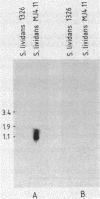
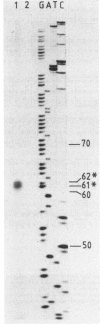
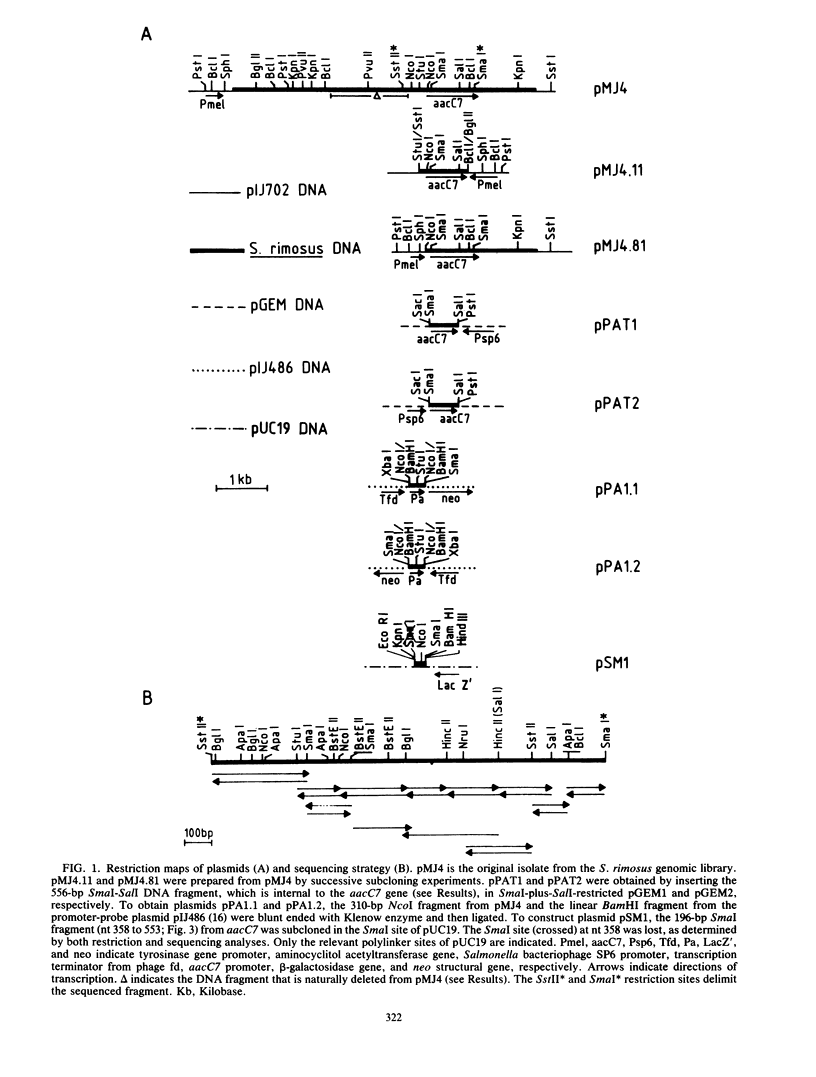
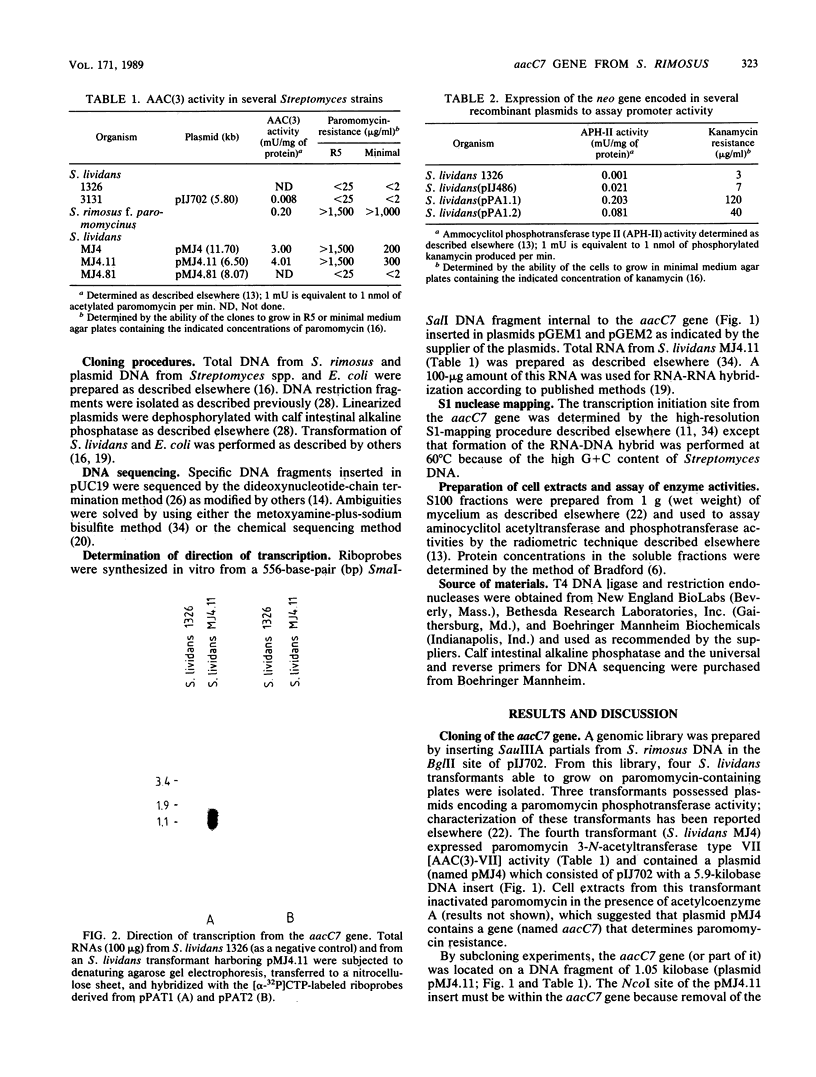
Abstract
A gene (aacC7) encoding an aminocyclitol 3-N-acetyltransferase type VII [AAC(3)-VII] from Streptomyces rimosus forma paramomycinus NRRL 2455 was cloned in the Streptomyces plasmid pIJ702 and expressed in Streptomyces lividans 1326. Subcloning experiments located the aacC7 structural gene on a 1.05-kilobase DNA sequence. The direction of transcription of aacC7 was determined by using riboprobes synthesized in vitro from a DNA fragment internal to the gene. A DNA segment encoding the AAC(3)-VII activity and comprising 1,495 base pairs was sequenced. The aacC7 gene was located in an open reading frame of 864 base pairs that encoded a polypeptide of Mr 31,070, consistent with the Mr (32,000) of the AAC(3)-VII enzyme as determined by physicochemical methods. High-resolution S1 nuclease mapping suggested that transcription starts at or near the A residue of the ATG initiator codon. A DNA fragment from the 5' region of aacC7 had promoter activity in the promoter-probe plasmid pIJ486. The -10 and -35 regions of this fragment showed limited sequence resemblance to other Streptomyces promoters. The primary structure of the AAC(3)-VII enzyme showed strong homology with those of the AAC(3)-III and AAC(3)-IV enzymes encoded by plasmids in gram-negative bacterial genera. Upstream of the aacC7 gene was an open reading frame of 357 nucleotides which did not appear to be involved in controlling the expression of the aacC7 gene.
Full text
PDF

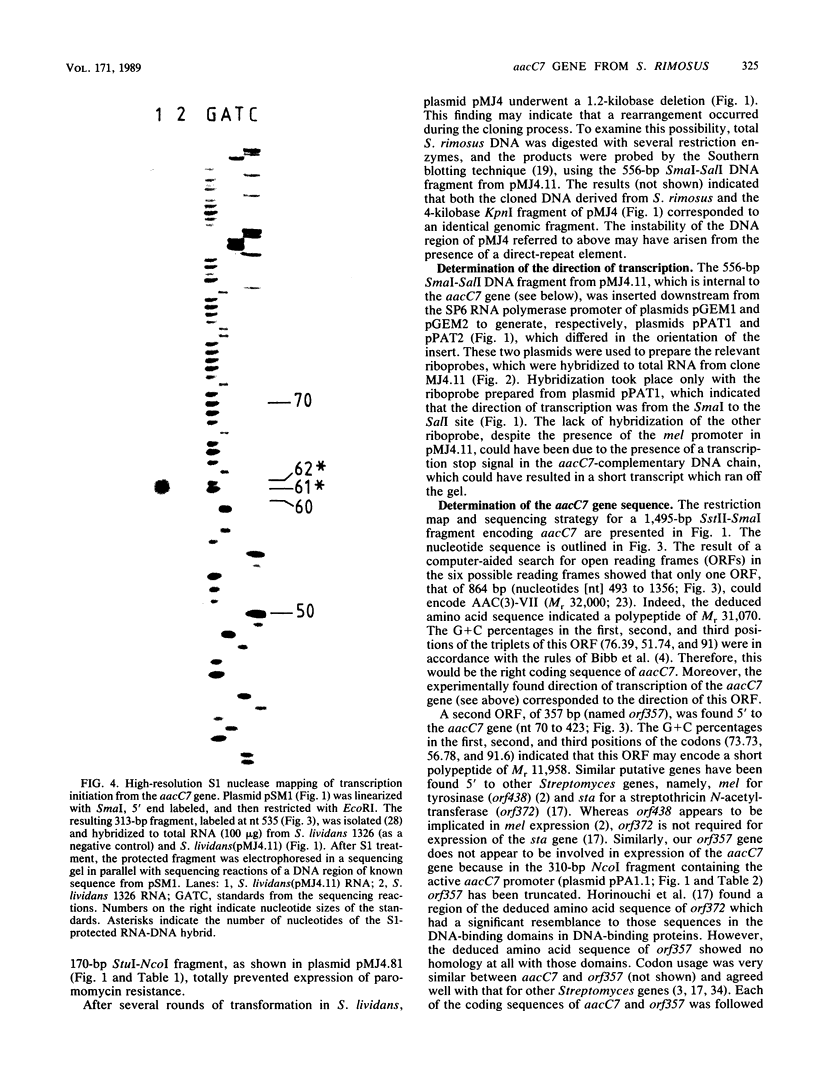



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allmansberger R., Bräu B., Piepersberg W. Genes for gentamicin-(3)-N-acetyl-transferases III and IV. II. Nucleotide sequences of three AAC(3)-III genes and evolutionary aspects. Mol Gen Genet. 1985;198(3):514–520. doi: 10.1007/BF00332949. [DOI] [PubMed] [Google Scholar]
- Bernan V., Filpula D., Herber W., Bibb M., Katz E. The nucleotide sequence of the tyrosinase gene from Streptomyces antibioticus and characterization of the gene product. Gene. 1985;37(1-3):101–110. doi: 10.1016/0378-1119(85)90262-8. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Bibb M. J., Ward J. M., Cohen S. N. Nucleotide sequences encoding and promoting expression of three antibiotic resistance genes indigenous to Streptomyces. Mol Gen Genet. 1985;199(1):26–36. doi: 10.1007/BF00327505. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Janssen G. R., Ward J. M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1985;38(1-3):215–226. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bräu B., Pilz U., Piepersberg W. Genes for gentamicin-(3)-N-acetyltransferases III and IV: I. Nucleotide sequence of the AAC(3)-IV gene and possible involvement of an IS140 element in its expression. Mol Gen Genet. 1984;193(1):179–187. doi: 10.1007/BF00327434. [DOI] [PubMed] [Google Scholar]
- Deng Z. X., Kieser T., Hopwood D. A. Activity of a Streptomyces transcriptional terminator in Escherichia coli. Nucleic Acids Res. 1987 Mar 25;15(6):2665–2675. doi: 10.1093/nar/15.6.2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distler J., Braun C., Ebert A., Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: analysis of a central region including the major resistance gene. Mol Gen Genet. 1987 Jun;208(1-2):204–210. doi: 10.1007/BF00330443. [DOI] [PubMed] [Google Scholar]
- Distler J., Ebert A., Mansouri K., Pissowotzki K., Stockmann M., Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1987 Oct 12;15(19):8041–8056. doi: 10.1093/nar/15.19.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaloro J., Treisman R., Kamen R. Transcription maps of polyoma virus-specific RNA: analysis by two-dimensional nuclease S1 gel mapping. Methods Enzymol. 1980;65(1):718–749. doi: 10.1016/s0076-6879(80)65070-8. [DOI] [PubMed] [Google Scholar]
- Gray G. S., Fitch W. M. Evolution of antibiotic resistance genes: the DNA sequence of a kanamycin resistance gene from Staphylococcus aureus. Mol Biol Evol. 1983 Dec;1(1):57–66. doi: 10.1093/oxfordjournals.molbev.a040298. [DOI] [PubMed] [Google Scholar]
- Haas M. J., Dowding J. E. Aminoglycoside-modifying enzymes. Methods Enzymol. 1975;43:611–628. doi: 10.1016/0076-6879(75)43124-x. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Heinzel P., Werbitzky O., Distler J., Piepersberg W. A second streptomycin resistance gene from Streptomyces griseus codes for streptomycin-3"-phosphotransferase. Relationships between antibiotic and protein kinases. Arch Microbiol. 1988;150(2):184–192. doi: 10.1007/BF00425160. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Furuya K., Nishiyama M., Suzuki H., Beppu T. Nucleotide sequence of the streptothricin acetyltransferase gene from Streptomyces lavendulae and its expression in heterologous hosts. J Bacteriol. 1987 May;169(5):1929–1937. doi: 10.1128/jb.169.5.1929-1937.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpartida F., Hopwood D. A. Physical and genetic characterisation of the gene cluster for the antibiotic actinorhodin in Streptomyces coelicolor A3(2). Mol Gen Genet. 1986 Oct;205(1):66–73. doi: 10.1007/BF02428033. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Piendl W., Böck A., Cundliffe E. Involvement of 16S ribosomal RNA in resistance of the aminoglycoside-producers Streptomyces tenjimariensis, Streptomyces tenebrarius and Micromonospora purpurea. Mol Gen Genet. 1984;197(1):24–29. doi: 10.1007/BF00327918. [DOI] [PubMed] [Google Scholar]
- Pulido D., Jiménez A. Optimization of gene expression in Streptomyces lividans by a transcription terminator. Nucleic Acids Res. 1987 May 26;15(10):4227–4240. doi: 10.1093/nar/15.10.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-González J. A., Jiménez A. Cloning and expression in Streptomyces lividans of a paromomycin phosphotransferase from Streptomyces rimosus Forma paromomycinus. Biochem Biophys Res Commun. 1984 Dec 28;125(3):895–901. doi: 10.1016/0006-291x(84)91367-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. J., Kieser T., Ward J. M., Hopwood D. A. Physical analysis of antibiotic-resistance genes from Streptomyces and their use in vector construction. Gene. 1982 Nov;20(1):51–62. doi: 10.1016/0378-1119(82)90086-5. [DOI] [PubMed] [Google Scholar]
- Vara J. A., Portela A., Ortín J., Jiménez A. Expression in mammalian cells of a gene from Streptomyces alboniger conferring puromycin resistance. Nucleic Acids Res. 1986 Jun 11;14(11):4617–4624. doi: 10.1093/nar/14.11.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vara J., Malpartida F., Hopwood D. A., Jiménez A. Cloning and expression of a puromycin N-acetyl transferase gene from Streptomyces alboniger in Streptomyces lividans and Escherichia coli. Gene. 1985;33(2):197–206. doi: 10.1016/0378-1119(85)90094-0. [DOI] [PubMed] [Google Scholar]
- Vögtli M., Hütter R. Characterisation of the hydroxystreptomycin phosphotransferase gene (sph) of Streptomyces glaucescens: nucleotide sequence and promoter analysis. Mol Gen Genet. 1987 Jun;208(1-2):195–203. doi: 10.1007/BF00330442. [DOI] [PubMed] [Google Scholar]
- Westpheling J., Ranes M., Losick R. RNA polymerase heterogeneity in Streptomyces coelicolor. Nature. 1985 Jan 3;313(5997):22–27. doi: 10.1038/313022a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yoshikawa A., Isono S., Sheback A., Isono K. Cloning and nucleotide sequencing of the genes rimI and rimJ which encode enzymes acetylating ribosomal proteins S18 and S5 of Escherichia coli K12. Mol Gen Genet. 1987 Oct;209(3):481–488. doi: 10.1007/BF00331153. [DOI] [PubMed] [Google Scholar]
- Zalacain M., González A., Guerrero M. C., Mattaliano R. J., Malpartida F., Jiménez A. Nucleotide sequence of the hygromycin B phosphotransferase gene from Streptomyces hygroscopicus. Nucleic Acids Res. 1986 Feb 25;14(4):1565–1581. doi: 10.1093/nar/14.4.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]