Abstract
BRCA2 is essential for DNA repair by homologous recombination via its interaction with RAD51, mediated by short motifs in the middle and at the C-terminus of its sequence. Here we report that a conserved 36-residue exon 27 sequence of human BRCA2 (BRCA2Exon 27) interacts with RAD51 through the specific recognition of oligomerised RAD51 ATPase domains. BRCA2Exon 27 binding stabilizes the RAD51 nucleoprotein filament against disassembly by BRC repeat 4. The protection is specific for RAD51 filaments formed on single-stranded DNA and is lost when BRCA2Exon 27 is phosphorylated on Ser3291. We propose that productive recombination results from the functional balance between the different modes of interaction with RAD51 of the BRC repeat and exon 27 regions of BRCA2. Our results further suggest a mechanism whereby CDK phosphorylation of BRCA2Exon 27 at the G2-M transition alters the balance in favor of RAD51 filament disassembly, thus terminating recombination.
Keywords: BRCA2, DNA repair, Homologous Recombination, Phosphorylation, RAD51
The inheritance of a mutated copy of the BRCA2 gene confers a high lifetime risk of developing breast, ovarian and other cancers1,2. This risk is explained by the fundamental role that the BRCA2 tumor suppressor plays in preserving the integrity of our DNA. BRCA2-deficient cells are defective in DNA repair3,4 and show an accumulation of chromosomal breaks5,6. BRCA2 has an essential role in the repair of double-strand (ds) DNA breaks by homologous recombination, via its ability to regulate the RecA-related recombinase RAD516-9. BRCA2 targets RAD51 to the site of damaged DNA and promotes formation of the RAD51-single stranded (ss) DNA nucleoprotein filament, the molecular species that performs the reactions of homology search and strand exchange underlying homologous recombination10-13. However, the precise mechanism by which BRCA2 directs RAD51-mediated homologous recombination upon DNA damage remains to be elucidated14.
The remarkable size of the human BRCA2 protein (3,418 amino acids) has so far precluded investigation of its recombination function in vitro. Studies have instead concentrated on the regions of BRCA2 that are known to function in homologous recombination (Figure 1a). Exon 11 of the BRCA2 gene codes for a large central region of the protein that houses eight ∼35 amino acid BRC repeats, six of which are known to bind RAD5115-17. Another RAD51-binding motif is present in the extreme C-terminus of BRCA2, coded for by exon 27 (BRCA2Exon 27). BRCA2Exon 27 is highly conserved in vertebrate BRCA2 proteins and is unrelated in sequence to the BRC repeats7,18. A large conserved C-terminal portion of BRCA2 can bind single-stranded DNA10; recent evidence indicates that this C-terminal domain localizes the BRCA2–RAD51 complex to ds-ssDNA junctions that are created at double-strand break loci in order to initiate nucleoprotein filament formation12.
Figure 1.
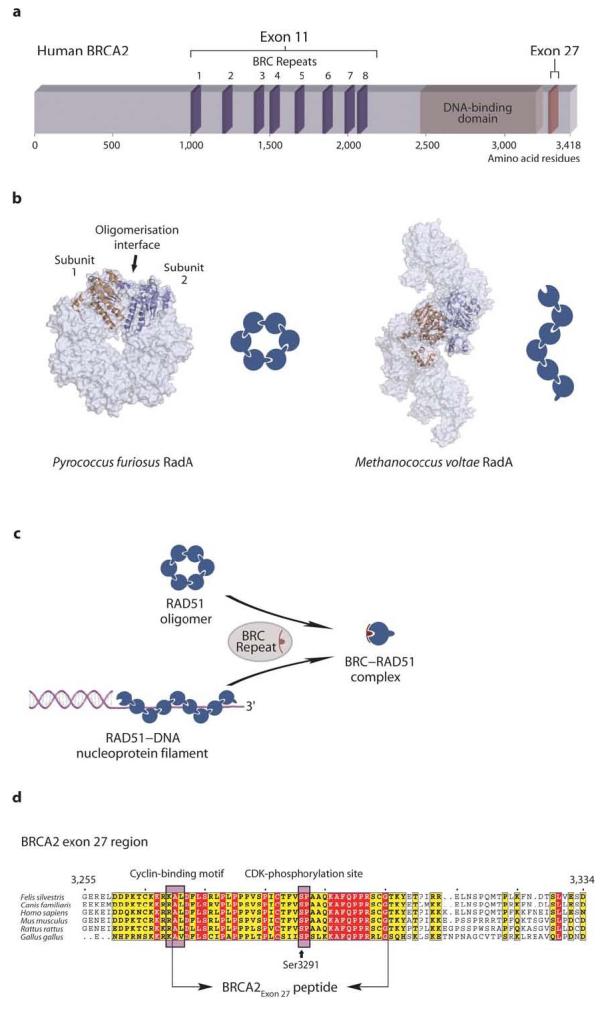
Structure and function of BRCA2 and RAD51 proteins. (a) Schematic diagram of the human BRCA2 protein. The RAD51-binding regions in the middle and C-terminus of the protein, as well as its DNA-binding domain, are indicated. (b) Recombinases of the RecA/RAD51/RadA family oligomerise to form rings or filaments. Representative examples of both arrangements are shown, as observed in the crystal structures of Pyrococcus furiosus RadA (PDB entry 1PZN) and Methanococcus voltae RadA (pdb entry 2F1H), respectively (adjacent subunits are shown in orange and blue). In the oligomeric forms, the recombinase protomers associate through an evolutionarily-conserved mechanism. (c) BRC repeat binding can disrupt RAD51 oligomers and nucleoprotein filaments, by sequestering RAD51 in a monomeric form. (d) Multiple sequence alignment of the RAD51-binding sequence coded by exon 27 of vertebrate BRCA2 (absolutely conserved residues are shown in red and partially conserved residues in yellow; numbering refers to the human BRCA2 sequence). The RAD51-binding sequence overlaps with a cyclin-binding motif that enables CDK-dependent phosphorylation of residue Ser3291 at the G2-M phase transition of the cell cycle. The extent of the BRCA2Exon 27 sequence used in this work is indicated. Please note that the schematic diagrams shown in this and subsequent figures are meant to illustrate the overall effect of BRCA2 peptides upon RAD51 oligomerisation. However, they are not intended to represent the actual number of protomers present in oligomeric RAD51 or the stoichiometry of the BRCA2Exon 27–RAD51 interaction.
The rationalization of BRCA2's activity in DNA repair requires a detailed functional understanding of RAD51-binding by the BRC repeat and exon 27 regions. Proteins of the RecA/RAD51/RadA family of recombinases assemble spontaneously in homo-oligomeric forms consisting of rings or filaments19-25 (Figure 1b). Individual BRC repeats can disrupt RAD51 oligomers and RAD51 nucleoprotein filaments through the formation of a 1:1 complex between monomeric RAD51 and the BRC repeat8,16,26 (Figure 1c). Intriguingly, BRC repeats interrupt RAD51 oligomerisation via a mechanism that mimics the mode of RAD51 self-association26. Potential roles for the disruptive function of BRC repeats include sequestration and transport of RAD51 to the site of DNA damage, remodeling of RAD51 oligomers into nucleoprotein filaments and disassembly of established nucleoprotein filaments11,12. Biochemical evidence for a further role of the BRC repeats in recombination has emerged through observations that under certain conditions BRC repeats can associate with RAD51 nucleoprotein filaments27, and that a large BRCA2 segment spanning all eight BRC repeats (BRCA2BRC1-8) can stimulate RAD51-mediated strand exchange in vitro11. Taken together, these observations provide a multi-faceted picture of BRC repeats' involvement in the process of recombinational repair. The ability of individual BRC repeats and the BRCA2BRC1-8 region to interact with RAD51 in ways that can interfere or be compatible with filament assembly11,16,27 (ORD & LP, unpublished observations) suggests that the different binding modes must be strictly controlled in cells during the different stages of the repair process. How the appropriate balance between the multiple functions of BRC repeats is achieved during recombination-mediated repair remains unknown.
The presence of a RAD51-binding site within the C-terminal exon 27 region of BRCA2 was originally identified by yeast two-hybrid analysis and later confirmed biochemically7,18,28 (Figure 1d). Exon 27 deletion induces an increased tumor incidence and reduced lifespan in mice29,30, hypersensitivity to DNA cross-linking agents and failure of RAD51 foci formation in cells31. These studies indicate that the exon 27 region is functionally relevant to the essential role of BRCA2 in preserving the integrity of the genome. Recent evidence shows that amino acid serine 3291 within exon 27 undergoes CDK-dependent phosphorylation at the G2-M phase transition and dephosphorylation upon DNA damage28. The finding that Ser3291 phosphorylation abrogates RAD51 binding as cells progress into mitosis suggests that the BRCA2 exon 27–RAD51 interaction is relevant to homologous recombination and that its disruption contributes towards the termination of recombination28. However, the lack of information regarding the mode of RAD51-binding by the exon 27 region of BRCA2 currently restricts our understanding of its function.
Here we report an experimental investigation of the interaction between a highly conserved exon 27 sequence of human BRCA2 (amino acids 3270-3305; 36 residues) and RAD51. In the rest of the paper, the BRCA2 exon 27 oligopeptide used here will be called simply BRCA2Exon 27. We find that BRCA2Exon 27 specifically recognizes and interacts with RAD51 oligomers and RAD51 nucleoprotein filaments. BRCA2Exon 27 protects RAD51 nucleoprotein filaments formed on ssDNA from disruption by the BRC repeats and this protective effect is lost upon Ser3291 phosphorylation. Taken together, these observations suggest that BRCA2Exon 27 modulates BRC repeat function in order to maintain active nucleoprotein filaments during recombination-mediated DNA repair. Phosphorylation of BRCA2Exon 27 at the G2-M transition might terminate recombination by altering the balance in favor of the BRC repeat-dependent disassembly of RAD51 nucleoprotein filaments. Our findings provide a novel insight into the molecular mechanism whereby BRCA2 regulates the recombination-mediated repair of DNA double-strand breaks by RAD51.
RESULTS
BRCA2Exon 27 binds to oligomeric RAD51
The function of the human BRCA2Exon 27 sequence was investigated using the well-characterized ability of BRC4 to disrupt RAD51 oligomers and RAD51 nucleoprotein filaments as a useful control16,26. RAD51 interactions were initially confirmed through streptavidin pull-down experiments of human RAD51 using biotinylated BRCA2Exon 27, BRCA2Exon 27-Ph (a BRCA2Exon 27 sequence phosphorylated at residue Ser3291) and BRC4 peptides. Whilst BRCA2Exon 27 and BRC4 were able to bind RAD51, no interaction was detected between BRCA2Exon 27-Ph and RAD51 (Figure 2a). No interaction was observed with the E. coli RAD51-orthologue RecA protein, thus confirming the specificity of the BRCA2Exon 27 binding (data not shown). These results recapitulate previous findings28 and confirm that the BRCA2 sequences chosen for this study are suitable for investigation of their complexes with RAD51.
Figure 2.
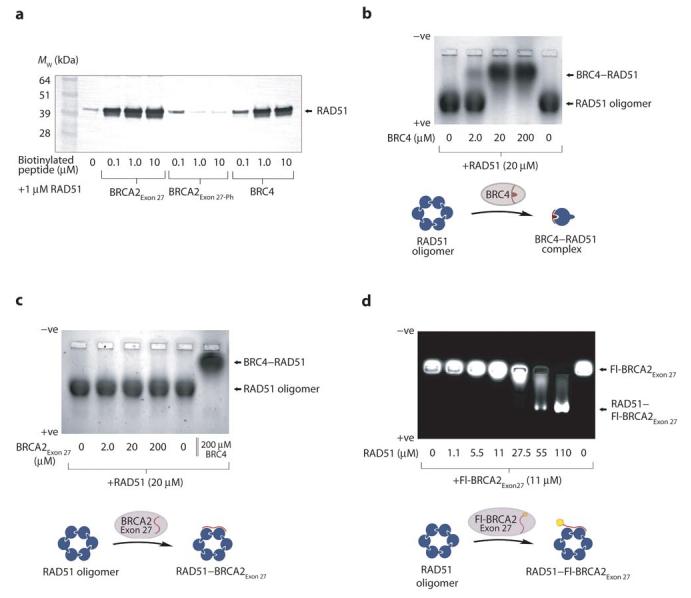
BRCA2Exon 27 binds to oligomeric RAD51. (a) Streptavidin pull-down of RAD51 with biotinylated BRCA2Exon 27, BRCA2Exon 27-Ph and BRC4 peptides, visualized by anti-RAD51 western blot. RAD51 binds to BRC4 and BRCA2Exon 27 but not to the BRCA2Exon 27 phosphorylated on Ser3291. (b) Native gel electrophoresis of RAD51 upon addition of increasing amounts of BRC4. (c) Native gel electrophoresis of RAD51 upon addition of increasing amounts of BRCA2Exon 27. (d) Native gel electrophoresis of fluorescein-labeled Fl-BRCA2Exon27 upon addition of increasing amounts of RAD51. A molar excess of RAD51 (lane 7) induces a complete shift of Fl-BRCA2Exon 27 to the position of oligomeric RAD51. The intermediate mobility of Fl-BRCA2Exon 27 in the presence of lower amounts of RAD51 (lanes 5-6) likely corresponds to RAD51–BRCA2Exon 27 complexes that have wholly or partially dissociated during electrophoresis.
BRC repeats are able to interfere with the oligomeric state of RAD51 by mimicking RAD51's self-association mechanism16,26. The lack of sequence similarity between BRCA2Exon 27 and the BRC repeat sequences suggests that their specific recognition of RAD51 is achieved via radically different mechanisms. We tested the effect of BRCA2Exon 27 on the oligomeric state of RAD51 by native gel electrophoresis. In isolation, recombinant RAD51 migrates as a single band corresponding to oligomeric species of net negative charge (Figure 2b). Pre-incubation of RAD51 with an equimolar amount of BRC4 caused a shift to a new species with reduced electrophoretic mobility, consistent with formation of a 1:1 BRC4–RAD51 complex (Figure 2b). The diminished mobility of the complex relative to free RAD51 is explained by the positively charged nature of the BRC4 peptide. In contrast, BRCA2Exon 27 had no effect upon the mobility of RAD51 (Figure 2c), suggesting that BRCA2Exon 27 binding does not alter the oligomeric state of RAD51. A fluorescently-labeled version of the BRCA2Exon 27 sequence (Fl-BRCA2Exon 27) was used for direct visualization of BRCA2Exon 27 binding to RAD51. Isolated Fl-BRCA2Exon 27 did not alter its position during gel electrophoresis (Figure 2d). However, the addition of RAD51 caused an increase in the electrophoretic mobility of Fl-BRCA2Exon 27 to the position of the RAD51 oligomer (Figure 2d), indicative of complex formation between BRCA2Exon 27 and RAD51.
To confirm these findings, BRCA2–RAD51 complexes were visualized through chemical cross-linking using BS3 (Figure 3a). RAD51 cross-linking produced molecular species corresponding to a series of multimers (Figure 3b), in agreement with its native oligomeric state. When pre-incubated with BRC4 a single new species was observed, corresponding to a 1:1 BRC4–RAD51 complex (Figure 3c), in keeping with the known functionality of BRC repeats16,26. Conversely, pre-incubation with Fl-BRCA2Exon 27 permitted the cross-linking of RAD51 oligomers. In addition, new species corresponding to the BRCA2Exon 27 peptide cross-linked to one or two RAD51 protomers were detected (Figure 3d). UV-visualization of Fl-BRCA2Exon 27 confirmed its cross-linking to the full spectrum of RAD51 oligomeric species (Figure 3e). These observations suggest that BRCA2Exon 27 interacts with RAD51 through a novel mechanism, distinct from that of the BRC repeats.
Figure 3.
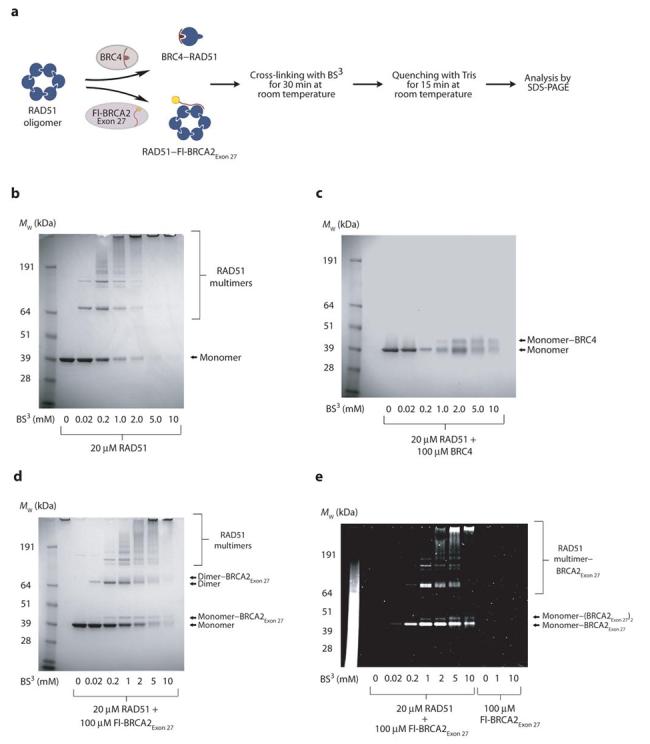
Cross-linking of RAD51, RAD51–BRC4 and RAD51–BRCA2Exon 27 complexes. (a) Complexes between RAD51 and BRC4 or BRCA2Exon 27 were cross-linked by BS3 and the resulting species were analyzed by SDS-PAGE. (b) The native oligomeric state of RAD51 is evident through the formation of several multimeric species. (c) An excess of BRC4 disassembles the RAD51 oligomer through the formation of a 1:1 complex between one BRC4 peptide and a RAD51 protomer. (d) SDS-PAGE of the cross-linked complex between RAD51 and Fl-BRCA2Exon 27 visualized by Coomassie staining. (e) SDS-PAGE of the cross-linked complex between RAD51 and Fl-BRCA2Exon 27, exposed to UV radiation to visualize the fluorescein-labeled peptide.
Specific recognition of oligomeric RAD51 by BRCA2Exon 27
The evidence so far indicated that BRCA2Exon 27 is capable of interacting with RAD51 without interfering with its oligomeric state. We next set out to investigate whether BRCA2Exon 27 binding actually relies on the oligomeric form of RAD51, or if BRCA2Exon 27 can bind to a single RAD51 protomer. This is particularly relevant if we consider that cellular RAD51 is likely to exist as either oligomers or monomers bound to the BRC repeats of BRCA213.
RAD51 oligomerisation depends upon the insertion of hydrophobic residues from the inter-domain linker of one protomer into matching pockets on the ATPase domain of the adjacent protomer19,26 (Figure 4a). Substitutions that invert the chemical nature of interface residues, such as mutation of Phe86 to glutamic acid (F86E), prevent RAD51 oligomerisation and thus create a constitutively monomeric form of RAD51. Indeed, an F86E RAD51 mutant fused to the Green Fluorescent Protein fails to oligomerise in vivo, whilst retaining an ability to bind BRCA213,26.
Figure 4.
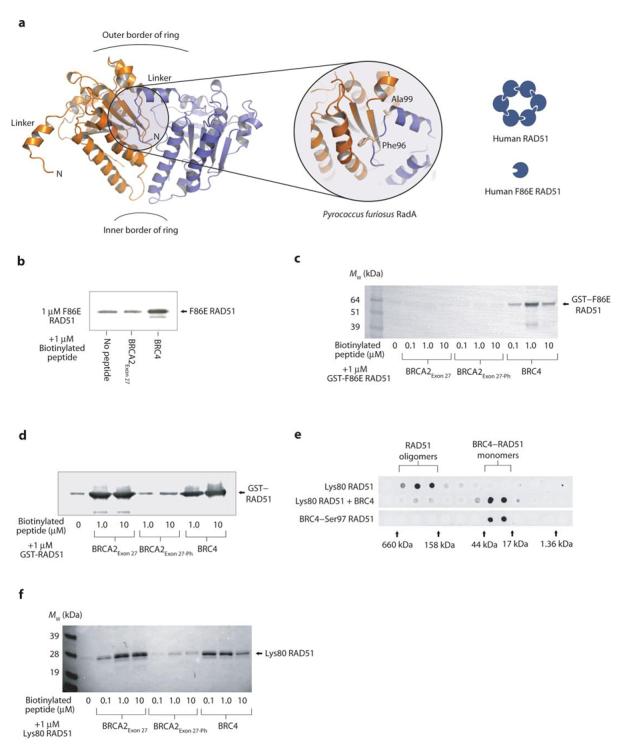
BRCA2Exon 27 binding requires an oligomeric RAD51 assembly. (a) The conserved oligomerisation interaction between the linker region of one protomer and the ATPase domain of the adjacent protomer is visualized in this view of two adjacent subunits of the Pyrococcus furiosus RadA ring structure (PDB entry 1PZN). Mutation of the interface residue Phe86, equivalent to Phe96 of P. furiosus RadA, provides a constitutively monomeric form of human RAD51. Furthermore, a construct of RAD51 containing residues 80-339 (Lys80 RAD51) lacks the N-terminal domain but retains the integrity of the oligomerisation interface. Streptavidin pull-downs of (b) F86E RAD51, (c) GST–F86E RAD51 fusion protein, and (d) GST–RAD51 fusion protein with biotinylated BRCA2Exon 27, BRCA2Exon 27-Ph and BRC4 peptides, visualized by anti-RAD51 western blot. (e) Gel filtration analysis of Lys80 RAD51, and Lys80 RAD51 incubated with excess BRC4, visualized by an anti-RAD51 dot-blot of eluted fractions. A control gel filtration run was performed using the previously described BRC4–S97 RAD51 fusion protein26, which approximates a monomeric complex between BRC4 and Lys80 RAD51. The elution positions of gel filtration standards are shown adjacent to the dot-blot. (f) Streptavidin pull-down of Lys80 RAD51 with biotinylated BRCA2Exon 27, BRCA2Exon 27-Ph and BRC4 peptides, visualized by anti-RAD51 western blot.
In order to determine whether the binding surface of BRCA2Exon 27 is contained within a single RAD51 protomer or spans adjacent protomers, we tested the ability of BRCA2Exon 27 to interact with a RAD51 mutant bearing the F86E substitution. As expected, the F86E RAD51 protein was retained on streptavidin beads when pre-incubated with biotinylated BRC4 (Figure 4b). However, the F86E RAD51 protein did not bind to BRCA2Exon 27 above the level of the control (Figure 4b). We confirmed these results using a GST–F86E RAD51 fusion protein, as we found that the presence of the tag improved the reduced stability of the mutant monomeric RAD51. As expected, BRC4 was able to bind to the GST–F86E RAD51 protein (Figure 4c). However, both BRCA2Exon 27 and BRCA2Exon 27-Ph failed to interact with the monomeric form of RAD51 (Figure 4c), whilst retaining binding to a GST–wild type RAD51 fusion protein (Figure 4d). This evidence indicates that BRCA2Exon 27 specifically recognizes the RAD51 oligomer and is incapable of associating with monomeric RAD51.
RAD51 ATPase domain is sufficient for BRCA2Exon 27 binding
RAD51 is a two-domain protein formed by a smaller, helical N-terminal domain linked to a larger, evolutionarily conserved, C-terminal ATPase domain. The dependency of the specific recognition by BRCA2Exon 27 on the oligomeric state of RAD51 suggests that the interaction is mediated by the ATPase domain. We designed an N-terminally truncated version of RAD51 (residues 80-339; Lys80 RAD51), that lacked the N-terminal domain but retained the linker region and ATPase domain necessary for self-association. The Lys80 RAD51 protein could be selectively precipitated by spermidine in a similar way to full-length RAD51 (data not shown) and its oligomeric status was confirmed by gel filtration analysis (Figure 4e). The interaction of BRC4 with the Lys80 RAD51 protein was not affected, as would be expected since the BRC4 binding site is located in the ATPase domain (Figure 4f). We found that BRCA2Exon 27 retained its ability to interact with the Lys80 RAD51 protein, and that the interaction was abrogated by the Ser3291 phosphorylated form of BRCA2Exon 27 (Figure 4f). Thus, the N-terminal domain of RAD51 is not required for the interaction with the exon 27 region of BRCA2. Taken together, the evidence demonstrates that the interaction with BRCA2Exon 27 is mediated by oligomeric ATPase domains of RAD51.
BRCA2Exon 27 binds to the RAD51 nucleoprotein filament
The effect of BRCA2Exon 27 on the RAD51 nucleoprotein filament was tested by electrophoretic mobility shift assay (EMSA) (Figure 5a). Adding increasing amounts of BRCA2Exon 27 to a RAD51 filament formed on φX174 virion ssDNA caused a minor retardation in mobility of the RAD51–DNA complex, with an upward smearing of the band (Figure 5b). As observed for free RAD51, the effect of BRCA2Exon 27 on the mobility of the RAD51–DNA complex was only slight. Thus, BRCA2Exon 27 binding does not eliminate the filament structure, in contrast to the ability of BRC4 to depolymerise RAD51 filaments.
Figure 5.
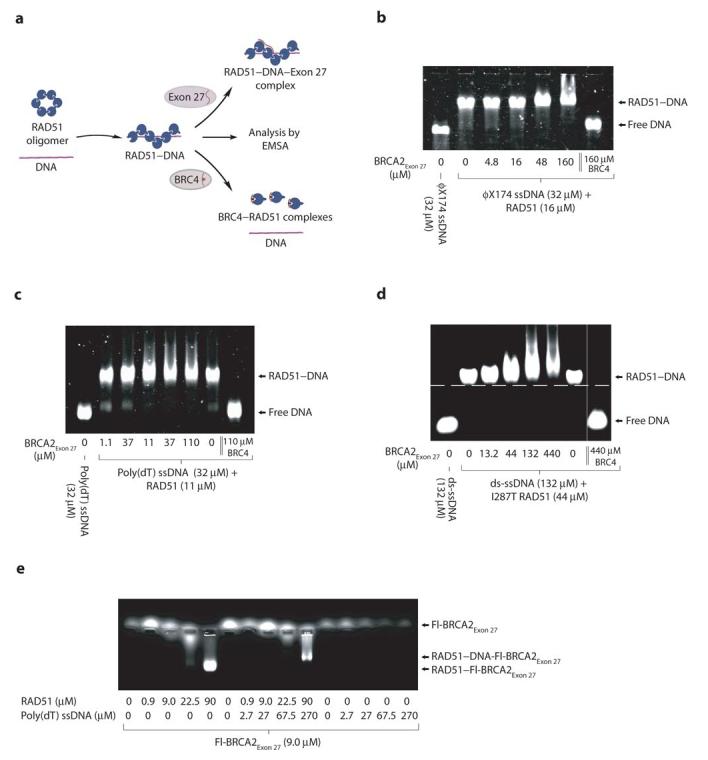
BRCA2Exon 27 binds to RAD51 nucleoprotein filaments. (a) Electrophoretic mobility shift assay testing the effect of BRCA2Exon 27 upon RAD51 nucleoprotein filaments formed on (b) φX174 virion ssDNA, (c) poly(dT) ssDNA, and (d) ds-ssDNA. In the case of the ds-ssDNA substrate, the I287T RAD51 protein was used. In each case, the RAD51 nucleoprotein filaments were also incubated separately with BRC4, as a control. (e) Electrophoretic mobility of fluorescein-labeled Fl-BRCA2Exon 27 upon the addition of RAD51, preformed nucleoprotein filaments between RAD51 and poly(dT) ssDNA, and poly(dT) ssDNA.
As the large size of the φX174 virion ssDNA might have masked the mobility shift induced by BRCA2Exon 27, we tested the effect of BRCA2Exon 27 upon RAD51 filaments formed on a shorter, linear poly(dT) ssDNA substrate. As observed for the φX174 virion ssDNA, BRCA2Exon 27 induced a slight retardation and upward smearing of the RAD51–poly(dT) complex (Figure 5c). In addition, we tested the effect of BRCA2Exon 27 on RAD51 bound to a 24 base-pair dsDNA substrate with a 24-base 3'-ssDNA tail, which mimics the ds-ssDNA junction formed at damaged loci in vivo. In order to form a stable complex with DNA, we used a RAD51 protein bearing a I287T mutation that is known to increase the affinity towards DNA of yeast Rad5132. BRCA2Exon 27 induced a clear retardation in the mobility of the RAD51–DNA complex (Figure 5d), thus demonstrating BRCA2Exon 27 binding to the nucleoprotein filament.
To further confirm the interaction of BRCA2Exon 27 with the RAD51 filament, EMSA was performed using the fluorescein-labeled Fl-BRCA2Exon 27 (Figure 5e). Isolated Fl-BRCA2Exon 27 did not display an appreciable electrophoretic mobility. The addition of increasing amounts of RAD51 induced a mobility shift of Fl-BRCA2Exon 27 to the position of the RAD51 oligomer (Figure 5e). Likewise, the addition of preformed RAD51–poly(dT) filament to Fl-BRCA2Exon 27 induced a change in its mobility to the position of the RAD51–DNA complex (Figure 5e). No mobility shift was observed upon the addition of DNA alone. These findings confirm that BRCA2Exon 27 can associate with the RAD51 oligomer and the RAD51 nucleoprotein filament.
BRCA2Exon 27 protects RAD51 filaments from BRC4 disassembly
The fact that the exon 27 and BRC repeat sequences of BRCA2 have different modes of interaction with RAD51 suggests that they might play complementary roles in regulating RAD51 activity during homologous recombination. We tested this hypothesis by incubating with BRCA2Exon 27 a RAD51 nucleoprotein filament formed on φX174 virion ssDNA, prior to the addition of BRC4 (Figure 6a). Interestingly, BRCA2Exon 27 exerted a moderate but clear effect in preventing the disruption of the RAD51 nucleoprotein filament by BRC4 (Figure 6b). Furthermore, the protection was lost when the nucleoprotein filament was incubated with BRCA2Exon 27-Ph (Figure 6c), suggesting that the observed effect is a consequence of the specific interaction between BRCA2Exon 27 and RAD51.
Figure 6.
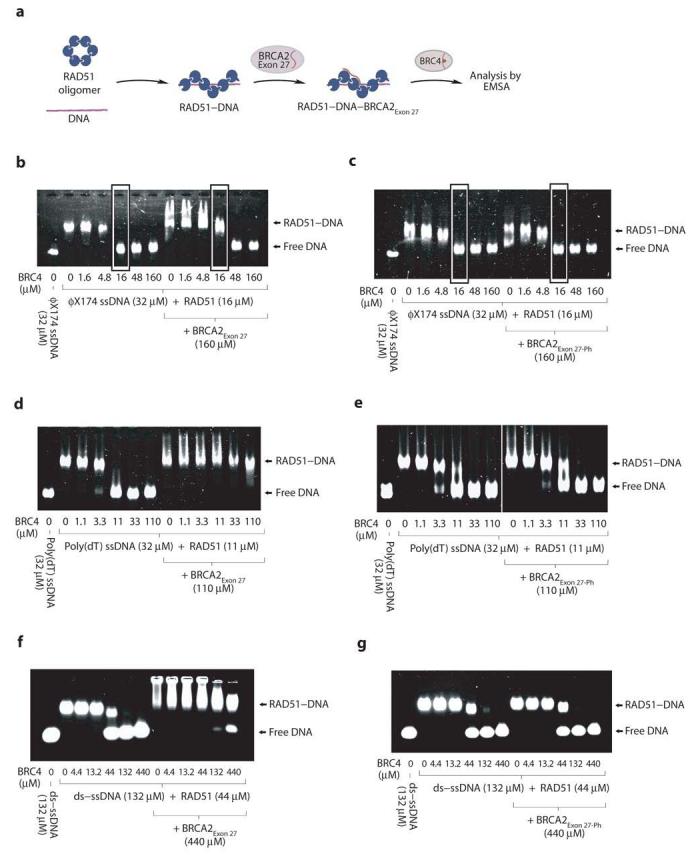
BRCA2Exon 27 protects RAD51 nucleoprotein filaments. (a) Electrophoretic mobility shift assay testing the ability of BRCA2Exon 27 and BRCA2Exon 27-Ph to protect RAD51 nucleoprotein filaments from disruption by BRC4. (b) Protection assay of RAD51 filaments formed on φX174 virion ssDNA using BRCA2Exon 27 and (c) BRCA2Exon 27-Ph. Lanes where the effect of BRCA2Exon 27 and BRCA2Exon 27-Ph is visible are highlighted by grey boxes. (d) Protection assay of RAD51 filaments formed on poly(dT) ssDNA using BRCA2Exon 27 and (e) BRCA2Exon 27-Ph. (f) Protection assay of filaments formed between I287T RAD51 and ds-ssDNA using BRCA2Exon 27 and (g) BRCA2Exon 27-Ph.
The protection afforded by BRCA2Exon 27 against BRC4-dependent dissolution of the RAD51–φX174 virion ssDNA filament was not complete and required an excess of BRCA2Exon 27 over BRC4. In order to minimize possible effects due to the secondary structure of the DNA substrate, we chose to repeat the experiment using the poly(dT) template. Strikingly, we found that BRCA2Exon 27 completely protected the RAD51–poly(dT) filament from an equimolar amount of BRC4 (Figure 6d). Pre-incubation with BRCA2Exon 27-Ph provided no protection (Figure 6e), confirming that the observed effect depends on the specific RAD51 recognition by BRCA2Exon 27. Moreover, BRCA2Exon 27 exerted a protective effect upon a nucleoprotein filament formed between the ds-ssDNA substrate and I287T RAD51 (Figure 6f), which was also lost upon Ser3291 phosphorylation (Figure 6g).
The protecting function of BRCA2Exon 27 was further tested through visualization of the nucleoprotein filament between RAD51 and poly(dT) by electron microscopy (Figure 7a). The presence of BRCA2Exon 27 did not cause disruption of the nucleoprotein filaments or macroscopic modification of its overall morphology (Figure 7b), suggesting that BRCA2Exon 27 does not alter its conformation. In contrast, the addition of BRC4 completely depolymerised the RAD51 nucleoprotein filaments (Figure 7c), as previously reported16. Upon pre-incubation with BRCA2Exon 27, the disruptive effect of BRC4 was entirely prevented and the RAD51 nucleoprotein filaments preserved their native morphology (Figure 7d). Thus, we have demonstrated by EMSA and electron microscopy that BRCA2Exon 27 protects RAD51–poly(dT) ssDNA nucleoprotein filaments from the known disruptive effect of BRC repeats.
Figure 7.
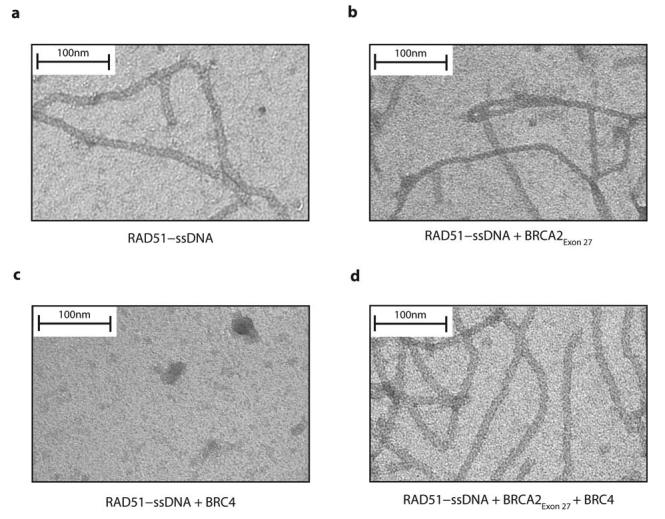
Electron microscopy of RAD51 nucleoprotein filaments. (a) RAD51–ssDNA filaments formed between RAD51 and poly(dT). (b) Incubation of pre-formed RAD51–poly(dT) filaments with BRCA2Exon 27. (c) Incubation of pre-formed RAD51–poly(dT) filaments with BRC4. (d) Incubation of RAD51–poly(dT) complexes with BRCA2Exon 27 prior to the addition of BRC4.
BRCA2Exon 27 protection is unique to RAD51–ssDNA filaments
We further dissected the specificity of the observed protective function of BRCA2Exon 27: in the absence of DNA, BRCA2Exon 27 was unable to prevent the conversion of oligomeric RAD51 to the 1:1 RAD51–BRC4 complex form (Figure 8a). The protective effect conferred upon RAD51–poly(dT) filaments was maintained when ATP was replaced by the non-hydrolysable nucleotide analogue AMP-PNP, indicating that the BRCA2Exon 27 function is independent of ATP hydrolysis (Figure 8b; compare with Figure 6d). However, we observed no protective effect of BRCA2Exon 27 when pre-incubated with RAD51 filaments containing either circular φX174 dsDNA or a linear, 1.2kbp dsDNA (Figure 8c, d). As RAD51 ATPase activity is known to be differentially activated by ssDNA and dsDNA33, we tested the effect of BRCA2Exon 27 on RAD51 filaments formed on the linear 1.2kbp dsDNA substrate in the presence of the non-hydrolysable nucleotide analogue AMP-PNP (Figure 8e). Whilst higher concentrations of BRC4 were required to dissolve the RAD51–dsDNA filaments in the presence of AMP-PNP, pre-incubation with BRCA2Exon 27 only mildly potentiated this effect (Figure 8e). Our data thus show that, whilst BRCA2Exon 27 can interact with RAD51 in the absence of DNA, its protective effect against disruption by BRC repeats is specific to nucleoprotein filaments formed on ssDNA. The specific action of BRCA2Exon 27 towards RAD51 complexes containing ssDNA is particularly interesting as the RAD51 filaments formed on ssDNA are thought to be the active filament form in recombination-mediated repair.
Figure 8.
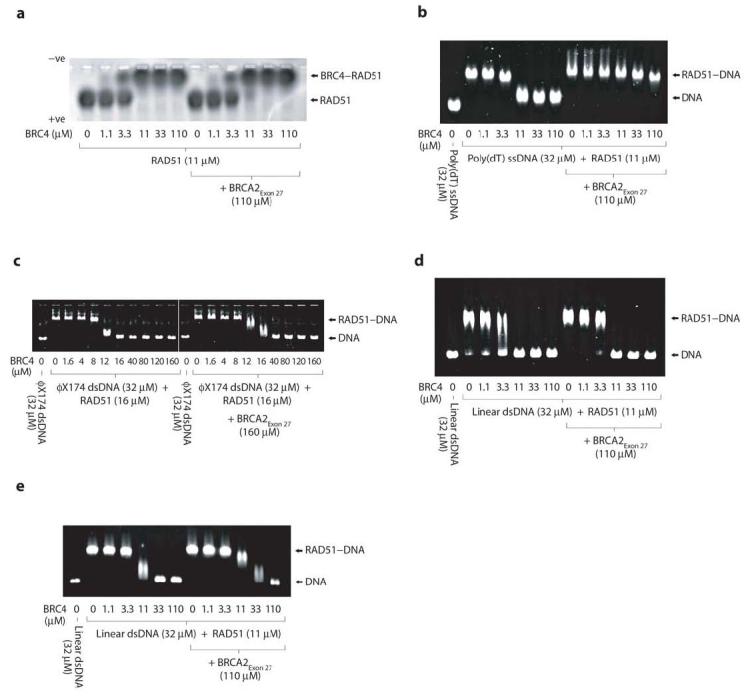
BRCA2Exon 27 protection is specific for ssDNA and independent of ATP hydrolysis. Electrophoretic mobility shift assay testing the ability of BRCA2Exon 27 to protect (a) the RAD51 oligomer in the absence of DNA, and nucleoprotein filaments formed between RAD51 and (b) poly(dT) ssDNA in the presence of AMP-PNP, (c) φX174 dsDNA in the presence of ATP, (d) 1.2kbp linear dsDNA in the presence of ATP and (e) 1.2kbp linear dsDNA in the presence of AMP-PNP. The dependence on nucleotide hydrolysis of the BRCA2Exon 27 protection effect can be assessed by comparing panel b with Figure 6d.
DISCUSSION
The BRCA2 tumor suppressor protein contains two distinct regions, unrelated by amino acid sequence and distant in location, that possess the ability to bind RAD51. Whereas the interaction between RAD51 and the BRC repeat sequences in the central region of BRCA2 has been well characterized, much less is known about the mode of interaction of RAD51 with the C-terminal binding site coded within the exon 27 of BRCA2, BRCA2Exon 27. Here we have provided a first account of the mechanism by which BRCA2Exon 27 interacts with RAD51. We find that a 36 amino-acid sequence within a highly conserved region of exon 27 specifically recognizes and interacts with RAD51 oligomers and RAD51 nucleoprotein filaments: unlike the BRC repeats, BRCA2Exon 27 does not bind to a monomeric RAD51 mutant and is unable to interfere with the oligomeric state of RAD51. Furthermore, we show that the ATPase domain of RAD51 is sufficient for the interaction with BRCA2Exon 27, whereas the N-terminal domain is not required for binding. These observations can be integrated in a model in which BRCA2Exon 27 occupies the groove formed by adjacent protomers of oligomeric RAD51 assemblies, making specific contacts with the ATPase domains of neighboring subunits. The observation that mutations of Ser3291 to alanine and glutamic acid abrogate RAD51 binding implicates the hydroxyl moiety of the serine in a crucial hydrogen bonding interaction, possibly required in order to maintain the exon 27 conformation necessary for RAD51 binding.
The conservative mode of RAD51 binding by BRCA2Exon 27 contrasts with the ability of BRC repeats to disrupt RAD51 oligomers and nucleoprotein filaments through the formation of 1:1 BRC:RAD51 complexes8,16,26. Thus, BRCA2 can bind RAD51 in multiple ways: via the disruptive action of BRC repeats with a potential RAD51 remodeling function, through a non-disruptive association of the BRC repeats with RAD51 filaments, and via the conservative interaction of the exon 27 region that stabilizes the RAD51–ssDNA nucleoprotein filament. We have further defined a functional interplay between RAD51-binding modes by demonstrating that BRCA2Exon 27 can protect RAD51 nucleoprotein filaments from disassembly by BRC repeats. The effect is specific to ssDNA as free RAD51 and RAD51 filaments formed on dsDNA experience no protection upon binding to BRCA2Exon 27. These observations suggest that once the BRC repeats have remodeled free oligomeric RAD51 into RAD51 nucleoprotein filaments, BRCA2Exon 27 might stabilize and protect the filaments from subsequent BRC repeat-dependent disruption. Interestingly, whilst BRC repeats have been identified in BRCA2 molecules from a wide range of eukaryotes, the C-terminal RAD51-binding motif is specific to vertebrate BRCA2 sequences. It is possible that the BRCA2Exon 27 sequence of vertebrate proteins might have evolved to protect RAD51 nucleoprotein filaments from the disruptive effects of their BRC repeats. In this respect, it will be interesting to compare the disruptive potency towards RAD51 nucleoprotein filaments of vertebrate BRC repeats with that of the single BRC repeat found in primitive BRCA2 proteins lacking the BRCA2Exon 27 RAD51-binding motif.
Homologous recombination is active during the S- and G2-phases of the cell cycle and is terminated at the G2-M transition in order to prepare for cellular entry into mitosis. As phosphorylation of Ser3291 at the G2-M transition abrogates the ability of BRCA2Exon 27 to bind RAD51, it has been proposed that this phosphorylation event is in part responsible for terminating recombination prior to mitosis28. Here, we present a mechanistic explanation for this observation. It has previously been reported that BRC repeats may interact with RAD51 in ways that can interfere or be compatible with its oligomeric state, and that both binding modes are likely to be relevant to recombination-mediated repair. Within the context of the housing exon 11 sequence, BRCA2BRC1-8 can stimulate strand exchange through a direct association with the RAD51 filament11. However, when present in excess, individual BRC repeats16,27 and the BRCA2BRC1-8 region (ORD & LP, unpublished observations) can disrupt RAD51 filaments. Remodeling cellular RAD51 into nucleoprotein filaments likely represents an important function of the BRC repeats, but it also raises the possibility that the BRC repeats may inadvertently disrupt RAD51 filaments that are engaged in repair. This highlights a requirement for a cellular mechanism that permits remodeling of free oligomeric RAD51 at the onset of repair, but protects established RAD51 filaments during active repair. The work presented here suggests that the exon 27 region of BRCA2 may fulfill such protective function: we find that BRCA2Exon 27 exercises a specific stabilizing effect on RAD51–ssDNA filaments against interference from BRC repeats. At the G2-M transition, CDK phosphorylation removes the ability of BRCA2Exon 27 to preserve the integrity of RAD51–ssDNA filaments from BRC repeat-dependent depolymerisation. We propose that this shift in balance between the disruptive and protective functions of BRCA2 induces filament disassembly upon cellular entry into mitosis, thus terminating recombination.
METHODS
Protein Expression and Purification
Full-length human RAD51, Glu18 I287T RAD51 and Lys80 RAD51 were expressed in BL21(DE3) cells as untagged polypeptides and purified by a previously published method34. GST–RAD51 and GST–F86E RAD51 were expressed in pGAT3 vectors35 in BL21(DE3) cells and purified by Ni-NTA (Qiagen). The GST-tag was removed from GST–F86E RAD51 by incubation with TEV protease (Invitrogen); F86E RAD51 was subsequently purified by incubation with Ni-NTA resin (Qiagen). Mutations were introduced to the RAD51 sequence by site-directed mutagenesis (Stratagene).
Peptide Synthesis
Peptides containing N-terminal biotin or fluorescein labels were synthesized by solid phase peptide synthesis (Alta Bioscience, University of Birmingham). BRCA2Exon 27 corresponds to ALDFLSRLPLPPPVSPICTFVSPAAQKAFQPPRSCG (human BRCA2 residues 3,270-3,305, BRCA2Exon 27-Ph contains a phosphorylated serine residue at position 3291, BRC4 corresponds to EKIKEPTLLGFHTASGKKVKIAKESLDKVKNLFDE (human BRCA2 residues 1,514-1,548). During early experimental work, the two cysteine residues of BRCA2Exon 27 were found to be highly susceptible to oxidation, which could be remedied by the presence of 20 mM DTT.
Biotin-Streptavidin Pull-down Assay
Biotinylated peptides at 0.1, 1 and 10 μM were incubated with 1 μM RAD51 (RAD51, GST–RAD51, GST–F86E RAD51, F86E RAD51 and Lys80 RAD51) in 50 mM Tris pH 8.0, 150 mM NaCl, 1% (v/v) Triton X-100, 10 mg ml−1 BSA, 20 mM DTT at 4°C for 3 hours. 100 μl streptavidin-iron oxide particles (Sigma-Aldrich) were equilibrated in this buffer and incubated with the protein-peptide complexes at 4°C for 3 hours. Complexes bound to the streptavidin-iron oxide particles were immobilized using a magnetic separation unit, thoroughly washed in 50 mM Tris pH 8.0, 500 mM NaCl, 1% (v/v) Triton X-100, 20 mM DTT and suspended in SDS-PAGE loading buffer. Samples were analyzed by SDS-PAGE and western blot using 2 μg rabbit polyclonal anti-RAD51 (H-92) IgG (for full length or GST-F86E RAD51) or goat anti-RAD51 (I-20) IgG (for Lys80 RAD51-ΔN) (Santa Cruz Biotech), and 2 μg alkaline phosphatase conjugated anti-rabbit IgG or anti-goat IgG (Promega). Blots were developed using SigmaFAST™ BCIP/NBT (5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium).
Gel Filtration Analysis
300 μl samples containing 10 μM Lys80 RAD51, 10 μM Lys80 RAD51 incubated with 100 μM BRC4, and 10 μM BRC4–Ser97 RAD51 fusion protein (as previously described26). Samples were loaded onto a Superdex 200 10/300 GL column (Amersham) equilibrated in 20 mM Tris pH 8.0, 150 mM KCl, 2 mM DTT. Control runs were performed using gel filtration standards (Bio-Rad). Following isocratic elution, 1 μl of each fraction was dotted onto nitrocellulose membrane (Invitrogen) and RAD51 was detected with 0.4 μg goat anti-RAD51 (C-20) IgG (Santa Cruz Biotech) and 0.08 μg HRP-conjugated anti-goat IgG (Promega), using the ECL advance detection system (Amersham).
Native Gel Electrophoresis
20 μM RAD51 was incubated with 0, 2, 20 and 200 μM BRC4 or BRCA2Exon 27 in 20 mM Hepes pH 7.5, 150 mM KCl, 20 mM DTT for 15 min at 4°C. Similarly, 11 μM Fl-BRCA2Exon 27 was incubated with 0, 1.1, 5.5, 11, 55 and 110 μM RAD51 in 20 mM Hepes pH 7.5, 150 mM KCl, 20 mM DTT for 15 min at 4°C. Samples were analyzed on a 1% (w/v) agarose gel in 0.5x TBE at 35 mV for 90 min at 4°C. The Fl-BRCA2Exon 27 peptide was visualized by exposure to UV radiation, and proteins were detected by staining with coomassie. For the protection experiments in the absence of DNA, 11 μM RAD51 was incubated with 110 μM BRCA2Exon 27 or BRCA2Exon 27-Ph prior to the addition of 0, 1.1, 3.3, 11, 33 and 110 μM BRC4. Samples were analyzed on a 0.3% (w/v) agarose gel in 1x TBE at 20 mV for 4 hours at 4°C.
Chemical Cross-Linking
Proteins and peptides were dialyzed in 20 mM Hepes pH 7.5, 150 mM KCl, 20 mM DTT. 20 μM RAD51 was incubated with 100 μM BRC4 or 100 μM Fl-BRCA2Exon 27 for 30 min at 4°C. Complexes were cross-linked by incubation with 0, 0.02, 0.2, 1, 2, 5 or 10 mM BS3 (Sigma-Aldrich) at room temperature for 30 min. Cross-linking was terminated by adding 80 mM Tris pH 8.0 for 15 min and analyzed by SDS-PAGE. The Fl-BRCA2Exon 27 peptide was detected by exposing the gel to UV radiation, protein was detected by staining with SimplyBlue™ SafeStain (Invitrogen).
Electrophoretic Mobility Shift Assay (EMSA)
RAD51 was incubated with φX174 virion ssDNA (NEB), φX174 dsDNA (NEB), poly(dT) ssDNA (observed length: 174-213 bases; GE Healthcare) and 1.2kbp linear dsDNA substrates (described in Supplementary Methods online; concentrations provided in figures) in 50 mM TEA pH 7.5, 2 mM ATP (or 0.5 mM AMP-PNP), 0.5 mM MgCl2, 2 mM DTT for 15 min at 4°C. BRCA2Exon 27 (or Fl-BRCA2Exon 27)or BRC4 were added (concentrations provided in figures) for a further 15 min at 4°C. For protection experiments, the protein–DNA complexes were incubated with BRCA2Exon 27 or BRCA2Exon 27-Ph for 15 min at 4°C prior to the addition of BRC4 and further incubation for 15 min at 4°C (concentrations provided in figures). Filaments containing φX174 ssDNA, φX174 dsDNA or 1.2kb dsDNA were analyzed by electrophoresis on a 0.3% (w/v) agarose gel in 1x TBE at 20 mV for 4 hours at 4°C. Filaments containing poly(dT) ssDNA were analysed on a 0.5% (w/v) agarose gel in 1x TBE at 20 mV for 4 hours at 4°C. DNA was detected by incubation with SYBR Gold (Invitrogen), and protein was detected by Coomassie staining; Fl- BRCA2Exon 27 was detected by exposure to UV.
Similar assays were performed using Glu18 I287T RAD51 and ds-ssDNA (a 24bp dsDNA with 24-base 3'-overhang; described in Supplementary Methods) using a modified method (concentrations provided in figures): protein–DNA complexes were formed in 40 mM Tris pH 7.5, 2 mM ATP, 1 mM MgCl2 and samples were analyzed on a 1% (w/v) agarose gel in 0.5x TBE at 30 mV for 2 hours at 4°C.
Electron microscopy
11 μM RAD51 was incubated with 32 μM poly(dT) ssDNA in 50 mM TEA pH 7.5, 2 mM ATP, 0.5 mM MgCl2, 2 mM DTT for 15 min at 4°C. 110 μM BRCA2Exon 27 or BRC4 were added for a further 15 min at 4°C; in protection experiments, 110 μM BRC4 was added following a 15 min incubation with 110 μM BRCA2Exon 27. Negative staining was performed using 0.1% (w/v) uranyl acetate, and grids were viewed in a CM-100 transmission electron microscope (Philips).
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Tom Blundell, Sebastian Klinge, Mahmud Shivji, Eeson Rajendra, Nabieh Ayoub and Ashok Venkitaraman for comments and advice. We thank Nicole Allcott and Takahiko Yamamoto for their contributions in the early stages of this work and Jeremy Skepper (Dept. of Anatomy, University of Cambridge) for assistance with electron microscopy. This work was supported by a Wellcome Trust Senior Fellowship in basic biomedical sciences to L.P. and a BBSRC PhD studentship to O.R.D.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare they have no competing financial interests.
References
- 1.Rahman N, Stratton M. The genetics of breast cancer susceptibility. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- 2.Nathanson KL, Wooster R, Weber BL, Nathanson KN. Breast cancer genetics: what we know and what we need. Nat Med. 2001;7:552–6. doi: 10.1038/87876. [DOI] [PubMed] [Google Scholar]
- 3.Connor F, et al. Tumorigenesis and a DNA repair defect in mice with a truncating BRCA2 mutation. Nat. Genet. 1997;17:423–430. doi: 10.1038/ng1297-423. [DOI] [PubMed] [Google Scholar]
- 4.Patel KJ, et al. Involvement of BRCA2 in DNA repair. Mol Cell. 1998:347–357. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 5.Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell. 2001;2:511–518. doi: 10.1016/s1097-2765(01)00174-5. [DOI] [PubMed] [Google Scholar]
- 6.Yu VP, et al. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;11:1400–1406. [PMC free article] [PubMed] [Google Scholar]
- 7.Sharan SK, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–10. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 8.Chen CF, Chen PL, Zhong Q, Sharp ZD, Lee WH. Expression of BRC repeats in breast cancer cells disrupts the BRCA2-Rad51 complex and leads to radiation hypersensitivity and loss of G(2)/M checkpoint control. J Biol Chem. 1999;274:32931–5. doi: 10.1074/jbc.274.46.32931. [DOI] [PubMed] [Google Scholar]
- 9.Yuan SS, et al. BRCA2 is required for ionizing radiation-induced assembly of Rad51 complex in vivo. Cancer Res. 1999;59:3547–51. [PubMed] [Google Scholar]
- 10.Yang H, et al. BRCA2 function in DNA binding and recombination from a BRCA2-DSS1-ssDNA structure. Science. 2002;297:1837–48. doi: 10.1126/science.297.5588.1837. [DOI] [PubMed] [Google Scholar]
- 11.Shivji MK, et al. A region of human BRCA2 containing multiple BRC repeats promotes RAD51-mediated strand exchange. Nucleic Acids Res. 2006;34:4000–11. doi: 10.1093/nar/gkl505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–7. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 13.Yu DS, et al. Dynamic control of Rad51 recombinase by self-association and interaction with BRCA2. Mol Cell. 2003;12:1029–41. doi: 10.1016/s1097-2765(03)00394-0. [DOI] [PubMed] [Google Scholar]
- 14.Pellegrini L, Venkitaraman A. Emerging functions of BRCA2 in DNA recombination. Trends Biochem Sci. 2004;29:310–6. doi: 10.1016/j.tibs.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Chen PL, et al. The BRC repeats in BRCA2 are critical for RAD51 binding and resistance to methyl methanesulfonate treatment. Proc Natl Acad Sci U S A. 1998;95:5287–92. doi: 10.1073/pnas.95.9.5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davies AA, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–82. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 17.Wong AK, Pero R, Ormonde PA, Tavtigian SV, Bartel PL. RAD51 interacts with the evolutionarily conserved BRC motifs in the human breast cancer susceptibility gene brca2. J Biol Chem. 1997;272:31941–4. doi: 10.1074/jbc.272.51.31941. [DOI] [PubMed] [Google Scholar]
- 18.Mizuta R, et al. RAB22 and RAB163/mouse BRCA2: proteins that specifically interact with the RAD51 protein. Proc Natl Acad Sci U S A. 1997;94:6927–32. doi: 10.1073/pnas.94.13.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shin DS, et al. Full-length archaeal Rad51 structure and mutants: mechanisms for RAD51 assembly and control by BRCA2. Embo J. 2003;22:4566–76. doi: 10.1093/emboj/cdg429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kinebuchi T, et al. Structural basis for octameric ring formation and DNA interaction of the human homologous-pairing protein dmc1. Mol Cell. 2004;14:363–74. doi: 10.1016/s1097-2765(04)00218-7. [DOI] [PubMed] [Google Scholar]
- 21.Conway AB, et al. Crystal structure of a Rad51 filament. Nat Struct Mol Biol. 2004 doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- 22.Wu Y, Qian X, He Y, Moya IA, Luo Y. Crystal structure of an ATPase-active form of Rad51 homolog from methanococcus voltae: insights into potassium-dependence. J Biol Chem. 2004 doi: 10.1074/jbc.M411093200. [DOI] [PubMed] [Google Scholar]
- 23.Wu Y, He Y, Moya IA, Qian X, Luo Y. Crystal Structure of Archaeal Recombinase RadA: A Snapshot of Its Extended Conformation. Molecular Cell. 2004;15:423–435. doi: 10.1016/j.molcel.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 24.Wyman C, Kanaar R. Homologous recombination: down to the wire. Curr Biol. 2004;14:R629–31. doi: 10.1016/j.cub.2004.07.049. [DOI] [PubMed] [Google Scholar]
- 25.Story RM, Weber IT, Steitz TA. The structure of the E. coli recA protein monomer and polymer. Nature. 1992;355:318–25. doi: 10.1038/355318a0. [DOI] [PubMed] [Google Scholar]
- 26.Pellegrini L, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–93. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 27.Galkin VE, et al. BRCA2 BRC motifs bind RAD51-DNA filaments. Proc Natl Acad Sci U S A. 2005;102:8537–42. doi: 10.1073/pnas.0407266102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esashi F, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 29.Donoho G, et al. Deletion of Brca2 exon 27 causes hypersensitivity to DNA crosslinks, chromosomal instability, and reduced life span in mice. Genes Chromosomes Cancer. 2003;36:317–31. doi: 10.1002/gcc.10148. [DOI] [PubMed] [Google Scholar]
- 30.McAllister KA, et al. Cancer Susceptibility of Mice with a Homozygous Deletion in the COOH-Terminal Domain of the Brca2 Gene. Cancer Res. 2002;62:990–994. [PubMed] [Google Scholar]
- 31.Atanassov BS, Barrett JC, Davis BJ. Homozygous germ line mutation in exon 27 of murine Brca2 disrupts the Fancd2-Brca2 pathway in the homologous recombination-mediated DNA interstrand cross-links' repair but does not affect meiosis. Genes Chromosomes Cancer. 2005;44:429–37. doi: 10.1002/gcc.20255. [DOI] [PubMed] [Google Scholar]
- 32.Fortin GS, Symington LS. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. Embo J. 2002;21:3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tombline G, Fishel R. Biochemical characterization of the human RAD51 protein. I. ATP hydrolysis. J Biol Chem. 2002;277:14417–25. doi: 10.1074/jbc.M109915200. [DOI] [PubMed] [Google Scholar]
- 34.Baumann P, Benson FE, Hajibagheri N, West SC. Purification of human Rad51 protein by selective spermidine precipitation. Mutat Res. 1997;384:65–72. doi: 10.1016/s0921-8777(97)00028-1. [DOI] [PubMed] [Google Scholar]
- 35.Peranen J, Rikkonen M, Hyvonen M, Kaariainen L. T7 vectors with modified T7lac promoter for the expression of proteins in E. coli. Anal Biochem. 1996;236:371–373. doi: 10.1006/abio.1996.0187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


