Abstract
With current techniques, genetic alterations of herpesviruses are difficult to perform, mostly because of the large size of their genomes. To solve this problem, we have designed a system that allows the cloning of any γ-herpesvirus in Escherichia coli onto an F factor-derived plasmid. Immortalized B cell lines were readily established with recombinant Epstein–Barr virus (EBV), demonstrating that the F factor-cloned EBV genome has all the characteristics of wild-type EBV. Because any genetic modification is possible in E. coli, this experimental approach opens the way to the genetic analysis of all EBV functions. Moreover, it is now feasible to generate attenuated EBV strains in vitro such that vaccine strains can be designed. Because we incorporated the genes for hygromycin resistance and green fluorescent protein onto the E. coli cloned EBV genome, the still open question of the EBV target cells other than B lymphocytes will be addressed.
Oncogenic viruses provide a unique model to understand neoplastic transformation of cells. The low complexity of some of these viruses, e.g., simian virus 40, has allowed their cloning and the generation of mutants, one prerequisite for the understanding of the role of viral proteins in the process of cell transformation.
Epstein–Barr virus (EBV) is one of the few viruses that has been shown to be oncogenic in humans, at least in immunodeficient patients. Most important, EBV infects human B lymphocytes in vitro, such that its immortalizing capacity can be investigated in a tractable system. Although cumbersome, genetic manipulation of EBV genes is possible (1–4) but conventional cloning techniques are not operative with the large genomes of herpesviruses. Traditionally, herpesvirus mutants are generated by homologous recombination in infected cells with DNA fragments or plasmids carrying the mutant allele as described almost 20 years ago (5–7). As a consequence, recombination between the herpesvirus genome and the mutant allele gives rise to a mixed population that consists of wild-type and mutant virus, such that their separation is necessary. This approach has been proven to be quite tedious with γ-herpesviruses, i.e., EBV, because no host cell fully supports the lytic, productive phase of these viruses. Thus, in the case of EBV, it is first essential to obtain an immortalized cell line latently infected with the mutant virus, which takes place often in combination with wild-type virus. To separate these viruses in a second step, the latently infected cell needs to support the lytic phase to produce infectious virions that are instrumental to establish another latently infected, immortalized B cell line carrying the viral mutant, exclusively. Because B cell immortalization is a prerequisite to establish a mutant EBV, this approach excludes the genetic analysis of genes that are essential for B cell immortalization in vitro (8–10).
To overcome these limitations, we cloned the complete EBV genome onto a prokaryotic replicon that also carries the genes for green fluorescent protein (GFP) and hygromycin resistance under the control of eukaryotic promoters. This technique allows the generation of viral mutants in E. coli, transfer of pure EBV mutant DNA into any cell, and the production of infectious virions carrying the mutant viral genome.
MATERIALS AND METHODS
Cells.
B95.8 is a lymphoblastoid cell line obtained by infection of marmoset monkey peripheral blood leukocytes with EBV (11). 293 is a human embryonic epithelial kidney cell line whose differentiation stage is not fully characterized (12). Raji is a human Burkitt’s cell line (13). B lymphocytes from peripheral blood buffy coats, adenoids, and tonsils were purified on a Ficoll cushion after T cell rosetting by using sheep erythrocytes as described (14). Cell lines were grown in RPMI 1640 medium/10% fetal calf serum (Life Technologies, Eggenstein, Germany).
Recombinant DNA Plasmids.
pMBO131 is an F factor-based prokaryotic replicon that carries the F factor origin of replication, the chloramphenicol-resistance gene, and the partitioning proteins A and B (15). The BamHI-HindIII fragment from the pTG76 plasmid carrying the hygromycin-resistance cassette (16) was inserted into the same sites in pMBO131 to yield p1898. The enhanced version of GFP (AscI-MluI fragment, Klenow filled-in) (eGFP, CLONTECH) was cloned into the HpaI site of the previous construct to produce the p1919. The EBV sequences that correspond to the segments located at both sites of the deletion in the B95.8 genome (17) were introduced into p1919 in several consecutive steps to provide a flanking region for homologous recombination with the B95.8 genome. To provide the right flank, a ClaI/AscI fragment spanning the nucleotide coordinates 149,930–159,880 of the B95.8 strain was cloned into the unique ClaI site of p1919 with the aid of synthetic ClaI linkers to yield the intermediate plasmid p1929. The left flank is a BsaBI/BsaBI fragment encompassing nucleotide coordinates 143,458–152,636, which was cloned into the unique PacI site of p1929 by using phosphorylated PacI linkers to obtain p1944. All plasmids were propagated in the DH5α E. coli strain (F−, Φ80dlacZΔM15, D(lacZYA-argF)U169, deoR, recA1, endA1, hsdR17(rk−, mk+), supE44, λ−thi-1, gyrA96, relA1).
DNA Transfections.
Transfections of cell lines with plasmid DNA were performed by electroporation or lipid micelles. B95.8 cells (107 cells) were washed in RPMI 1640 without fetal calf serum, resuspended in 250 μl of the same medium, and placed with the plasmid DNA in 0.4-cm gap electroporation cuvettes. Cells were transfected by using an electroporator (Bio-Rad) at 230 V and 960 μF. 293 cells were placed in Optimem minimal medium (Life Technologies) for 2 hr and incubated for 4 hr with DNA embedded in lipid micelles (Lipofectamine, Life Technologies).
Hygromycin Selection.
One day after transfection, cells were plated in 96-well cluster plates (4,000 cells per well) and hygromycin (Calbiochem) was added to the culture medium (100 μg/ml). Cells were fed weekly with fresh RPMI 1640 containing 10% fetal calf serum with the same hygromycin concentration.
Plasmid Rescue in E. coli.
Circular DNA molecules were extracted from F factor-positive B95.8 clones by using a denaturation–renaturation method as described (18). After extraction, DNA was introduced into E. coli DH10B strain (F−, mcrA, Δ(mrr-hsdRMS-mcrBC), Φ80dlacZΔM15, ΔlacX74, deoR, recA1, endA1, araD139, Δ(ara, leu)7697, galU, galK, λ−, rpsL, nupG) by electroporation (1,800 V, 25 μF, 100 Ω). Cells were plated on agar plates containing 15 μg/ml of chloramphenicol.
Infections.
Primary B cells (2 × 107) were infected with filtered (0.45-μm pore size) supernatants from B95.8/F factor-transfected 293 cells in which the lytic cycle had been induced by transfecting an expression plasmid encoding BZLF1 (19, 20). B cells were then plated in 96-well cluster plates (2 × 107) and fed once a week with RPMI 1640 containing 10% fetal calf serum.
Southern Blot Analysis.
Cells were lysed in 1% SDS, proteins were digested overnight in proteinase K (Boehringer Mannheim) (50 μg/ml final concentration), and cellular DNA was extracted by using a modified Hirt procedure: the extraction medium was made 0.3 M NaCl, thoroughly mixed, and spun down at 7,000 rpm in a swinging-bucket rotor. Supernatants then were precipitated with 2 volumes of ethanol. Pellets were washed in 70% ethanol and resuspended in TE (Tris 10 mM/EDTA 5 mM). Ten micrograms of DNA was digested with restriction enzymes, separated on agarose gel, and blotted onto a Hybond N+ nylon membrane (Amersham) after depurination in 0.25 M HCl using an alkaline transfer procedure. Blots were hybridized overnight with a 32P-radiolabeled probe in “Church” buffer (7% SDS/1 mM EDTA/0.5 Na2HPO4). Blots were washed in 1% SDS/0.2 SSC and exposed by using intensifying screens.
Gardella Gel Analysis.
Gardella gel electrophoresis were performed as described previously (21, 22) followed by Southern blot hybridization.
Immunostaining.
Cells were fixed 15 min in pure acetone and incubated 30 min with a mouse mAb (Chemicon) against the viral capsid antigen (dilution 1:1,000 in PBS 5% fetal calf serum) in a moist chamber at 37°C. After three washings, a sheep anti-mouse antibody coupled to the Cy5 fluorochrome was applied 30 min at 37°C to the cells. After further washings and embedding in glycerol 10% PBS, immunostainings were evaluated by using an Axiovert-inverted epifluorescence microscope (Zeiss).
RESULTS
Cloning of the B95.8 EBV DNA in E. coli.
To address the problem of genetic manipulations of EBV, we cloned the genome of the B95.8 strain onto an F factor-based replicon in E. coli. F factor is a endogenous low-copy plasmid (one to two copies per cell) of E. coli that is about 94 kbp in size. Derivatives of the F factor replicon can encompass large, foreign DNA fragments that serve as, e.g., bacterial artificial chromosomes. The 6.5-kbp pMBO131 F factor plasmid (15), which encodes chloramphenicol resistance, was used as a basic F factor construct to add the genes for hygromycin resistance for selection in eukaryotic cells together with GFP as a phenotypic marker. To introduce this F factor construct into the EBV genome, the strategy depicted in Fig. 1 was employed. To target the modified F factor construct right into the site of the deletion in the B95.8 genome, flanking regions of approximately 9 and 5 kbp in length were added as shown in Fig. 1 to promote homologous recombination events. The final p1944 F factor construct was linearized via its flanking NotI sites, transfected into the B95.8 marmoset cell line infected with B95.8 virus, and cells were selected for resistance against hygromycin. Three to 6 weeks after transfection, 120 clones grew out and 100 were tested for integration of the F factor in genomic B95.8 DNA by Gardella gel analysis, using the F factor plasmid as a probe. A total of 40 clones showed two distinct signals corresponding to the linear and circular forms of viral DNA, indicating that the F plasmid had recombined successfully with the endogenous B95.8 genome. The signal corresponding to unit-length virion DNA further indicated that spontaneous lytic DNA replication of the hybrid molecules occurred as expected for B95.8 cells (Fig. 2). These 40 positive clones also expressed GFP as indicated by their bright green color after exposure to UV light (data not shown). We rescued the hybrid B95.8/F factor plasmids from these B95.8 cells into the E. coli strain DH10B via electroporation. After selection for chloramphenicol resistance, colonies were expanded and their plasmid DNA was analyzed with numerous restriction enzymes. E. coli clones obtained with DNAs from seven independent, hygromycin-resistant B95.8 cell clones showed restriction fragments indicative of a perfect integration of the F factor plasmid into the site of the B95.8 deletion (Fig. 3).
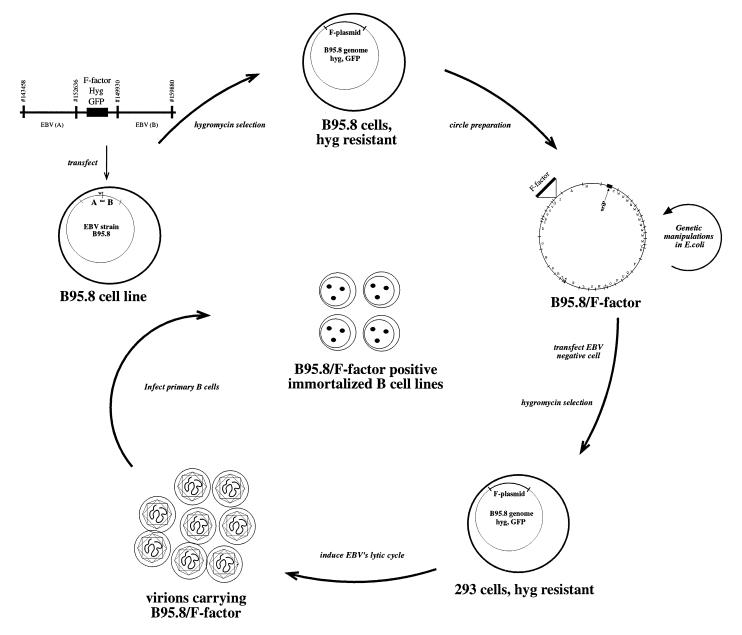
Figure 1.
Schematic overview of the EBV shuttle system. A linearized DNA fragment consisting of an F factor plasmid and two flanking regions. A and B of EBV are transfected into the B95.8 cell line, which is latently infected with EBV. Homologous recombinations occur via the regions A and B to generate a B95.8 cointegrate that encompasses the gene for hygromycin resistance (hyg) and green fluorescent protein (GFP) together with the F factor replicon. Cells that contain such a cointegrate B95.8/F factor survive under hygromycin selection. Preparation of circular DNA from these cells and its transfection into an appropriate E. coli strain establishes the B95.8/F factor molecule in E. coli for further genetic modifications. The B95.8/F factor DNA can be amplified and isolated from E. coli in microgram quantities to be used for transfection into EBV-negative cells, i.e., 293. Upon hygromycin selection, cell lines that carry the B95.8/F factor molecule as extrachromosomal copies can be established. Induction of the lytic phase of EBV’s life cycle yields viruses that carry the B95.8/F factor molecule as genetic information. Infection of primary B cells leads to their immortalization and B95.8/F factor-positive B cell lines.
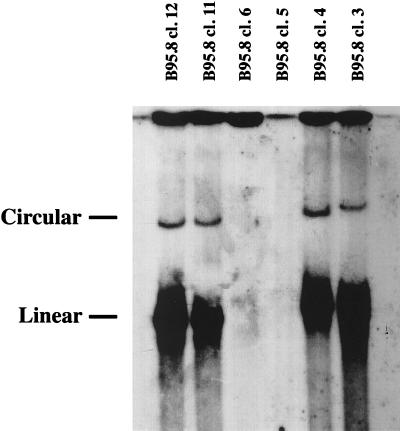
Figure 2.
Gardella gel analysis of individual, hygromycin-resistant B95.8 cell clones that were generated after transfection of the linearized F factor plasmid DNA shown in Fig. 1. After Southern blotting the blot was probed with an F factor-specific probe. Four of seven hygromycin-resistant cell lines showed clear signals, indicating a successful recombination between endogenous, wild-type B95.8 DNA molecules and the transfected linearized plasmid DNA carrying the F factor replicon. The two discrete bands correspond to the circular and linear forms of the B95.8/F factor DNA molecules. Linear EBV molecules are indicative of spontaneous lytic EBV replication, which is common in B95.8 cells.
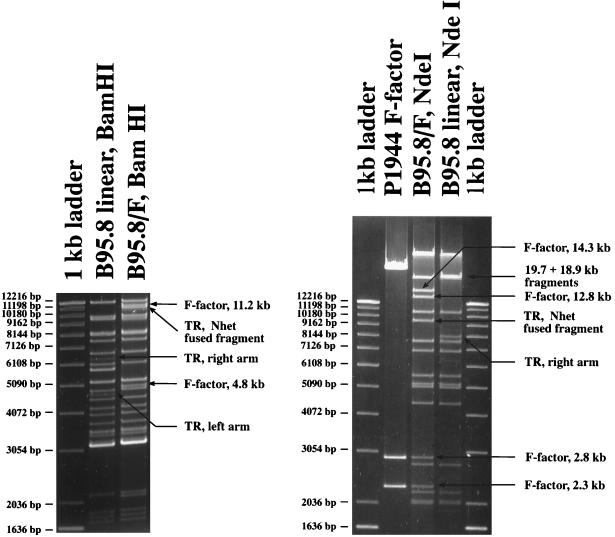
Figure 3.
Restriction fragment analysis of B95.8/F factor DNA in comparison with B95.8 virion DNA. Circular B95.8/F factor DNA molecules were extracted from the hygromycin-resistant B95.8 cell lines and rescued in E. coli strain DH10B. DNA was purified from the chloramphenicol-resistant E. coli clones and digested with NdeI and BamHI. The restriction pattern of the B95.8/F factor DNA was compared with the one obtained with linear wild-type B95.8 DNA extracted from viral capsids. Both restriction patterns were identical with the exception of distinct DNA fragments. New fragments were generated by the integration of the F factor into the B95.8/F factor molecule (11.2 and 4.8 kbp in size after digestion with BamHI; 14.3, 12.8, 2.8, and 2.3 kbp in size after digestion with NdeI). Fragments that represent the genomic terminal fragments in wild-type B95.8 virion DNA are fused in B95.8/F factor DNA to form a new terminal fusion fragment. One of the terminal fragments in B95.8 DNA generated by digestion with NdeI is too small to be visible on this gel.
Establishment of 293 Cell Clones Producing Infectious Virions.
To establish the envisioned EBV shuttle system, the B95.8/F factor DNA needed to be stably introduced into an EBV-negative cell line that would maintain a latent infection but support the lytic phase of EBV to generate infectious virions at will. Toward this aim, 293 cells were transfected with three, independently isolated but otherwise identical B95.8/F factor plasmid DNAs isolated from the cell lines B95.8 cl.4, cl.11, and cl.12 (Fig. 2), and selected for resistance against hygromycin. The outgrowing clones were analyzed for the presence of the viral B95.8/F factor hybrid DNA molecules by using the Gardella gel technique and exposure to UV light to detect GFP expression. Fourteen independent cell lines proved to be positive in Gardella gel analysis (Fig. 5 and data not shown), which also expressed GFP (Fig. 4). Most of the 14 cell lines contained circular but also linear forms of the B95.8/F factor DNA, indicating that a small proportion of B95.8/F factor carrying 293 cells spontaneously supported the lytic cycle of EBV (293 B95.8/F V, 293 B95.8/F VI, 293 B95.8/F XI in Fig. 5 (Fig. 5) and data not shown).
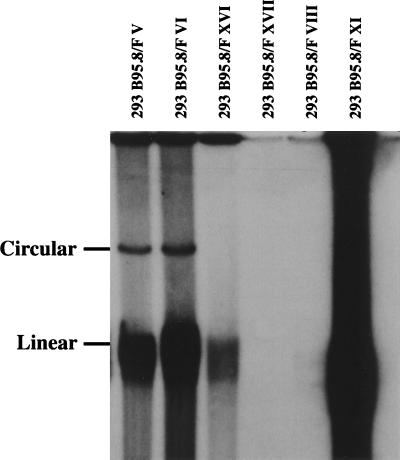
Figure 5.
Gardella gel analysis of hygromycin-resistant 293 cell lines transfected with the B95.8/F factor plasmid DNA. After Southern blotting the blot was probed with an F factor-specific probe. Half of the tested clones proved to be positive after hybridization. As already observed in the B95.8/F factor clones, spontaneous replication occurs in these positive cell lines as demonstrated by the presence of linear genomic virion DNA.
Figure 4.
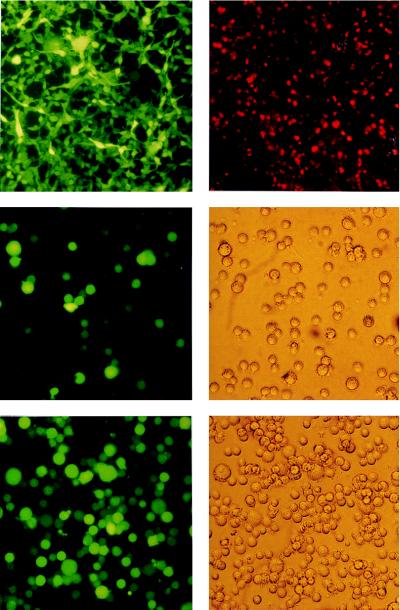
(Top Left) GFP expression in 293 B95.8/F factor-positive cell lines. Living cells were examined with an inverse microscope under UV light. In both cases, the bright green color of all cells demonstrates a high level of expression of the GFP gene, which is part of the F factor backbone, as shown schematically in Fig. 1. (Top Right) Expression of the viral capsid antigen (VCA) in 293 carrying the B95.8/F factor DNA after induction of the lytic cycle. Fixed cells were incubated with an antibody against VCA and a second anti-mouse antibody coupled to the Cy5 fluorochrome. Stained cells were exposed to UV light. (Middle) GFP expression in cells incubated with supernatants from induced 293 cell lines carrying the B95.8/F factor DNA. Raji cells (1 × 105) were incubated with 0.5 ml of supernatant from BZLF1-transfected 293 cells carrying the B95.8/F factor. Approximately 50% of the cells were GFP-positive, indicating a virus titer of at least 105 infectious viruses per ml. GFP fluorescence was investigated 48 hr after infection (Left). (Middle Right) Phase-contrast light microscopy. (Bottom) Primary human B cells were infected with supernatants from BZLF1 transfected 293 cells carrying the B95.8/F factor. As a consequence, immortalized B cell lines were generated that were investigated for GFP expression about 6 weeks after infection. (Bottom Left) UV light exposure. (Bottom Right) Phase-contrast light microscopy.
To induce the lytic cycle in the majority of the cells, the 293 clones were transfected with an expression plasmid encoding the viral transactivator BZLF1 (20). A strong increase in the number of viral linear molecules could be identified after Gardella gel analysis (data not shown), and the presence of viral capsid antigens was visualized by immunostaining with a specific antibody directed against viral capsid antigen (Fig. 4). To test whether the transfected 293 cells produced mature and infectious virions, we incubated Raji cells with supernatants from the induced 293 cells carrying the B95.8/F factor DNA. As revealed by direct visualization of the Raji cells under UV light, nearly 100% of the Raji cells expressed GFP, supporting the infectious nature of the virions produced (Fig. 4).
Generation of Immortalized B Cell Lines with the B95.8/F Factor Virus.
The ability to immortalize human B cells in vitro is unique to EBV. To see whether the B95.8/F factor DNA molecule has this property, we infected primary B cells with the virus stocks. B cells were prepared from four different buffy coat blood samples, three different tonsils, and two adenoids and infected with supernatants from induced 293 cells carrying the recombinant B95.8/F factor virus. In addition, we cocultivated four B cell preparations with irradiated, noninduced B95.8/F factor carrying 293 cells. One night after infection, cells were plated in 96-well cluster plates that contained an irradiated feeder layer as described (1). In all cases, immortalized B cells grew out in all wells of the 96-well cluster plates after 2 (adenoids) to 6 weeks (peripheral B cells). Many infected cells showed a moderate to high expression of the GFP protein (Fig. 4). DNA was extracted from 20 of these immortalized B cell lines, digested with BamHI, and hybridized with an F factor-specific probe. All cell lines tested were found to carry the F factor DNA (Fig. 6 and data not shown).
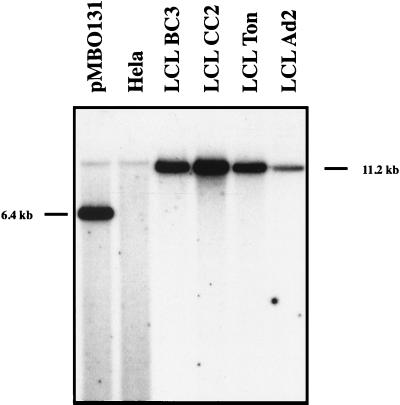
Figure 6.
Southern blot analysis of immortalized B cell lines that were established with virus stocks derived from 293 cells carrying the B95.8/F factor molecule. Total cell DNA from the immortalized B cell lines was digested with BamHI, separated by electrophoresis, blotted, and hybridized with a probe specific for the pMBO131 plasmid. DNAs extracted from the EBV-negative cell line HeLa served as a negative control. The positive control consisted of 100 pg of the pMBO plasmid lacking the genes for GFP and hygromycin resistance, which was digested together with 10 μg HeLa DNA. All cell lines tested were found to carry the F factor, although the number of B95.8/F factor copies varied in the immortalized B cell lines, as expected.
DISCUSSION
To understand EBV’s immortalizing functions genetically, we had a system in mind that would allow any possible mutation of herpesvirus genomes. Toward this end, we cloned the B95.8 EBV DNA onto a F factor-based replicon and show here that such DNA molecules can be propagated stably in E. coli and, when introduced into EBV-negative 293 cells, establish a latent infection in eukaryotic cells. Moreover, 293 cells carrying the recombinant B95.8/F factor DNA support the lytic cycle and release infectious virions that fully retain the immortalizing capacity of wild-type EBV. Thus, it is now possible to modify any EBV gene in E. coli as has been demonstrated multiple times by us with F factor-based mini-EBV plasmids and mouse cytomegalovirus, more recently (23–26). Having introduced such a mutation in E. coli, one can easily obtain a pure and otherwise fully functional virion DNA molecule in contrast to the current approach in which helper virus is required (1–3). Moreover, even genes and cis-acting elements involved in lytic or latent DNA replication, encapsidation of genomic viral DNA, or viral infectivity are now accessible to genetic analyses.
A specific problem in the field of EBV viruses is the difficulty in testing for mutations that do not completely inhibit the immortalizing potential of EBV but have auxiliary functions (1, 10, 23). This is because a direct evaluation of the concentration of mutant virus in the virus stocks was impossible. Now this problem is easily solved because the expression of GFP in the infected cells provides a reproducible means to evaluate the concentration of infectious particles in pure virus stocks and to quantify the effect of a given mutation. After transfection of an expression plasmid for BZLF1, virus stocks obtained from 293 cells carrying B95.8/F factor are in the order of 105 infectious particles per milliliter (Fig. 4).
In the past, genetic analyses of herpesviruses proved to be a particularly tedious task mainly because of the large size of their genomes. On the other hand, herpesviruses provide a unique opportunity to incorporate large pieces of foreign DNA into their genomes. This is in contrast to the majority of viral vectors used today in gene therapy approaches, e.g., retroviruses, adenoviruses, or others. Very large genes or even complete loci could be introduced in EBV-derived vectors that minimally encompass the cis- and trans-acting elements of EBV required for DNA amplification (oriLyt), packaging (TR), and maintenance of the vector in the recipient cell (oriP and EBNA1) (1, 10, 27–29). To use EBV-derived vectors in gene transduction, a helper cell line must provide the trans-acting factors that normally are not present on EBV-derived vectors. The generation of such a helper cell line is now feasible because a deletion of the packaging signals on the recombinant B95.8/F factor molecule will generate a helper virus genome that itself is unable to become encapsidated but will provide all functions in trans required for packing.
Posttransplant lymphoproliferative disorders are thought to be a direct consequence of the ability of EBV to immortalize B cells. The role of EBV in other diseases such as nasopharyngeal carcinoma or Hodgkin’s disease is unclear to date, but it is highly probable that EBV contributes to the acquisition of the malignant phenotype. Therefore, vaccination against EBV should allow prevention of these diseases. Vaccination against EBV by using immunogenic peptides, for example, is currently under study but we think that our shuttle system could provide an interesting alternative to these approaches. Generation of attenuated EBV strains that, for example, lack LMP1 or EBNA2, would impede the development of neoplasms but would not necessarily alter the clinical latency that is established in the majority of infected individuals. Alternatively, construction of EBV vaccine strains that are sensitive to pharmaceutical drugs is perfectly feasible.
In conclusion, we have designed a system that permits cloning and mutation of herpesvirus genomes in general. The experimental approach used in this work is equally applicable to any large DNA virus genomes.
Acknowledgments
We thank the members of the Hammerschmidt laboratory for many stimulating discussions and critical reading of the manuscript. Our research was supported by Grant CA70723 from the National Institutes of Health, Grant Ha1354/3-1 from the Deutsche Forschungsgemeinschaft, Grant 10-1016-Ze from the Deutsche Krebshilfe, and institutional grants.
ABBREVIATIONS
- EBV
Epstein–Barr virus
- GFP
green fluorescent protein
References
- 1.Hammerschmidt W, Sugden B. Nature (London) 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 2.Tomkinson B, Robertson E, Yalamanchili R, Longnecker R, Kieff E. J Virol. 1993;67:7298–7306. doi: 10.1128/jvi.67.12.7298-7306.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kempkes B, Spitkovssky D, Jansen-Dürr P, Ellwart J W, Delecluse H-J, Rottenberger C, Kremmer E, Bornkamm G W, Hammerschmidt W. EMBO J. 1995;14:88–96. doi: 10.1002/j.1460-2075.1995.tb06978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen J I, Wang F, Mannick J, Kieff E. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smiley J. Nature (London) 1980;285:333–335. doi: 10.1038/285333a0. [DOI] [PubMed] [Google Scholar]
- 6.Post L E, Roizman B. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 7.Mocarski E S, Post L E, Roizman B. Cell. 1980;22:243–255. doi: 10.1016/0092-8674(80)90172-5. [DOI] [PubMed] [Google Scholar]
- 8.Tomkinson B, Robertson E, Kieff E. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaye K M, Izumi K M, Kieff E. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kempkes B, Pich D, Zeidler R, Hammerschmidt W. Proc Natl Acad Sci USA. 1995;92:5875–5879. doi: 10.1073/pnas.92.13.5875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller G, Shope T, Hermann L, Stitt D, Lipman M. Proc Natl Acad Sci USA. 1972;69:383–387. doi: 10.1073/pnas.69.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 13.Pulvertaft R J V. Lancet. 1964;I:238–240. [Google Scholar]
- 14.Zeidler R, Meissner P, Eissner G, Lazis S, Hammerschmidt W. Cancer Res. 1996;56:5610–5614. [PubMed] [Google Scholar]
- 15.O’Connor M, Peifer M, Bender W. Science. 1989;244:1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- 16.Giordano T J, McAllister W T. Gene. 1990;88:285–288. doi: 10.1016/0378-1119(90)90045-s. [DOI] [PubMed] [Google Scholar]
- 17.Baer R, Bankier A T, Biggin M D, Deininger P L, Farrell P J, Gibson T J, Hatfull G, Hudson G S, Satchwell S C, Seguin C, Tufnell P S, Barell B G. Nature (London) 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 18.Griffin B E, Björck E, Bjursell G, Lindahl T. J Virol. 1981;40:11–19. doi: 10.1128/jvi.40.1.11-19.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Countryman, J. & Miller, G. (1985) Proc. Natl. Acad. Sci. USA 4085–4089. [DOI] [PMC free article] [PubMed]
- 20.Hammerschmidt W, Sugden B. Cell. 1988;55:427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- 21.Gardella T, Medveczky P, Sairenji T, Mulder C. J Virol. 1984;50:248–254. doi: 10.1128/jvi.50.1.248-254.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delecluse H-J, Schüller S, Hammerschmidt W. EMBO J. 1993;12:3277–3286. doi: 10.1002/j.1460-2075.1993.tb05997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brielmeier M, Mautner J, Laux G, Hammerschmidt W. J Gen Virol. 1996;77:2807–2818. doi: 10.1099/0022-1317-77-11-2807. [DOI] [PubMed] [Google Scholar]
- 24.Kilger E, Kieser A, Baumann M, Hammerschmidt W. EMBO J. 1998;17:1700–1709. doi: 10.1093/emboj/17.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zimber-Strobl U, Kempkes B, Marschall G, Zeidler R, Van Kooten C, Banchereau J, Bornkamm G W, Hammerschmidt W. EMBO J. 1996;15:7070–7080. [PMC free article] [PubMed] [Google Scholar]
- 26.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Banerjee S, Livanos E, Vos J M. Nat Med. 1995;1:1303–1308. doi: 10.1038/nm1295-1303. [DOI] [PubMed] [Google Scholar]
- 28.Wang S, Vos J M. J Virol. 1996;70:8422–8430. doi: 10.1128/jvi.70.12.8422-8430.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kempkes B, Pich D, Zeidler R, Sugden B, Hammerschmidt W. J Virol. 1995;69:231–238. doi: 10.1128/jvi.69.1.231-238.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]