on either SC-2% glucose (Glu) or after different times of growth in SC-2% galactose (Gal), double digested with XhoI and SpeI, separated by gel electrophoresis and analyzed by Southern. Quantification of DSBs (1.4 plus 2.4-kb bands), SCR (4.7-kb band) and IC-BIR (2.9 minus 4.7-kb bands) related to total plasmid DNA is shown. The 3.3-kb band expected from IC-BIR events reaching the end of the template is not detected, indicating that either this does not occur, or the intermediate is quickly processed.
Therefore, we decided to determine the efficiency of SCR versus IC-BIR in the repair of replication-born DSBs. Southern analysis of the kinetics of repair after HO-induction was used to determine the relative intensity of SCR and IC-BIR specific fragments. As mentioned earlier, the 2.4 and 1.4-kb bands indicate DSBs (Figure 1A), the 4.7-kb band is a specific indicator of SCR events, and IC-BIR could be estimated by the difference between the 2.9 and 4.7-kb bands. It is worth noting that a difference in intensity between these two bands cannot be due to different hybridization efficiency of both fragments, as the probe used hybridizes equally with the two leu2 alleles, regardless of the presence of the HO site (data not shown). Following this procedure, we found that the maximum peak of broken molecules was obtained after 3 h of HO induction. After this time, DSBs started to decrease (Figure 2B) as a consequence of both effective repair and DNA end processing, as once the 5′ end of DSBs are resected the DNA molecules change their electrophoretic mobility. SCR and IC-BIR products were detectable after 1 h, the levels of SCR being continuously increasing until reaching a plateau around 2%. Instead, IC-BIR peaked at 3 h to levels of ∼0.5% to later decrease to undetectable levels. These results are in agreement with previous observations indicating that SCR is the major DSB recombinational repair pathway detected in these constructs (8).
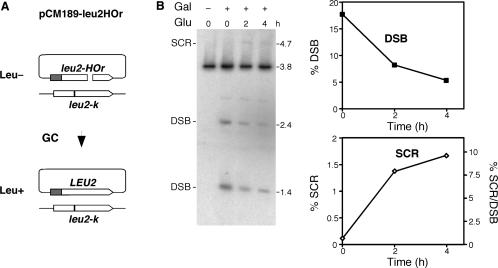
In this construct, as well as in most available recombination systems, only a portion of recombination events can be scored. Thus, gene conversions occurring either with the sister chromatid (equal or unequal) or with the intrachromatid repeat are undetectable. We asked whether the SCR events detected in our study represented a significant fraction of total HR or, on the contrary, were rare events. For this, we measured gene conversion induced at the 21-bp HO site. This can be scored genetically as Leu+ events, using the pCM189-leu2HOr plasmid (which contains the leu2HOr allele but no repeats) in a strain carrying a leu2-k allele at the endogenous chromosomal locus (25) (Figure 3A). In this case, induction of HO cleavage for 2 h resulted in a recombination frequency of 0.2%. We then used pRS316-TINV to measure SCR in these same conditions; these are 2 h of HO induction followed by the addition of glucose in order to stop further cleavage. As can be seen in Figure 3B, SCR reaches levels of 1.6% after 4 h of cleavage repression, which is 8-fold higher than the value observed for gene conversion measured with the pCM189leu2HOr/leu2-k system (Figure 3A). In addition, relating SCR to initial cleavage, we can estimate that above 9% of the DSBs are repaired by unequal SCR within 4 h (Figure 3B). These results are in agreement with SCR being a major recombinational repair pathway.
Figure 3.
Relevance of SCR in the repair of replication-born DSBs. (A) Scheme of pCM189-leu2HOr/leu2-k recombination system used to measure gene conversion (GC) induced at the 21-bp HO site. A DSB induced at the HO site of pCM189-leu2HOr can be repaired by GC using the leu2-k chromosomal allele rendering Leu+ colonies that can be scored genetically. (B) Kinetics of DSB disappearance and SCR-product formation in the TINV system after HO expression is repressed. Cleavage was induced by 2 h growth in SC-2% galactose (Gal+), and then repressed by growth in SC-2% glucose (Glu). Samples for Southern analyses were taken 0, 2 and 4 h after HO repression. A non-induced sample was also included as a negative control (Gal−). Quantification of DSBs, SCR products and SCR relative to initial DSBs is shown. Other details as in Figure 2.
The ability of our assay to study repair of replication-born DSBs by HR with either the sister chromatid or an ectopic repeat, permits establishing the importance of partner choice (sister versus ectopic sequence), and the specific genetic requirements for each type of HR event (14). It is important to notice, nevertheless, that in these molecular assays, both SCR and IC-BIR could require additional recombination events to produce a viable recombination product. For SCR, an additional intramolecular reciprocal exchange or a SSA event would be required to resolve the dicentric dimer generated by SCR, whereas in IC-BIR an SSA event would be needed for healing the two free DNA ends generated by DNA synthesis (24,26). However, SCR products, even though leading to dicentric plasmids, remain stable under conditions of recurrent cleavage, while IC-BIR fragments disappear presumably due to DNA resection occurring during SSA (Figure 2B).
Repair of replication-born DSBs in the absence of Rad51- and Rad54-mediated strand exchange
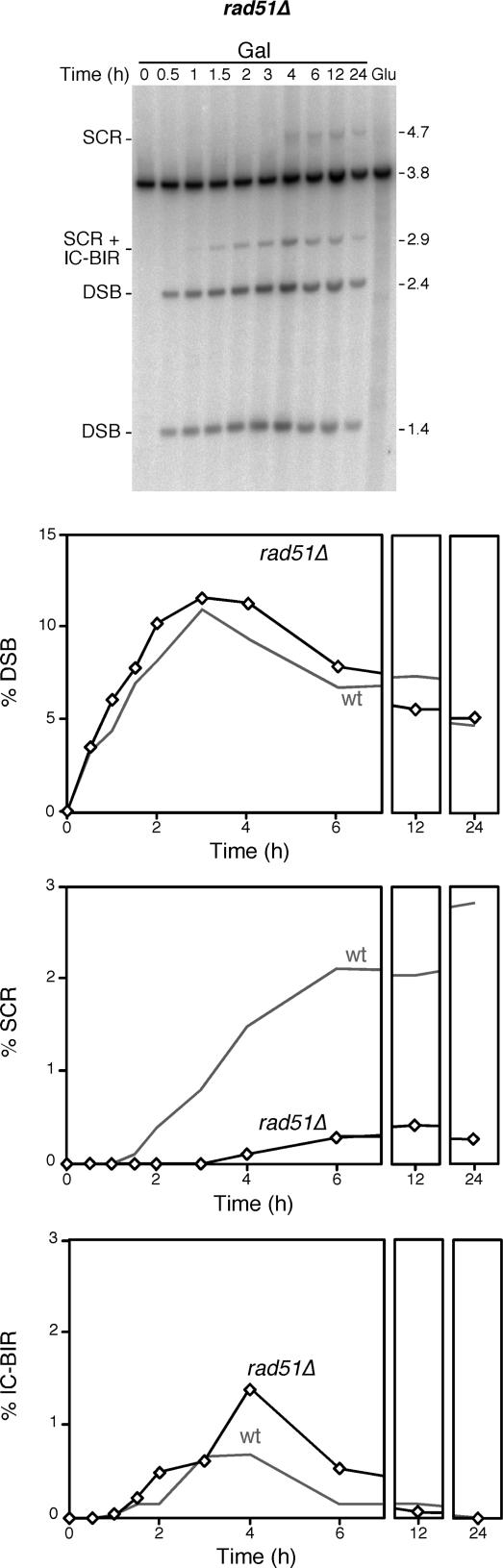
Aimed at establishing the genetic basis of DSB-repair events specifically initiated during replication, we first determined the role of the strand-exchange protein Rad51 on SCR in comparison with IC-BIR. As can be seen in Figure 4, the accumulation of DSBs after HO induction by galactose was pretty similar in rad51Δ versus wild-type cells. As we have previously shown, recombination with the sister chromatid was dramatically decreased in rad51Δ, consistent with the idea that it occurs by a strand-exchange-dependent mechanism, as it is SCE. Instead, IC-BIR products accumulate in rad51Δ at frequencies that double those of the wild type, consistent with the observation that BIR events can occur in the absence of Rad51-mediated strand-exchange (19,27,28). This result could be explained in two ways, either Rad51 is required for the preference for the sister chromatid in DSB repair, SCR events being channeled to ICR in rad51Δ mutants, or ICR events become evident in rad51Δ, as a higher proportion of them occur by BIR. Keeping in mind that in other two-ended DSB-induced recombination systems, BIR events can only be detected in rad51Δ background (19), we favor the latter hypothesis. In this sense, it is worth noting that initial intermediates of either BIR (IC-BIR as detected in this study) or the recombination events occurring by the standard DSB-repair model are not different, the diversification into each pathway presumably occurring later if BIR extension is impeded by the barrier formed by Rad51-dependent invasion of the other DSB end (29).
Figure 4.
Role of Rad51 in the repair of replication-born DSBs via SCR and IC-BIR. Kinetics and quantification of HO induction and DSB repair in a rad51Δ strain. Wild-type control data are taken from Figure 2. Other details as in Figure 2.
Two DNA-dependent ATPases of the SNF-SWI family of proteins are involved in DSB repair in S. cerevisiae, Rad54 and its paralog Tid1/Rdh54. As expected from the known activity of Rad54 in Rad51-dependent strand exchange (30,31), identical results to rad51Δ were found for DSB accumulation, SCR and IC-BIR events in rad54Δ cells (Figure 5). tid1Δ/rdh54Δ mutants showed a slightly higher accumulation of DSBs than wild-type cells, a modest decrease in SCR and only a delay in IC-BIR events (Figure 5). These results are in agreement with a role of Tid1 in HR different to that of its paralog Rad54, and indicate that Tid1/Rdh54 does not play an essential role by itself in SCR or IC-BIR. Furthermore, in contrast to rad54Δ and rad51Δ, tid1Δ does not cause better efficiency of IC-BIR events, consistent with the idea that Tid1 does not participate in the formation or stabilization of Rad51 nucleoprotein filaments that could interfere with BIR events (29).
Figure 5.
Role of Rad54 and Tid1/Rdh54 paralogs in DSB repair by SCR and IC-BIR. Kinetics and quantification of HO induction and DSB repair in rad54Δ and tid1Δ. Wild-type control data are taken from Figure 2. Other details as in Figure 2.
Role of Rad59 in SCR and IC-BIR induced by a site-specific replication-born DSB
Rad59, which is homologous to the N-terminus of Rad52, was identified in a screening for mutants with reduced recombination between inverted repeats in a rad51Δ background (32). The effect of rad59 null mutations per se on HR is very subtle, especially when compared to rad52Δ cells, in inverted-repeat and ectopic recombination systems (4,25,26,32,33). Similarly, a reduction in BIR in rad59Δ is only observed when combined with rad51Δ (28). In addition, allelic recombination is unaffected or even increased in the absence of Rad59 (28,32), in contrast to the strong reduction observed in rad51Δ. However, deletion-recombination between direct repeats, which is independent of Rad51, depends on Rad59 (33–35), especially if the repeats are short (36). These data led to the idea of Rad59 acting specifically in Rad51-independent recombination. In particular, rad59Δ cells show no defect in DSB-induced SCR and BIR when Rad51 is present (20,28,37). We decided to test if Rad59 could play a role in the repair of replication-born DSBs by SCR in comparison with IC-BIR, even in the presence of Rad51. As can be seen in Figure 6A, rad59Δ had little impact on the overall accumulation of DSBs, but it caused a strong reduction in both SCR and IC-BIR. We previously reported a weaker defect of rad59Δ on SCR in a system in which the wild-type levels of SCR were lower (around 1%) (8), which limited our ability to detect the strong defect observed in this study. Our study, therefore, provides molecular evidence for an implication of Rad59 in Rad51-dependent DSB recombinational repair. Analysis of rad51Δ rad59Δ double mutants indicated that, whereas DSBs accumulated at similar levels as in rad59Δ cells, SCR was completely abolished (Figure 6B). Instead, rad51Δ was epistatic to rad59Δ for IC-BIR, the IC-BIR level at 4h of HO induction being only slightly lower in rad51Δ rad59Δ as compared to rad51Δ cells. In contrast, rad52Δ completely abolished all HR events (Figure 6B). This unexpected result indicates that, in the absence of Rad51, the contribution of Rad59 is minor for IC-BIR but essential for SCR.
Figure 6.
Role of Rad59 in the repair of replication-born DSBs via SCR and IC-BIR. (A) Kinetics and quantification of HO induction and DSB repair in a rad59Δ strain. (B) Physical analysis of DSB repair by SCR and IC-BIR in rad52Δ and in the rad51Δ rad59Δ double mutant. Only three time points after HO induction were analyzed: 0 (control), 4 (maximum for IC-BIR) and 24 (maximum for SCR) h. Wild-type control data are taken from Figure 2. Other details as in Figure 2.
DISCUSSION
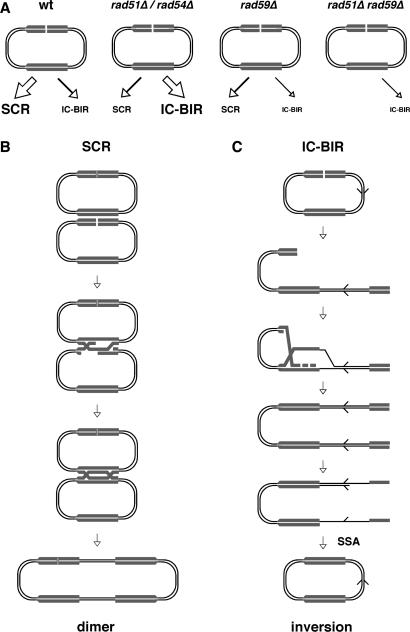
This study provides a molecular characterization of the genetic requirements of repair by SCR, in competition with IC-BIR, of DSBs specifically arising during replication (Figure 7). This is possible because of two features of the plasmid used in this study (pRS316-TINV): a 21-bp HO site, which allows specific formation of DSBs during replication in one of the sister chromatids (14); and the presence of inverted repeats, which allows detection of both SCR and IC-BIR events. The study confirms that SCR induced by replication-born DSBs is dependent on Rad51 and Rad54, which is in agreement with the idea that SCR occurs by the standard strand-exchange-dependent DSB-repair (DSBR) mechanism (SCE). Instead, observation of IC-BIR in the absence of Rad51 is consistent with its ability to occur also by a strand-exchange-independent mechanism. The rad59Δ mutant is affected in both SCR and IC-BIR, suggesting that Rad59 functions in the repair of replication-born DSBs by HR even in the presence of Rad51. In contrast, the tid1Δ mutant shows no detectable SCR or IC-BIR defects.
Figure 7.
Repair by SCR and IC-BIR in pRS316-TINV. (A) Levels of SCR and IC-BIR in different genetic backgrounds. (B) SCR occurs by the standard DSB-repair mechanism resulting in the formation of a dimer. (C) IC-BIR followed by SSA can result in an inversion. Invasion of only one side of the DSB is shown for simplification.
SCR is reduced and IC-BIR stimulated in the absence of Rad51 or Rad54
Although genetic detection of SCR is not trivial, several systems have been developed for its study, based on the analysis of recombination between heteroallelic direct repeats (10,13,38,39). Some of these direct repeat-based assays permit the study of DSB-induced events by introducing an endonuclease-recognition site within the repeats but do not measure SCR at a molecular level (9,12). Here, we take advantage of the fact that the pRS316-TINV system permits induction and physical detection of SCR (8). Since both chromatids are identical, our system, as well as any other developed so far, can only measure events involving a reciprocal exchange (Figure 2, SCE) or extensive DNA synthesis (Figure 2, SC-BIR). Although we only analyze unequal SCR events, we have shown that it is a valid estimate of total SCR in these assays (8) (data not shown).
SCR is strongly dependent on Rad51 (Figure 7A), in agreement with previous reports (8,12). This suggests that it occurs by crossover, this is SCE, rather than by BIR (Figure 7B), as crossover requires the strand-exchange activity of Rad51 while BIR can also occur in the absence of Rad51 (19,26). In contrast, IC-BIR kinetics was stimulated in the rad51Δ mutant (Figure 7A). This could be explained if BIR events are favored in rad51Δ cells, regardless of whether originally detected genetic BIR events (19) could occur via a complex mechanism also involving SSA between Ty elements (40). In this sense, it is worth noting that the fact that in two-ended recombination systems BIR is stimulated in the absence of Rad51 is compatible with the observation that BIR depends on Rad51 when only one end of the DSB is present (20,41). As previously proposed, in two-ended recombination events, the Rad51/DNA nucleoprotein filament resulting from invasion of the second DSB end would be a barrier that impedes progression of the DNA synthesis required for BIR (29).
The distinction between Rad51-dependent and -independent events is evident in the case of recombination-mediated telomere maintenance (42). In the absence of telomerase two types of survivors, regarding telomere structure, appear, both of which require Rad52 and, therefore, HR. However, while one type is Rad51 dependent, the other depends on Rad59 but not on Rad51. Since a shortened telomere is analogous to a one-ended DSB, both types of survivors are likely to arise by BIR, the different structure reflecting differences between Rad51-dependent and -independent BIR.
Invasion on the sister chromatid seems to be dependent on Rad51, since SCR is severely reduced in rad51Δ strains, regardless of whether it could also occur by BIR. In this sense, it has been proposed that Rad51 could be more necessary for inter- than for intra-molecular recombination (7,16). In our inverted repeat assay, a chromatid may be relaxed locally by a DSB in one repeat, facilitating strand invasion on the other repeat even in the absence of Rad51, whereas invasion on the intact sister chromatid allele would still be highly Rad51-dependent.
As expected, our study reveals that Rad54 is required for SCR and not for IC-BIR, in agreement with its role in Rad51-dependent recombination (28,30,31,37). Instead, Tid1 plays no evident role in either SCR or IC-BIR. It has been previously suggested that Tid1 acts specifically in allelic recombination occurring between homologous chromosomes (17,18). Moreover, in meiotic return-to-growth experiments, Rad54 has been shown to be required for SCR, whereas this is not the case for Tid1 (43).
Molecular evidence that inversions can occur by BIR-SSA
The fact that in contrast to allelic recombination, inversions between inverted repeats are not significantly reduced by rad51Δ (44), suggested that they are not necessarily produced via reciprocal exchange in rad51Δ cells, but presumably by a double event consisting of BIR followed by SSA (24) (Figure 7C), a model supported by different genetic data (4,26,33,45,46), but yet not demonstrated. While mitotic recombination is rarely associated with crossover, this model predicts that half of the products are resolved giving rise to the inversion, which is consistent with the increase in the proportion of inversions observed in rad51Δ cells (4,32,44,45). The efficient kinetics of IC-BIR in rad51Δ versus wild-type cells observed in this study provides molecular evidence for inversions between repeats occurring by BIR followed by SSA in rad51Δ cells. This represents, indeed, a direct physical evidence for BIR, which has been previously observed only by genetic or PCR analysis (19,20,41). Interestingly, the fact that IC-BIR is also observed to some extent in wild-type suggests that it may also occur when Rad51 is present.
Post-replicative repair of DSBs by SCR and IC-BIR is mediated by Rad59
In contrast to the previous reports (20,28,37), we find that both SCR and IC-BIR are strongly and equally affected by the rad59Δ mutation in a RAD51 background (Figure 7A). This indicates that Rad59 participates in all HR events scored with the pRS316-TINV system regardless of the presence of Rad51. Keeping in mind the homology of Rad59 with Rad52 and the fact that both proteins have been shown to interact and to share several biochemical properties, it is likely that Rad59 functions in the initial annealing step, in concert with Rad52 (34,35,47,48). Therefore, although genetically scored inverted-repeat recombination is not strongly affected in the absence of Rad59 (4,25,26,32), this molecular kinetic analysis uncovers a prominent role of Rad59 in the repair of DSBs induced during replication.
Interestingly, in rad51Δ cells, the rad59Δ mutation, while completely abolishing SCR, reduces the kinetics of IC-BIR only slightly. This effect of rad59Δ is much smaller than that previously observed for BIR in the absence of Rad51 (28). However in those studies, as in ours, BIR in rad51Δ rad59Δ still remains above wild-type levels. Other genetic studies showed either a synergistic defect of rad51Δ and rad59Δ or an epistatic effect of rad59Δ over rad51Δ for inverted repeat recombination (25,26,32). The difference between our physical studies and previously reported genetic studies could rely on the possibility that, in the absence of Rad51, Rad59 could be required for the SSA step following BIR (33,36), rather than to a defect in the BIR reaction itself. Therefore, our data are consistent with previous observations of HR not being completely abolished in rad51Δ rad59Δ, as compared with rad52Δ (26,28,32,33), and suggest that the residual HR in rad51 rad59 cells occurs by BIR.
CONCLUDING REMARKS
In summary, we present a molecular analysis of the role carried out by the main HR proteins in the repair of DSBs generated during replication either by SCR or by IC-BIR. We provide molecular evidence demonstrating that, in the absence of strand exchange, recombination between inverted repeats can still occur by a BIR mechanism that, if followed by SSA, could result in the previously genetically detected inversions. In addition, our study reveals that Rad59 plays an important role in SCR and BIR, even in the presence of Rad51, which suggests that the functional relevance of recombinational repair proteins may vary depending on the nature of the DSBs, opening new perspectives for a more complete understanding of the role of HR in post-replicative repair.
ACKNOWLEDGEMENTS
We thank F. Prado and R. Wellinger for reading the manuscript, D. Haun for style supervision and J. Escalante for technical support. This work was supported by grants from the Ministry of Science of Education of Spain (SAF2003-00204 and BFU2006-05260) and Junta de Andalucía (CVI102 and CVI624). F.C.-L. was the recipient of a pre-doctoral training grant from the Spanish Ministry of Health. Funding to pay the Open Access publication charges for this article was provided by the Spanish Ministry of Science and Education (grant BFU2006-05260).
Conflict of interest statement. None declared.
REFERENCES
- 1.Resnick MA, Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol. Gen. Genet. 1976;143:119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- 2.Lisby M, Rothstein R, Mortensen UH. Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl Acad. Sci. USA. 2001;98:8276–8282. doi: 10.1073/pnas.121006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aylon Y, Kupiec M. Cell cycle-dependent regulation of double-strand break repair: a role for the CDK. Cell Cycle. 2005;4:259–261. [PubMed] [Google Scholar]
- 4.Ira G, Haber JE. Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 2002;22:6384–6392. doi: 10.1128/MCB.22.18.6384-6392.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 6.Michel B, Flores MJ, Viguera E, Grompone G, Seigneur M, Bidnenko V. Rescue of arrested replication forks by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8181–8188. doi: 10.1073/pnas.111008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gonzalez-Barrera S, Cortes-Ledesma F, Wellinger RE, Aguilera A. Equal sister chromatid exchange is a major mechanism of double-strand break repair in yeast. Mol. Cell. 2003;11:1661–1671. doi: 10.1016/s1097-2765(03)00183-7. [DOI] [PubMed] [Google Scholar]
- 9.Johnson RD, Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sonoda E, Sasaki MS, Morrison C, Yamaguchi-Iwai Y, Takata M, Takeda S. Sister chromatid exchanges are mediated by homologous recombination in vertebrate cells. Mol. Cell. Biol. 1999;19:5166–5169. doi: 10.1128/mcb.19.7.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fasullo M, Giallanza P, Dong Z, Cera C, Bennett T. Saccharomyces cerevisiae rad51 mutants are defective in DNA damage-associated sister chromatid exchanges but exhibit increased rates of homology-directed translocations. Genetics. 2001;158:959–972. doi: 10.1093/genetics/158.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fasullo MT, Davis RW. Recombinational substrates designed to study recombination between unique and repetitive sequences in vivo. Proc. Natl Acad. Sci. USA. 1987;84:6215–6219. doi: 10.1073/pnas.84.17.6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortes-Ledesma F, Aguilera A. Double-strand breaks arising by replication through a nick are repaired by cohesin-dependent sister-chromatid exchange. EMBO Rep. 2006;7:919–926. doi: 10.1038/sj.embor.7400774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sung P, Trujillo KM, Van Komen S. Recombination factors of Saccharomyces cerevisiae. Mutat. Res. 2000;451:257–275. doi: 10.1016/s0027-5107(00)00054-3. [DOI] [PubMed] [Google Scholar]
- 16.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein HL. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shinohara M, Shita-Yamaguchi E, Buerstedde JM, Shinagawa H, Ogawa H, Shinohara A. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malkova A, Ivanov EL, Haber JE. Double-strand break repair in the absence of RAD51 in yeast: a possible role for break-induced DNA replication. Proc. Natl Acad. Sci. USA. 1996;93:7131–7136. doi: 10.1073/pnas.93.14.7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis AP, Symington LS. RAD51-dependent break-induced replication in yeast. Mol. Cell. Biol. 2004;24:2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fishman-Lobell J, Rudin N, Haber JE. Two alternative pathways of double-strand break repair that are kinetically separable and independently modulated. Mol. Cell. Biol. 1992;12:1292–1303. doi: 10.1128/mcb.12.3.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin FL, Sperle K, Sternberg N. Intermolecular recombination between DNAs introduced into mouse L cells is mediated by a nonconservative pathway that leads to crossover products. Mol. Cell. Biol. 1990;10:103–112. doi: 10.1128/mcb.10.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartsch S, Kang LE, Symington LS. RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol. Cell. Biol. 2000;20:1194–1205. doi: 10.1128/mcb.20.4.1194-1205.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gonzalez-Barrera S, Garcia-Rubio M, Aguilera A. Transcription and double-strand breaks induce similar mitotic recombination events in Saccharomyces cerevisiae. Genetics. 2002;162:603–614. doi: 10.1093/genetics/162.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malagon F, Aguilera A. Yeast spt6-140 mutation, affecting chromatin and transcription, preferentially increases recombination in which Rad51p-mediated strand exchange is dispensable. Genetics. 2001;158:597–611. doi: 10.1093/genetics/158.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malkova A, Signon L, Schaefer CB, Naylor ML, Theis JF, Newlon CS, Haber JE. RAD51-independent break-induced replication to repair a broken chromosome depends on a distant enhancer site. Genes Dev. 2001;15:1055–1060. doi: 10.1101/gad.875901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Signon L, Malkova A, Naylor ML, Klein H, Haber JE. Genetic requirements for RAD51- and RAD54-independent break-induced replication repair of a chromosomal double-strand break. Mol. Cell. Biol. 2001;21:2048–2056. doi: 10.1128/MCB.21.6.2048-2056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aguilera A. Double-strand break repair: are Rad51/RecA–DNA joints barriers to DNA replication? Trends Genet. 2001;17:318–321. doi: 10.1016/s0168-9525(01)02309-5. [DOI] [PubMed] [Google Scholar]
- 30.Petukhova G, Stratton S, Sung P. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature. 1998;393:91–94. doi: 10.1038/30037. [DOI] [PubMed] [Google Scholar]
- 31.Rattray AJ, Symington LS. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bai Y, Symington LS. A Rad52 homolog is required for RAD51-independent mitotic recombination in Saccharomyces cerevisiae. Genes Dev. 1996;10:2025–2037. doi: 10.1101/gad.10.16.2025. [DOI] [PubMed] [Google Scholar]
- 33.Jablonovich Z, Liefshitz B, Steinlauf R, Kupiec M. Characterization of the role played by the RAD59 gene of Saccharomyces cerevisiae in ectopic recombination. Curr. Genet. 1999;36:13–20. doi: 10.1007/s002940050467. [DOI] [PubMed] [Google Scholar]
- 34.Bai Y, Davis AP, Symington LS. A novel allele of RAD52 that causes severe DNA repair and recombination deficiencies only in the absence of RAD51 or RAD59. Genetics. 1999;153:1117–1130. doi: 10.1093/genetics/153.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Davis AP, Symington LS. The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics. 2001;159:515–525. doi: 10.1093/genetics/159.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugawara N, Ira G, Haber JE. DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dong Z, Fasullo M. Multiple recombination pathways for sister chromatid exchange in Saccharomyces cerevisiae: role of RAD1 and the RAD52 epistasis group genes. Nucleic Acids Res. 2003;31:2576–2585. doi: 10.1093/nar/gkg352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackson JA, Fink GR. Gene conversion between duplicated genetic elements in yeast. Nature. 1981;292:306–311. doi: 10.1038/292306a0. [DOI] [PubMed] [Google Scholar]
- 39.Szostak JW, Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980;284:426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- 40.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu. Rev. Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 41.Malkova A, Naylor ML, Yamaguchi M, Ira G, Haber JE. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol. Cell. Biol. 2005;25:933–944. doi: 10.1128/MCB.25.3.933-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.VanHulle K, Lemoine FJ, Narayanan V, Downing B, Hull K, McCullough C, Bellinger M, Lobachev K, Petes TD, et al. Inverted DNA repeats channel repair of distant double-strand breaks into chromatid fusions and chromosomal rearrangements. Mol. Cell. Biol. 2007;27:2601–2614. doi: 10.1128/MCB.01740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arbel A, Zenvirth D, Simchen G. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1. EMBO J. 1999;18:2648–2658. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rattray AJ, Symington LS. Use of a chromosomal inverted repeat to demonstrate that the RAD51 and RAD52 genes of Saccharomyces cerevisiae have different roles in mitotic recombination. Genetics. 1994;138:587–595. doi: 10.1093/genetics/138.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang LE, Symington LS. Aberrant double-strand break repair in rad51 mutants of Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:9162–9172. doi: 10.1128/mcb.20.24.9162-9172.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rattray AJ, McGill CB, Shafer BK, Strathern JN. Fidelity of mitotic double-strand-break repair in Saccharomyces cerevisiae: a role for SAE2/COM1. Genetics. 2001;158:109–122. doi: 10.1093/genetics/158.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cortes-Ledesma F, Malagon F, Aguilera A. A novel yeast mutation, rad52-L89F, causes a specific defect in Rad51-independent recombination that correlates with a reduced ability of Rad52-L89F to interact with Rad59. Genetics. 2004;168:553–557. doi: 10.1534/genetics.104.030551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petukhova G, Stratton SA, Sung P. Single strand DNA binding and annealing activities in the yeast recombination factor Rad59. J. Biol. Chem. 1999;274:33839–33842. doi: 10.1074/jbc.274.48.33839. [DOI] [PubMed] [Google Scholar]