Abstract
The expression of ribosomal protein (rp) genes is regulated at multiple levels. In yeast, two genes are autoregulated by feedback effects of the protein on pre-mRNA splicing. Here, we have investigated whether similar mechanisms occur in eukaryotes with more complicated and highly regulated splicing patterns. Comparisons of the sequences of ribosomal protein S13 gene (RPS13) among mammals and birds revealed that intron 1 is more conserved than the other introns. Transfection of HEK 293 cells with a minigene-expressing ribosomal protein S13 showed that the presence of intron 1 reduced expression by a factor of four. Ribosomal protein S13 was found to inhibit excision of intron 1 from rpS13 pre-mRNA fragment in vitro. This protein was shown to be able to specifically bind the fragment and to confer protection against ribonuclease cleavage at sequences near the 5′ and 3′ splice sites. The results suggest that overproduction of rpS13 in mammalian cells interferes with splicing of its own pre-mRNA by a feedback mechanism.
INTRODUCTION
Apart from their functions in ribosome assembly (1) and mRNA translation (2,3), many ribosomal proteins are involved in a variety of extraribosomal processes (4). There are 79 different mammalian ribosomal proteins, 34 of which have bacterial counterparts (5). Human ribosomal protein genes are distributed widely through the genome and, except for ribosomal protein (rp) S4, each protein is encoded by a single gene (5). The level of expression of these genes depends on various factors such as the cell type, the phase of the cell cycle, ambient conditions, etc. (6,7). Fine regulation of ribosomal protein biosynthesis is necessary for the vitality of individual cells and whole organisms, and impairments of this regulation may lead to critical consequences such as malignant transformation (8,9), apoptosis (10,11), developmental malformations (12,13) and so on.
It is known that the expression of ribosomal protein genes in eukaryotes can be regulated at the levels of transcription (5,14), translation (15) and splicing (16,17). In addition, the content of ribosomal proteins in eukaryotic cells is controlled via changes in the degradation rate of newly synthesized proteins (18). The transcriptional and translational levels of the regulation are coordinated by common regulatory elements in the genes (5) and mRNA (19) of ribosomal proteins. The regulation of ribosomal protein biosynthesis at the splicing level is less well documented. In yeast, at least two genes are regulated via a feedback mechanism in which ribosomal proteins bind their own pre-mRNAs, preventing their splicing (16,17,20). As for humans, indications for the regulation of expression via splicing were recently obtained for two rp genes. So, we demonstrated that rpS26 suppresses splicing of its own pre-mRNA in vitro (21). Binding of this protein to its cognate pre-mRNA may prevent the recognition of the splicing signals by components of the splicing machinery and thus interfere with spliceosome assembly. However, whether rpS26 biosynthesis is regulated this way in vivo is still unknown. A similar pathway may be realized in the regulation of human rpL3 biosynthesis (22). It was shown that rpL3 overexpression caused an increase in its aberrantly spliced mRNA that contained intron 3 and was degraded by the nonsense-mediated decay (NMD) pathway (23). A similar aberrantly spliced isoform of mRNA containing unremoved intron 1 was found also for rpL12 from Caenorhabditis elegans (24). However, a direct implication of rpL3 and rpL12 in their biosynthesis regulation was not proven.
Autoregulation appears to be a potent mechanism by which the level of each individual ribosomal protein in the cell could be controlled independently, which may be crucial for the extraribosomal functions of ribosomal proteins. To test this possibility further, we have examined human rpS13. This protein is a homologue of prokaryotic rpS15, which binds to the central domain of the 16S rRNA and promotes the binding of neighbouring proteins in the 30S ribosomal subunit (25). Its yeast counterpart is indispensable for the initial stage of pre-18S rRNA processing (1). The extraribosomal functions of human rpS13 are still unclear; nevertheless there are data suggesting that rpS13 (along with rpL23) promotes multidrug resistance in gastric cancer cells by suppressing drug-induced apoptosis (26). Remarkably, of the 17 human EST records containing the sequence corresponding to RPS13 intron 1 in GenBank, at least two (GenBank accession no CD050699 and BI756632) are derived from rpS13 mRNA containing retained intron 1. This suggested to us that the protein might be involved in autoregulation of its expression via splicing, so we have tested this possibility both in vivo and in vitro.
In the current study we have analysed the extent of conservation of intron 1 within RPS13 gene in mammals and examined role of human rpS13 in its own gene expression regulation. Our first results showed that the presence of the first intron in an rpS13 minigene reduced the level of mRNA expressed in cell culture. The possibility that there was a protein-dependent inhibition of splicing was confirmed by in vitro splicing experiments. Remarkably, the rpS13-binding sites on the pre-mRNA turned out to be located closely to the splice sites that allowed us to suggest a feedback mechanism for the RPS13 gene expression regulation at the splicing step.
MATERIALS AND METHODS
PCR and plasmid constructions
DNA sequences containing a coding part of rpS13 cDNA were PCR amplified using total HeLa cDNA and a pair of primers, S13F and S13R. The PCR product was digested with PstI and BamHI and cloned in the vector pECFP-N1 (Clontech, USA), producing pECFP-S13. The DNA fragment containing a coding part of rpS13 cDNA with inserted intron 1 was obtained in two stages. Initially, a DNA fragment containing intron 1 of RPS13 gene flanked with exon sequences was PCR amplified using genomic HeLa DNA and a pair of primers, S13F and S13Rint1. Then a second DNA fragment containing a coding part of rpS13 cDNA excluding the sequence of exon 1 was PCR amplified using total HeLa cDNA and primers S13Fint1 and S13R. PCR amplification with the use of two of these PCR products as a template and primers S13F and S13R produced the desired DNA fragment. This DNA fragment was digested with PstI and BamHI and cloned in the vector pECFP-N1 at the same sites of restriction producing pECFP-S13-int1. All recombinant plasmids were verified by sequencing. The oligonucleotides used in this work are listed in Table 1.
Table 1.
Oligodeoxynucleotides used in the study
| Name | Sequence (5′to 3′) | Description |
|---|---|---|
| S13F | GTTCTGCAGGTGGAGGTTCAATGGGTCGCATGCATGCTC | pECFP-S13 construction |
| S13R | CCAGGATCCAATGCGACCAGGGCAGAGGC | pECFP-S13 construction |
| S13Rint1 | GACTGGGACAGGCCCTTCCTGGGGAGAGAAGC | pECFP-S13-int1 construction |
| S13Fint1 | CACCTCGAGACTGCTTCTCTCCCCAGGAAGGGCCTGTCCCAGTC | pECFP-S13-int1 construction |
| preS13F | GCTAATACGACTCACTATAGGGCTTTCGTTGCCTGATCG | RPS13-specific |
| preS13R | CGCGCTACTTACAGTG | RPS13-specific |
| preS16F | TAATACGACTCACTATAGGGCCTTTTCCGGTTGCGGC | RPS16-specific |
| preS16R | GGATCCCAGCACCTTG | RPS16-specific |
| preS17F | TAATACGACTCACTATAGGGACCAAGGACCCGCCAACATGGTAGGTGTTT | RPS17-specific |
| preS17R | CCCGACTCACCCTGCTATC | RPS17-specific |
| preS26F | GGATCCTAATACGACTCACTATAGGGGTGAGTCTTCTTGCGTGGTGAG | RPS26-specific |
| preS26R | AATAGGCTGCACGTGGCCGC | RPS26-specific |
| S13Fmut | CATGCTAGAGGGTGAGCTCGGGGCATC | RPS13-specific, mutant |
| S13Rmut | CAGGCCGGGCCTGGGGAGAGAAGCAGT | RPS13-specific, mutant |
Cell culture transfection and real-time PCR
Human HEK 293 cells were routinely maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) containing 10% of fetal calf serum. For transient transfections, 4 × 105 cells (70–80% confluent) were transfected with 1 μg of plasmid DNA with the use of jetPEI transfection reagent (Polyplus-transfection, France), according to the manufacturer's instructions. After 18 h (when the level of transfection detected by blue fluorescence was ∼10–15%), total RNA was isolated using TRI reagent (Sigma, USA). Reverse transcription was performed using 2 μg of the RNA, 0.5 μg (pN)6 and M-MLV RT (Promega, USA) in according to the protocol of the enzyme manufacturer. Real-time PCR was performed on a Corbett Rotor-Gene 3000 (Corbett, Australia) using the Platinum SYBR Green qPCR SuperMix (Invitrogen, USA) according to the manual of the manufacturer. For standardization of the quantity of RNA in the samples taken for the real-time PCR assay a pair of primers specific to GAPDH cDNA was used. Dispersal of amount of GAPDH cDNA in the samples that reflects total amount of RNA was ±25% from average value. Experiments with cell transfection and real-time PCR were repeated independently for three times.
Protein-binding assay
The templates for synthesis of fragments of ribosomal proteins pre-mRNAs by T7 RNA polymerase were obtained by means of PCR using genomic HeLa DNA and pairs of primers, namely, preS13F and preS13R for rpS13 pre-mRNA fragment—S13INT, preS16F and preS16R for rpS16 pre-mRNA, preS17F and preS17R for rpS17 pre-mRNA and preS26F and preS26R for rpS26 pre-mRNA. To prepare DNA template for synthesis of mutant S13INT (S13INTm), two mutant semi-templates were initially obtained by PCR using primers preS13F and S13Rmut and S13Fmut and preS13R. These semi-templates were annealed and used in the second PCR step with primers preS13F and preS13R. All PCR products were gel-purified and sequenced. Respective RNA transcripts were synthesized in 32P-labelled (27) and non-labelled (28) variants. Binding of 32P-labelled S13INT (or the pre-mRNA fragments) with recombinant rpS10 (29), S13 (30) and S16 (31) was carried out in buffer (20 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 200 mM KCl, 2.5 mM 2-mercaptoethanol and 0.05% Triton X-100) at 0°C for 1 h. Reaction mixtures were filtered on a nitrocellulose membrane with pores diameter 0.45 μm. Binding was measured as the ratio of radioactivity trapped on the filter to the total radioactivity in the reaction mixture. All experiments on proteins binding were at least triplicated. The relative error in measurements was less than 15%.
Immunoprecipitation
Recombinant rpS13 (10 pmol) was pre-incubated in 30 μl of buffer A (50 mM Tris–HCl, pH 7.5, 300 mM NaCl, 1 mM EDTA and 0.1% Triton X-100) containing 50% HeLa nuclear extract (‘CCCC’, Belgium) at 30°C for 5 min. A mixture containing 32P-labelled RNA transcripts (S13INT and fragments of pre-mRNAs of rpS17 and rpS26, 0.5 fmol, 2000–4000 c.p.m. of each) was pre-incubated in 20 μl of the same buffer at 30°C for 10 min. Then the pre-incubated rpS13 solution and the mixture of the RNA transcripts were united and incubated at 20°C for 1 h. Rabbit antibodies against human rpS13 purified from specific antiserum by affinity chromatography on a column with immobilized recombinant rpS13 were bound to 20 μl of protein G-sepharose resin (Sigma, USA) and the resin was washed with buffer A for five times. The washed resin was added to the mixture containing rpS13 and RNA transcripts and the mixture was incubated at 0°C for 1 h under gentle agitation. Finally, the resin was washed with buffer A for five times, then bound proteins were eluted with 2% SDS, and RNA transcripts were isolated by phenol deproteination and analysed on a 6% denaturing gel.
Splicing in vitro
Splicing in vitro was performed as described in (32). Where mentioned, reaction mixtures contained appropriate amount of recombinant ribosomal proteins. The reaction samples were run on a 6% denaturing polyacrylamide gel. The fixed and dried gels were then exposed to a phosphorimager screen of Molecular Imager FX Pro equipment (Bio-Rad, USA).
Footprinting assay
Footprinting was performed according to protocol (33) with minor modifications. A complex of S13INT with rpS13 was formed by incubation of 1 pmol of RNA and 30 pmol of protein in 10 μl of buffer (20 mM HEPES–KOH, pH 7.5, 150 mM KCl, 8 mM MgCl2, 10 mM DTT and 0.4 μg/μl total Escherichia coli tRNA) at 20°C for 15 min. Then reaction mixtures were incubated at the same conditions in presence of either RNase T1 (0.5 U) or RNase T2 (0.2 U) or RNase V1 (0.05 U). RNAs from the reaction mixtures were isolated by phenol deproteinization. Nucleotides 5′ to cleaved phosphodiester bonds were determined by reverse transcription–primer extension as described in (34).
RESULTS
Intron 1 in the gene coding for rpS13 in mammals and birds is more conserved than other introns
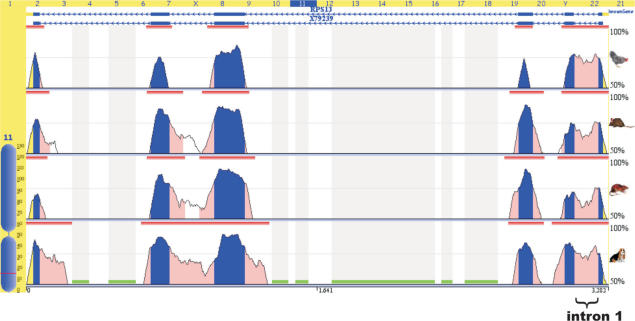
Alignment and comparative analysis of the human gene coding for rpS13 with homologous genes in the genomes of some mammals (Canis familiaris, Rattus norvegicus, Mus musculus) and birds (Gallus gallus) performed in the Evolutionary Conserved Regions (ECR) Browser (http://ecrbrowser.dcode.org) (35) revealed that intron 1 within this gene is particularly well conserved (Figure 1). This suggests that the region of the gene including intron 1 is functionally significant and may contain regulatory sequence elements. We have investigated the possibility that such regulation could take place at the level of pre-mRNA splicing.
Figure 1.
Intron 1 in RPS13 gene of mammals and birds is evolutionary conserved. Generated by the ECR browser (35) graphic plot of genomic alignment and comparative analysis with the genes coding for rpS13 in dog, mouse, rat and chicken. Smooth peaks represent levels of sequence identity. Exons coloured in blue, introns in pink. Intron 1 is marked by brace.
Intron-dependent reduction in rpS13 mRNA in HEK 293 cells
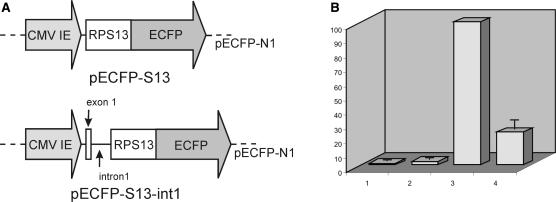
To investigate the functional importance of intron 1 in the pre-mRNA of rpS13, HEK 293 cells were transiently transfected with plasmid constructs coding for human rpS13 fused with a fluorescent reporter protein, eCFP (Figure 2A). The first construction (pECFP-S13) contained the coding part of rpS13 cDNA inserted into the vector pECFP-N1 in frame with the eCFP gene. The second construction (pECFP-S13-int1) was similar to pECFP-S13 but contained RPS13 intron 1 inserted in the coding part of rpS13 cDNA between sequences corresponding to exons 1 and 2. As a control, cells were transfected with the original vector pECFP-N1. The rpS13-eCFP fusion protein produced in the cells transfected with pECFP-S13 and pECFP-S13-int1 was nuclear and nucleolar, as expected for ribosomal proteins overexpressed in eukaryotic cells (36,37). The relative amount of the mRNA coding for rpS13 (both endogenous and plasmid derived) in the cell culture was determined by real-time PCR (Figure 2B). As expected, the levels of rpS13 mRNA in the cell cultures treated with plasmids coding for rpS13 (either pECFP-S13 or pECFP-S13-int1) were much higher than in the cell cultures treated with the original vector or in the untreated culture. It means that in the cells transfected with plasmids coding for rpS13, the pool of mRNA coding for rpS13 consists largely of the plasmid-derived mRNA. Remarkably, the level of rpS13 mRNA in cells transfected with pECFP-S13-int1 was about four times less than that in the cells transfected with the intronless construction pECFP-S13, even though the levels of GAPDH mRNA were about the same. The result is consistent with the hypothesis that rpS13 might regulate the level of protein expression via feedback on splicing and it suggests that the target is intron 1, while the result does not exclude both other intron-dependent mechanisms and possible effect of rpS13 on splicing of some other pre-mRNAs.
Figure 2.
Overexpression of rpS13 in mammalian cells suppresses the expression of mRNA from a minigene containing intron 1. (A) Diagrams of vectors pECFP-S13 and pECFP-S13-int1. (B) Relative amount of mRNA coding for rpS13 (both endogenous and plasmid-derived) in HEK 293 cells. Non-transfected cells (column 1), cells transfected with pECFP-N1 (column 2), cells transfected with pECFP-S13 (accepted as 100 arbitrary units, column 3) and cells transfected with pECFP-S13-int 1 (column 4). Experiments made in triplicate; error bars are shown. For standardization of the quantity of RNA in the samples primers specific to GAPDH cDNA were used. Dispersal of amount of GAPDH cDNA in the samples that reflects total amount of RNA was ±25% from average value.
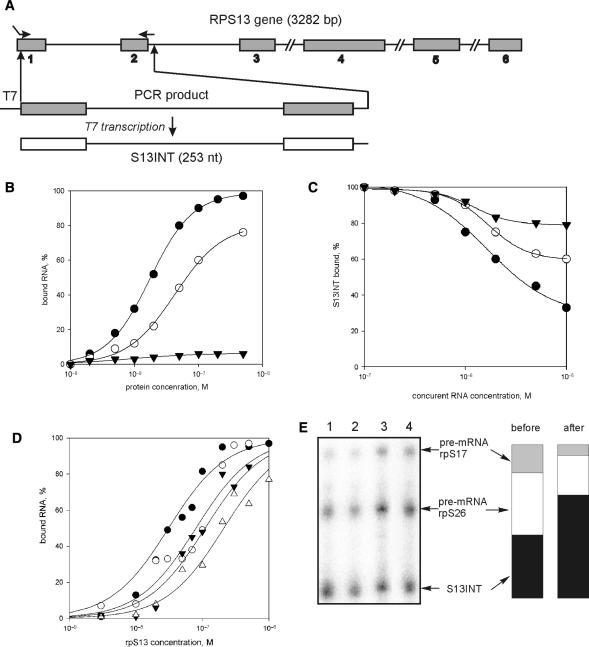
Ribosomal protein S13 binds specifically to pre-mRNA containing intron 1 and inhibits splicing of intron 1 in vitro
To test whether rpS13 could bind directly to the first intron and to study the effect of the protein on splicing in vitro, we made a transcript containing intron 1 and flanking exon sequences (transcript S13INT) (Figure 3A). The ability of recombinant ribosomal proteins S13, S10 and S16 to bind 32P-labelled RNA was followed by nitrocellulose binding (Figure 3B). To reduce the risk of non-specific binding due to electrostatic interaction between the positively charged ribosomal proteins and negatively charged RNA, protein binding was measured in 250 mM KCl. The apparent association constant for rpS13 was 5.0 × 107 M−1, 4-fold higher than the constant for rpS16; rpS10 showed little binding to S13INT. Unlabelled S13INT, used as a competitor, effectively displaced labelled S13INT from the complex with rpS13 whereas non-specific RNA competitors (a fragment of adenovirus major late (AdML) RNA or poly(AU)) were much weaker (Figure 3C).
Figure 3.
rpS13 preferentially binds to its RNA containing the first intron (S13INT). (A) Schematic diagram of human RPS13 gene showing the location of the exons (numbered shadowed boxes) and a scheme of S13INT synthesis. (B) Isotherms of binding of 32P-labelled S13INT to rpS13 (filled circles), rpS16 (open circles) and rpS10 (filled triangles). (C) Displacement of 32P-labelled S13INT from its complex with rpS13 by competition with unlabelled RNAs: S13INT (filled circles), AdML (open circles) and poly (AU) (filled triangles). (D) Isotherms of binding of various 32P-labelled pre-mRNA fragments containing intron 1 to rpS13: S13INT (filled circles), S13INTm (open circles), rpS16 pre-mRNA fragment (filled triangles) and rpS26 pre-mRNA fragment (open triangles). (E) Immunoprecipitation of rpS13 associated with 32P-labelled RNA in nuclear extract. The mixture of S13INT, rpS17 and rpS26 RNA was separated by denaturing PAGE after isolation on beads either charged with antibodies against rpS13 (lane 1), or uncharged (unspecific sorbtion, lane 2); lane 3, aliquot of the initial mixture; lane 4, aliquot of the sample containing RNAs unbound to the beads charged with antibodies. Columns illustrate the proportion of each RNA transcript in the initial mixture (‘before’) and in the mixture isolated from nuclear extract (‘after’).
The ability of rpS13 to bind to other intron-containing RNAs was examined with transcripts corresponding to portions of rpS16 and rpS26 pre-mRNAs of comparable length (Figure 3D). The affinity of rpS13 for the non-cognate pre-mRNAs was much less as compared with that for S13INT, indicating the specificity of interaction of rpS13 with S13INT. The affinity of rpS13 for a fragment of the cognate pre-mRNA was also tested in HeLa nuclear extract using immunoprecipitation. For this purpose, recombinant rpS13 was incubated in nuclear extract with 32P-labelled S13INT and transcripts derived from rpS17, and rpS26 pre-mRNAs. RpS13 was immunoprecipitated, and the transcripts thereby recovered were separated on a polyacrylamide gel. The results are presented in Figure. 3E. The proportion of S13INT was higher after immunoselection. Therefore, we conclude that rpS13 discriminates between these alternative RNA sequences in favour of its own pre-mRNA.
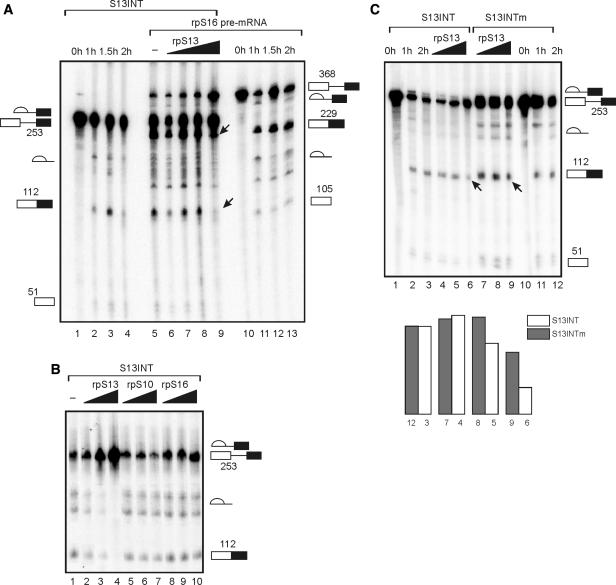
To establish whether the binding of rpS13 to S13INT affected splicing in vitro, reactions were done with two labelled substrates, S13INT and a fragment of rpS16 pre-mRNA, in the same reaction mixture. Incubation of S13INT under splicing conditions resulted in the formation of a 112 nt-long product corresponding to the spliced exons (Figure 4A). In vitro splicing of rpS16 pre-mRNA produced RNA of 229 nt (spliced exons) and splicing intermediates (Figure 4A). The addition of increasing concentrations of rpS13 to reaction mixtures containing both the pre-mRNAs caused complete inhibition of splicing of S13INT at a protein concentration of 8 μM, whereas the suppression of splicing of the non-cognate rpS16 pre-mRNA was much weaker (Figure 4A). The non-cognate ribosomal proteins rpS10 and rpS16 barely affected S13INT splicing (Figure 4B). This implies that rpS13 is able to suppress splicing of the cognate pre-mRNA in vitro by a feedback mechanism.
Figure 4.
rpS13 specifically inhibits in vitro splicing of the S13INT transcript. (A) Effect of rpS13 on splicing of S13INT and rpS16 pre-mRNA fragment. Products of splicing were separated on 6% denaturing PAAG. Time course reactions of the transcripts alone (lanes 1–4 and 10–13) and 2 h incubation under splicing conditions of the transcripts taken together in the presence of rpS13 at concentrations of 0.3 μM (lane 6), 1 μM (lane 7), 3 μM (lane 8), 8 μM (lane 9) or without the protein (lane 5). Positions of RNAs on the gel and their lengths are denoted on left (for S13INT) and on right (for rpS16 pre-mRNA). Arrows on the gel (lane 9) indicate presence of the mRNA product for rpS16 pre-mRNA splicing and its absence for S13INT. (B) Effect of rpS13, rpS16 and rpS10 on the S13INT splicing. Products of S13INT splicing after 2 h incubation under splicing conditions in the presence of rpS13 (lanes 2–4), rpS10 (lanes 5–7) and rpS16 (lanes 8–10) at concentrations of 1 μM (lanes 2, 5, 8), 5 μM (lanes 3, 6, 9), 8 μM (lanes 4, 7, 10) or without protein (lane 1), separated on 6% denaturing gel. The positions of the RNAs are indicated on right. (C) Comparison of effects of rpS13 on the in vitro splicing of S13INT transcript and of the transcript mutated in the vicinity of both the 5′ and 3′ splice sites (S13INTm). Lanes 1–3 and 10–12—time-course reactions. Splicing products after 2 h incubation of S13INT (lanes 4–6) and S13INTm (lanes 7–9) under splicing conditions at the presence of rpS13 at the concentration 1 μM (lanes 4 and 7), 3 μM (lanes 5 and 8), 6 μM (lanes 6 and 9). Arrows indicate presence of the mRNA product with S13INTm (lane 9) and its absence with S13INT (lane 6). The diagram below represents quantification of the gel data. Each column represents the ratio of the intensities of bands for the mRNA product and the pre-mRNA on the respective lane on the gel in arbitrary units. The height of column is proportional to the splicing efficiency. White columns are for S13INT, grey ones are for S13INTm. Columns are grouped in pairs for splicing reactions carried out under the same conditions.
Binding site of rpS13 on its pre-mRNA fragment locates closely to the 5′- and 3′- splice sites
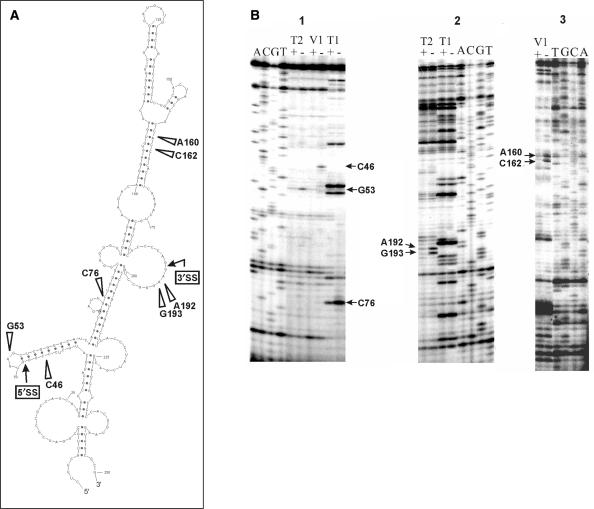
To ascertain how rpS13 binding may prevent splicing of the S13INT transcript, we attempted to establish the rpS13-binding site using an enzymatic footprinting experiment. RNA footprints detect either the direct protection of phosphodiester bonds by the bound protein or rearrangements of the RNA spatial structure caused by protein binding. A prediction of the S13INT secondary structure using Sfold 2.0 (http://sfold.wadsworth.org/index.pl) (38) revealed that folded S13INT represents an ensemble of molecules with similar tightly folded structures formed with several extended stems and few loops. An ensemble centroid with a minimum free energy (ΔG37) equal to −88.5 kcal/mol is shown in Figure 5A. Taking into account the high (65%) GC-content of the S13INT sequence, we infer that a structure at least similar to this would be formed by pre-mRNA before addition to nuclear extract. Therefore, we carried out footprinting of free RNA and the RNA in the binary complex (rpS13 • S13INT) under conditions when more than 90% of the RNA was bound to the protein. Three RNases, T1, T2 and V1, were employed in the experiments. The positions of cleavages identified by primer extension analysis using two different primers are presented in Figure 5B. The footprinting data generally supported the secondary structure proposed for S13INT. Seven phosphodiester bonds within S13INT changed their accessibility to RNases due to rpS13 binding (Figure 5A). Two of them (cleavages 5′ to C46 and G53) are located in the vicinity of the 5′ splice site, another two (cleavages 5′ to A192 and G193) are close to the 3′ splice site and three sites (cleavages 5′ to C76 and to neighbouring residues C162 and A164) are intronic, somewhat distant from the splice sites.
Figure 5.
rpS13 binds to S13INT in the vicinity of the splice sites. Footprinting of S13INT transcript in the complex with rpS13. (A) Ensemble centroid of S13INT calculated by Sfold 2.0 (38). Positions of rpS13 protection from RNase cleavage are shown by arrows marked with the respective nucleotide positions. Boxes with arrows indicate positions of the 5′ and 3′ splice sites. (B) Primer extension analysis of the S13INT transcript cleaved with RNAses T1, T2 and V1 free (−) or in the complex with rpS13 (+). Lanes A, C, G and T correspond to the sequencing reactions. Positions of the nucleotides protected by the protein from RNase cleavage (footprints) are marked by arrows. Primers complementary to positions 103–116 (panel 1) and 235–250 (panels 2 and 3) of S13INT transcript were used in the study.
To test whether the sites affected by rpS13 could be involved in protein binding, we introduced mutations in S13INT close to these regions. The trinucleotides CCC (positions 44–46) and AAG (positions 191–193) were replaced with AGA and CCC, respectively, (S13INTm). The affinity of rpS13 for S13INTm was substantially smaller than that for S13INT (Figure 3D), indicating that the mutations really disturbed the protein-binding site. S13INTm was reactive in splicing in vitro and it generated the same splicing pattern as S13INT, but it was less sensitive to inhibition by rpS13 (Figure 4C). These results suggest that rpS13 inhibits splicing of its own pre-mRNA due to its binding close to the splicing sites in intron 1.
DISCUSSION
Ribosomal proteins are very abundant cellular proteins that are capable of binding to different RNA species. Recent investigations have demonstrated that eukaryotic ribosomal proteins are associated with newly transcribed RNA Pol II transcripts (39,40). Thus, the presence of 27 ribosomal proteins in sites of active transcription in Drosophila salivary gland cells was shown by means of chromosomal immunostaining and in situ hybridization (39). Method of chromatin immunoprecipitation revealed that ribosomal proteins in Saccharomyces cerevisiae associate with nascent RNP complexes within nucleus (40). Ribosomal proteins were also found in spliceosomal complexes in human cells (41). Although, it was assumed in these cases that nascent transcripts were bound non-specifically, the example of human rpS13 suggests that there are also specific interactions between ribosomal proteins and their pre-mRNAs that may have functional significance.
Earlier, it was shown that introns in a few eukaryotic ribosomal protein genes participate in regulation of expression (22,24). These introns, as a rule, are short and bear conserved regulatory elements. Our search of the RPS13 gene demonstrated that intron 1 is conserved within the genomes of higher eukaryotes (Figure 1). This is consistent with a role in the regulation of expression.
The constructions pECFP-S13 and pECFP-S13-int1 used for the in vivo experiments differed in the presence of intron 1 of RPS13 gene in the latter construction. Cells transfected with pECFP-S13-int1 overproduced rpS13-eCFP fusion protein, indicating that splicing of the premature transcript produced from pECFP-S13-int1 was correct. The 4-fold decrease in rpS13 mRNA as compared with those in the cells transfected with pECFP-S13 suggested a connection between the presence of intron 1 in the premature transcript and the level of rpS13 mRNA in the cells. One possibility is that rpS13 inhibited splicing of intron 1, decreasing the total rpS13 mRNA level in the transfected cells. However, other intron-dependent pathways remained possible too; one example might be targeting by rpS13 of nuclease activity to the pre-mRNA. Due to its high positive charge, rpS13, like other ribosomal proteins, would be expected to bind readily to RNA. Actually, the in vitro binding assays showed that rpS13 was able to bind to non-cognate pre-RNAs, but this binding was much weaker than its binding to cognate pre-mRNA containing intron 1. Moreover, rpS13 was able to inhibit splicing in vitro of this fragment, whereas other ribosomal proteins did not do so; at the same time, rpS13 had little effect on the splicing of non-cognate pre-mRNAs. Hence, it seemed probable that rpS13 would interact with regions in the pre-mRNA that are necessary for splicing.
The footprinting experiments with S13INT in the complex with rpS13 revealed changes in accessibility to RNases for several internucleotide phosphodiester bonds that are located both in regions of the RNA close to 5′ and 3′ splice sites and in the vicinity of them (Figure 5). Taking into account that ribosomal proteins upon their binding to rRNA, as a rule, do not introduce major changes into its structure (42,43) we consider it more likely that the footprints arose from protein binding rather than from S13INT structural rearrangements. Either the regions of S13INT containing the 5′ and 3′ splice sites are spatially drawn together and form the rpS13-binding site or several protein molecules bind to different splice sites.
Although, the binding sites for rpS13 have not been determined, its prokaryotic homologue, rpS15, has been shown to recognize RNA structure rather than specific sequences. In particular, it recognizes on 16S rRNA an imperfect double helix with a three-way junction at its base and G-U/G-C base pairs separated from the junction by one helical turn (42,44,45). Remarkably, the region recognized in S13INTby rpS13 contains a helix formed by nucleotides from 39 to 64, with base pairs G48–U55 and G49–C54 approximately one turn from the RNA three-way junction (Figure 5A). The 5′ splice site in S13INT is located within this helix. Moreover, we found that disruption of this helix structure by mutations in positions 44–46 (together with mutations in positions 191–193) reduces the rpS13 binding (Figure 3D). The similarity with the binding site of rpS15 is consistent with the possibility that the footprints (at least close to the 5′ splice site) arose from rpS13 binding to corresponding sites in S13INT.
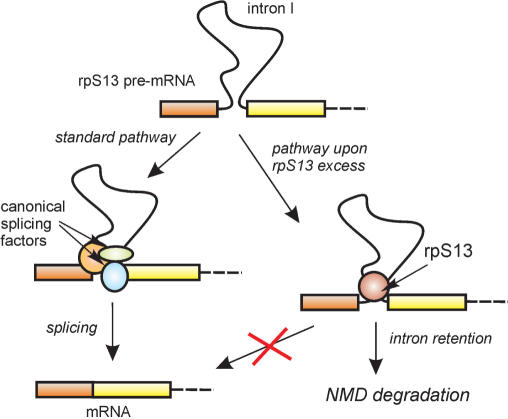
Our findings suggest that the inhibition of splicing of the rpS13 pre-mRNA by an increase in rpS13 concentration results from the binding of rpS13 to the pre-mRNA near the splicing signals within intron 1. The consequences of this binding for splicing reactions are unknown. However, if splicing of intron 1 is blocked, intron-retaining RNA may be formed, as described for some other eukaryotic ribosomal protein mRNAs (22,24), and it may be degraded by NMD (Figure 6). Such a mechanism is likely to work most efficiently with terminal introns adjacent to short terminal exons, in which impediments to splicing are to lead to either skipping or the activation of cryptic sites. Interestingly, both E. coli (46,47) and Thermus thermophilus (48) rpS15 proteins have a regulatory function. The latter binds to its own mRNA and represses its own translation by a feedback mechanism (48).
Figure 6.
Schematic representation of possible pathways for autoregulation of the biosynthesis of rpS13 via splicing of intron 1.
The autoregulation of expression by eukaryotic ribosomal proteins appears to operate by a variety of mechanisms. Thus, rpL12 of C. elegans inhibits constitutive splicing of its own pre-mRNA and stimulates alternative splicing in intron 2 (24), whereas human rpS26 inhibited both constitutive and alternative splicing of its intron 1 (21). A rise in ribosomal protein L1 concentration in oocytes of Xenopus laevis leads to the formation and rapid degradation of rpL1 mRNA isoforms containing unremoved introns 2 and 3 (49), and rpL30 of S. cerevisiae inhibits not only splicing of its own pre-mRNA by binding to the intron, but also translation of its own mRNA (20). Feedback autoregulation may have evolved independently for some or all ribosomal proteins, but it would be unsurprising if the major process subject to regulation was splicing. The subtlety and complexity of splicing suggest that the process might be unusually delicate and susceptible to perturbation.
ACKNOWLEDGEMENTS
This work was supported by grant from the Russian Fund for Basic Research (No. 05-04-48393-a), grant from Siberian Brunch of the Russian Academy of Sciences for young scientist projects competition in honour of M.A. Lavrentiev (No. 92) to A.V.I. and EMBO Short Term Fellowship (ASTF 8-2005) to A.A.M. Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ferreira-Cerca S, Poll G, Gleizes PE, Tschochner H, Milkereit P. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell. 2005;20:263–275. doi: 10.1016/j.molcel.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 2.Bulygin K, Chavatte L, Frolova L, Karpova G, Favre A. The first position of a codon placed in the A site of the human 80S ribosome contacts nucleotide C1696 of the 18S rRNA as well as proteins S2, S3, S3a, S30, and S15. Biochemistry. 2005;44:2153–2162. doi: 10.1021/bi0487802. [DOI] [PubMed] [Google Scholar]
- 3.Graifer D, Molotkov M, Styazhkina V, Demeshkina N, Bulygin K, Eremina A, Ivanov A, Laletina E, Ven’yaminova A, et al. Variable and conserved elements of human ribosomes surrounding the mRNA at the decoding and upstream sites. Nucleic Acids Res. 2004;32:3282–3293. doi: 10.1093/nar/gkh657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wool IG. Extraribosomal functions of ribosomal proteins. Trends Biochem. Sci. 1996;21:164–165. [PubMed] [Google Scholar]
- 5.Perry RP. The architecture of mammalian ribosomal protein promoters. BMC Evol. Biol. 2005;5:15. doi: 10.1186/1471-2148-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Angelastro JM, Klimaschewski L, Tang S, Vitolo OV, Weissman TA, Donlin LT, Shelanski ML, Greene LA. Identification of diverse nerve growth factor-regulated genes by serial analysis of gene expression (SAGE) profiling. Proc. Natl Acad. Sci. USA. 2000;97:10424–10429. doi: 10.1073/pnas.97.19.10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bortoluzzi S, d’Alessi F, Romualdi C, Danieli GA. Differential expression of genes coding for ribosomal proteins in different human tissues. Bioinformatics. 2001;17:1152–1157. doi: 10.1093/bioinformatics/17.12.1152. [DOI] [PubMed] [Google Scholar]
- 8.Lian Z, Liu J, Li L, Li X, Tufan NL, Wu MC, Wang HY, Arbuthnot P, Kew M, et al. Human S15a expression is upregulated by hepatitis B virus X protein. Mol. Carcinog. 2004;40:34–46. doi: 10.1002/mc.20012. [DOI] [PubMed] [Google Scholar]
- 9.Vaarala MH, Porvari KS, Kyllonen AP, Mustonen MV, Lukkarinen O, Vihko PT. Several genes encoding ribosomal proteins are overexpressed in prostate-cancer cell lines: confirmation of L7a and L37 overexpression in prostate-cancer tissue samples. Int. J. Cancer. 1998;78:27–32. doi: 10.1002/(sici)1097-0215(19980925)78:1<27::aid-ijc6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 10.Jang CY, Lee JY, Kim J. RpS3, a DNA repair endonuclease and ribosomal protein, is involved in apoptosis. FEBS Lett. 2004;560:81–85. doi: 10.1016/S0014-5793(04)00074-2. [DOI] [PubMed] [Google Scholar]
- 11.Khanna N, Sen S, Sharma H, Singh N. S29 ribosomal protein induces apoptosis in H520 cells and sensitizes them to chemotherapy. Biochem. Biophys. Res. Commun. 2003;304:26–35. doi: 10.1016/s0006-291x(03)00532-1. [DOI] [PubMed] [Google Scholar]
- 12.Marygold SJ, Coelho CM, Leevers SJ. Genetic analysis of RpL38 and RpL5, two minute genes located in the centric heterochromatin of chromosome 2 of Drosophila melanogaster. Genetics. 2005;169:683–695. doi: 10.1534/genetics.104.034124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torok I, Herrmann-Horle D, Kiss I, Tick G, Speer G, Schmitt R, Mechler BM. Down-regulation of RpS21, a putative translation initiation factor interacting with P40, produces viable minute imagos and larval lethality with overgrown hematopoietic organs and imaginal discs. Mol. Cell. Biol. 1999;19:2308–2321. doi: 10.1128/mcb.19.3.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Sohn JH, Warner JR. Autoregulation in the biosynthesis of ribosomes. Mol. Cell. Biol. 2003;23:699–707. doi: 10.1128/MCB.23.2.699-707.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levy S, Avni D, Hariharan N, Perry RP, Meyuhas O. Oligopyrimidine tract at the 5′ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc. Natl Acad. Sci. USA. 1991;88:3319–3323. doi: 10.1073/pnas.88.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fewell SW, Woolford JL., Jr Ribosomal protein S14 of Saccharomyces cerevisiae regulates its expression by binding to RPS14B pre-mRNA and to 18S rRNA. Mol. Cell. Biol. 1999;19:826–834. doi: 10.1128/mcb.19.1.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vilardell J, Chartrand P, Singer RH, Warner JR. The odyssey of a regulated transcript. RNA. 2000;6:1773–1780. doi: 10.1017/s135583820000145x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warner JR, Mitra G, Schwindinger WF, Studeny M, Fried HM. Saccharomyces cerevisiae coordinates accumulation of yeast ribosomal proteins by modulating mRNA splicing, translational initiation, and protein turnover. Mol. Cell. Biol. 1985;5:1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avni D, Shama S, Loreni F, Meyuhas O. Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: characterization of the translational cis-regulatory element. Mol. Cell. Biol. 1994;14:3822–3833. doi: 10.1128/mcb.14.6.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dabeva MD, Warner JR. Ribosomal protein L32 of Saccharomyces cerevisiae regulates both splicing and translation of its own transcript. J. Biol. Chem. 1993;268:19669–19674. [PubMed] [Google Scholar]
- 21.Ivanov AV, Malygin AA, Karpova GG. Human ribosomal protein S26 suppresses the splicing of its pre-mRNA. Biochim. Biophys. Acta. 2005;1727:134–140. doi: 10.1016/j.bbaexp.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 22.Cuccurese M, Russo G, Russo A, Pietropaolo C. Alternative splicing and nonsense-mediated mRNA decay regulate mammalian ribosomal gene expression. Nucleic Acids Res. 2005;33:5965–5977. doi: 10.1093/nar/gki905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 24.Mitrovich QM, Anderson P. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 2000;14:2173–2184. doi: 10.1101/gad.819900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jagannathan I, Culver GM. Assembly of the central domain of the 30S ribosomal subunit: roles for the primary binding ribosomal proteins S15 and S8. J. Mol. Biol. 2003;330:373–383. doi: 10.1016/s0022-2836(03)00586-2. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y, Zhai H, Wang X, Han Z, Liu C, Lan M, Du J, Guo C, Zhang Y, et al. Ribosomal proteins S13 and L23 promote multidrug resistance in gastric cancer cells by suppressing drug-induced apoptosis. Exp. Cell Res. 2004;296:337–346. doi: 10.1016/j.yexcr.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F. Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc. Natl Acad. Sci. USA. 2003;100:4114–4119. doi: 10.1073/pnas.0633863100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malygin A, Baranovskaya O, Ivanov A, Karpova G. Expression and purification of human ribosomal proteins S3, S5, S10, S19, and S26. Protein Expr. Purif. 2003;28:57–62. doi: 10.1016/s1046-5928(02)00652-6. [DOI] [PubMed] [Google Scholar]
- 30.Malygin A, Parakhnevitch N, Karpova G. Human ribosomal protein S13: cloning, expression, refolding, and structural stability. Biochim. Biophys. Acta. 2005;1747:93–97. doi: 10.1016/j.bbapap.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Parakhnevitch NM, Malygin AA, Karpova GG. Recombinant human ribosomal protein S16: expression, purification, refolding, and structural stability. Biochemistry (Mosc.) 2005;70:777–781. doi: 10.1007/s10541-005-0183-3. [DOI] [PubMed] [Google Scholar]
- 32.Venables JP, Elliott DJ, Makarova OV, Makarov EM, Cooke HJ, Eperon IC. RBMY, a probable human spermatogenesis factor, and other hnRNP G proteins interact with Tra2beta and affect splicing. Hum. Mol. Genet. 2000;9:685–694. doi: 10.1093/hmg/9.5.685. [DOI] [PubMed] [Google Scholar]
- 33.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wollenzien PL. Isolation and identification of RNA cross-links. Methods Enzymol. 1988;164:319–329. doi: 10.1016/s0076-6879(88)64052-3. [DOI] [PubMed] [Google Scholar]
- 35.Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic Acids Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antoine M, Reimers K, Wirz W, Gressner AM, Muller R, Kiefer P. Identification of an unconventional nuclear localization signal in human ribosomal protein S2. Biochem. Biophys. Res. Commun. 2005;335:146–153. doi: 10.1016/j.bbrc.2005.07.069. [DOI] [PubMed] [Google Scholar]
- 37.Rosorius O, Fries B, Stauber RH, Hirschmann N, Bevec D, Hauber J. Human ribosomal protein L5 contains defined nuclear localization and export signals. J. Biol. Chem. 2000;275:12061–12068. doi: 10.1074/jbc.275.16.12061. [DOI] [PubMed] [Google Scholar]
- 38.Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brogna S, Sato TA, Rosbash M. Ribosome components are associated with sites of transcription. Mol. Cell. 2002;10:93–104. [PubMed] [Google Scholar]
- 40.Schroder PA, Moore MJ. Association of ribosomal proteins with nascent transcripts in S. cerevisiae. RNA. 2005;11:1521–1529. doi: 10.1261/rna.2134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Luhrmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. doi: 10.1126/science.1077783. [DOI] [PubMed] [Google Scholar]
- 42.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 43.Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 44.Nikulin A, Eliseikina I, Tishchenko S, Nevskaya N, Davydova N, Platonova O, Piendl W, Selmer M, Liljas A, et al. Crystal structure of the S15±rRNA complex. Nat. Struct. Biol. 2000;7:273–277. doi: 10.1038/74028. [DOI] [PubMed] [Google Scholar]
- 45.Serganov A, BeÂnard L, Portier C, Ennifar E, Garber M, Ehresmann B, Ehresmann C. Role of conserved nucleotides in building the 16S rRNA binding site for ribosomal protein S15. J. Mol. Biol. 2001;305:785–803. doi: 10.1006/jmbi.2000.4354. [DOI] [PubMed] [Google Scholar]
- 46.Philippe C, BeÂnard L, Portier C, Westhof E, Ehresmann B, Ehresmann C. Molecular dissection of the pseudoknot governing the translational regulation of Escherichia coli ribosomal protein S15. Nucleic Acids Res. 1995;23:18–28. doi: 10.1093/nar/23.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Serganov A, Ennifar E, Portier C, Ehresmann B, Ehresmann C. Do mRNA and rRNA binding sites of E. coli ribosomal protein S15 share common structural determinants? J. Mol. Biol. 2002;320:963–978. doi: 10.1016/s0022-2836(02)00553-3. [DOI] [PubMed] [Google Scholar]
- 48.Serganov A, Polonskaia A, Ehresmann B, Ehresmann C, Patel DJ. Ribosomal protein S15 represses its own translation via adaptation of an rRNA-like fold within its mRNA. EMBO J. 2003;22:1898–1908. doi: 10.1093/emboj/cdg170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caffarelli E, Fragapane P, Gehring C, Bozzoni I. The accumulation of mature RNA for the Xenopus laevis ribosomal protein L1 is controlled at the level of splicing and turnover of the precursor RNA. EMBO J. 1987;6:3493–3498. doi: 10.1002/j.1460-2075.1987.tb02674.x. [DOI] [PMC free article] [PubMed] [Google Scholar]