Abstract
Kaposi’s sarcoma (KS) develops in a variety of clinical states and is the most common tumor seen in patients with HIV-1 infection. KS develops as a multifocal mucocutaneous disease with subsequent spread to visceral organs, and it has been argued to be a benign proliferation caused by its multifocality at initial presentation, lack of aneuploidy, and spontaneous regression upon withdrawal of immunosuppressive agents in iatrogenically induced disease. We wished to determine whether KS lesions are clonal, indicative of a true neoplasm. Also, we tested whether multifocal KS lesions are clonally related, derived from a common progenitor cell or of independent cellular origin. We studied the X-chromosome inactivation pattern of the human androgen receptor gene in tumor biopsies of women with KS. This procedure tests for the clonality of a tissue specimen, a hallmark of neoplasia. Each specimen was microdissected to minimize normal cell contamination. Of 12 evaluable cases, 10 were HIV-seropositive and 2 were HIV-seronegative. Twenty-four biopsies from the 12 patients were examined. Five cases were consistent with individual KS lesions being clonal. In two cases, multiple KS specimens derived from the individual patients had different androgen receptor alleles inactivated, proving unequivocally that these KS lesions arose independently from distinct transformed cells. In seven cases, only a polyclonal pattern of inactivation was observed, whereas two others had tumor areas of both clonal and polyclonal inactivation patterns. These findings suggest that KS can be a clonal neoplasm, and in some of the cases multiple KS lesions in a given patient can arise from independent cellular origins and acquire clonal characteristics. The polyclonal inactivation pattern observed in other KS lesions may represent a premalignant stage or false negative results.
Keywords: clonality/HIV/androgen receptor
The classical form of Kaposi’s sarcoma (KS) is a rare vascular tumor affecting elderly men of Mediterranean and eastern European Jewish ancestry (1). It is predominantly localized to the lower extremities and has an indolent course. A second form of KS is found among young black males in Central and Eastern Africa (2). This form often involves the lymph nodes, invades deeper organs, and spreads to visceral sites resulting in high morbidity and mortality (2). KS is also observed in renal transplant recipients (3). In this form, the tumor can regress upon withdrawal of the drugs used to prevent organ rejection. The fourth (epidemic) form of KS is seen in patients infected with HIV-1 (4, 5) and is predominantly found in homosexual or bisexual men where it develops in 15–30% of patients during the course of infection (4, 5). KS is the most common tumor seen in patients with HIV-1 infection and can become widely disseminated with the involvement of visceral organs.
Histopathologically, KS is indistinguishable in its various forms. All show evidence of the KS herpes virus (KSHV or HHV-8), suggesting a common pathogenesis (6). The tumor is localized to the dermis and is characterized by a proliferation of aberrant vascular structures lined by spindle cells, which are the neoplastic cells (5). Bundles of spindle cells within the vascular structures predominate in advanced (nodular) stages of the disease. Other features of the tumor include extravasated red blood cells in the vascular structures, deposition of hemosiderin within the tissue macrophages, and the infiltration of lymphocytes and plasma cells.
Clinically, several features of KS provoke questions regarding its malignant nature. (i) KS often presents as multiple, widely disseminating lesions on the skin and mucus membranes without evidence of a primary tumor. (ii) KS can regress spontaneously, even in severely immunocompromised patients. (iii) The clinical course of KS is influenced by environmental factors, and KS often progresses rapidly in association with infectious complications such as cytomegalovirus and pneumocystis carinii pneumonia (5). KS has been reported to develop following glucocorticoid use in patients with HIV infection and collagen vascular diseases (7–12). In addition, glucocorticoid use is associated with progression of existing KS, and withdrawal of glucocorticoids can lead to tumor regression (11, 12). (iv) The tumor does not show aneuploidy, a hallmark of neoplasia (13).
Recently, immortalized tumor cell lines have been isolated from patients with HIV-1-related KS (KSY-1) and the HIV negative, classic form of KS (KS-SLK) by independent investigators from the United States and Israel (14, 15). These cell lines show a common deletion in the short arm of chromosome 3. Both cell lines grow efficiently in immunocompromised mice, thus documenting that KS is a true malignancy. However, the possibility that KS begins as a benign proliferative process with eventual transformation to a true malignancy as a result of single or multiple genetic alterations cannot be discounted.
A benign proliferative lesion can be distinguished from a neoplasm because the latter originates from a single cell and gives rise to a clonal tumor (16), whereas a benign nonneoplastic cellular proliferation necessarily would be polyclonal. The clonality of B and T cell lymphoid neoplasia has been determined by studying the pattern of Ig T cell receptor gene rearrangements (17). In other neoplasms, the pattern of X-chromosome inactivation has been widely used (18, 19). This methodology is based on the fact that in each female cell, one copy of the X-chromosome is randomly inactivated by DNA methylation (19). This event occurs around the time of implantation, and the inactivation is permanent for the life of that cell and its daughter cells. Because of the random nature of the inactivation, nearly equal numbers of the cells in normal female tissues contain inactive maternal or paternal X-chromosomes. However, in clonally derived tumors, all cells have either the maternal or paternal X-chromosome inactivated. The active and inactive X-chromosomes can be distinguished based on their structural and biochemical characteristics. Exon 1 of the human androgen receptor (AR) gene, which resides on the X-chromosome, contains five methylation-sensitive restriction enzyme sites (three HhaI and two HpaII sites) within 100 bp 5′ to a polymorphic CAG repeat region (19). The CAG repeat polymorphism can be used to distinguish between the maternal and paternal X-chromosome sequences in heterozygous females, and the methylation status can also distinguish between the active and inactive allele. In this study, we have expanded our preliminary results (20) to examine the clonal nature of KS in HIV seropositive and seronegative women.
METHODS
A total of 28 women with KS were identified. Of the 28 cases, DNA isolated from specimens of 3 patients failed to amplify by PCR, 1 case had insufficient tumor, and 12 cases were noninformative caused by overlapping AR alleles or the presence of two copies of the same AR allele (homozygous). The remaining 12 women with KS comprised the evaluable cases; 4 of these had multiple biopsies. Study subjects were identified from the Los Angeles area through the County-wide Cancer Registry or were referred for their care to either the Los Angeles County–University of Southern California Medical Center, the University of Southern California/Norris Cancer Hospital, University of Miami Medical Center, or Mount Sinai Hospital, New York. Of the 12 evaluable patients, 2 women were HIV-1 seronegative and 10 were HIV-1 seropositive.
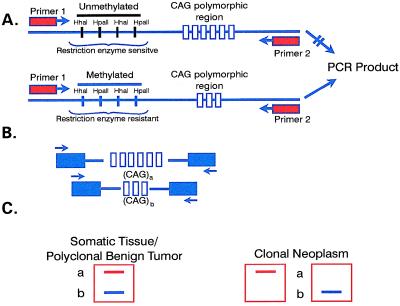
Cutaneous tumor biopsies were obtained from all cases. The basis for the experimental design (Fig. 1) is as follows: oligonucleotide primers were designed to flank both the 5′ methylation-sensitive restriction enzyme sites and the polymorphic CAG repeat in the AR gene. By incubation of a sample DNA with methylation-sensitive restriction enzymes, the sites on the active (unmethylated) AR allele was cleaved, whereas the sites on the inactive (methylated) AR allele was not be cleaved. Subsequent amplification by PCR by using these primers yields a product from the uncleaved (methylated) AR allele on the inactive X-chromosome. Undigested DNA from the tumor specimen was used as a control, which upon amplification should yield products from both the active and inactive alleles. It should be noted that it is common to observe multiple bands from a given allele on PCR amplification of short tandem repeats such as dinucleotide CA or trinucleotide CAG repeats caused by Taq DNA polymerase slippage.
Figure 1.
Schema of clonality analysis based on x-chromosome inactivation of the AR. (A) Top allele contains unmethylated restriction sites that will undergo cutting of DNA with the methylation-sensitive enzymes and thus fail to amplify during PCR. Bottom allele contains methylated restriction sites, and thus DNA will not be cut with methylation-sensitive enzymes, resulting in amplification during PCR. (B) Polymorphism of CAG repeats of the AR. (C) Amplification of AR allele from somatic cells with specific primers flanking the CAG repeat will result in two products of different size. In a clonally derived cell population, the same allele is inactivated by methylation in all the cells. Amplification of only the endonuclease insensitive allele can occur leading to the product size of one of the two alleles.
Formalin-fixed paraffin embedded cutaneous KS biopsies were used for DNA analysis. Five serial sections of 8 μm thickness each were stained with hematoxylin/eosin. A representative slide was cover-slipped to identify tumor-containing regions with minimal infiltration of mononuclear cells and marked for microscopic dissection. KS regions were microdissected with a sterile scalpel under the microscope from three to four serial uncovered and stained sections. The KS tissue was pooled, and DNA extracted with proteinase K by using methods described previously (21, 22). The DNA was divided into two equal aliquots for either digestion with 1 to 2 units each of methylation-sensitive restriction enzymes HhaI or mock digestion with the restriction enzyme buffer alone for 90 min at 37°C. The samples were then amplified by using two series of PCR with nested primers specific for exon 1 of the AR gene.
The primary PCR used primers 5′-GTGCGCGAAGTGATCCAGAA-3′ and 5′-TCTGGGACGCAACCTCTCTC-3′, and the PCR was performed in a 25 μl standard reaction consisting of 17 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1.5 min. The secondary PCR was performed with inside primers 5′-AGAGGCCGCGAGCGCAGCACCTC-3′ and 5′-GCTGTGAAGGTTGCTGTTCCTCAT-3′ by using 0.8–1 μl of the primary PCR product as the template, 2 μCi (1 Ci = 37 GBq) of [α-32P]dCTP, and consisted of 28 cycles at 94°C for 1 min, 66°C for 1 min, and 72°C for 1.5 min. The radioactively labeled secondary PCR products were analyzed by electrophoresis on 5% denaturing polyacrylamide gels and subjected to autoradiography.
Pathologic review of all microdissected regions was performed by one of us (B.N.) blindly and without the knowledge of PCR results. Several variables were examined and scored. These included the degree of blood cell extravasation, hemosiderin deposition, and overall tumor grade, each of which was scored from 0 to 4. Lack of lymphocyte infiltration was also scored from 0 to 4, with 4 showing the least infiltration. These variables were examined alone and collectively as predictors of clonality. The scores for clonal and nonclonal samples were compared by using the nonparametric Wilcoxon rank sum test (23).
RESULTS
Characteristics of the 12 patients heterozygous for the exon 1 CAG repeat of the AR gene are shown in Table 1. Three patients had a history of glucocorticoid use prior to the development of KS; 10 were HIV seropositive. Eight of the 10 HIV seropositive patients had advanced disease, including visceral involvement in 2. Twenty-four different biopsy specimens were obtained. In some biopsies, more than one discrete tumor site was microdissected and analyzed. A total of 41 different regions from 24 biopsies were studied.
Table 1.
Patient demographics (n = 12)
| Characteristic | Value |
|---|---|
| Age (years) | |
| Median | 49 |
| Range | 27–89 |
| Race | |
| Caucasian | 4 |
| Hispanic | 3 |
| African American | 5 |
| KS risk factors | |
| Glucocorticoid use | 2 |
| HIV infection | 9 |
| Both HIV infection and glucocorticoid use | 1 |
| CD4+ lymphocyte counts | |
| Median | 486 |
| Range | 2–1835 |
| Tumor burden (no. of mucocutaneous lesions) | |
| <25 | 5 |
| >50 | 7 |
| Visceral involvement | 2 |
Table 2 shows X-chromosome inactivation results for the 12 patients. The two women with HIV-negative KS each had two biopsies analyzed. In one (case 2), tumor regions from one biopsy showed amplification of a single AR allele, indicating clonal derivation from a single cell. The second biopsy from this patient showed amplification of both AR alleles, indicating a polyclonal origin. In the other case (case 1), both biopsies showed evidence of polyclonality.
Table 2.
Results of clonality analysis of KS biopsies
| No. of | No. of | Monoclonal KS
|
Polyclonal KS | |||
|---|---|---|---|---|---|---|
| HIV | biopsies | microdissected | Shorter allele | Larger allele | retention of | |
| Case | status | examined | regions examined | retained | retained | both alleles |
| 1 | − | 2 | 2 | 0 | 0 | 2 (2 biopsies) |
| 2 | − | 2 | 4 | 0 | 3 (1 biopsy) | 1 |
| 3 | + | 1 | 3 | 0 | 0 | 4 |
| 4 | + | 1 | 1 | 1 | 0 | 0 |
| 5 | + | 3 | 3 | 0 | 2 (2 biopsies) | NE |
| 6 | + | 1 | 2 | 1 | 1 | 0 |
| 7 | + | 9 | 21 | 1 (1 biopsy) | 5 (4 biopsies) | 15 |
| 8 | + | 1 | 1 | 0 | 0 | 1 |
| 9 | + | 1 | 1 | 0 | 0 | 1 |
| 10 | + | 1 | 1 | 0 | 0 | 1 |
| 11 | + | 1 | 1 | 0 | 0 | 1 |
| 12 | + | 1 | 1 | 0 | 0 | 1 |
Formalin-fixed KS lesions were microscopically examined to select tumor areas and microdissected. In several biopsies, more than one tumor region was suitable for microdissection. In one case (no. 5), the amplification product from one of the biopsies was not evaluable (NE). Shorter or longer allele retained indicates the length of the respective allele amplified by PCR following digestion with a methylation-sensitive enzyme.
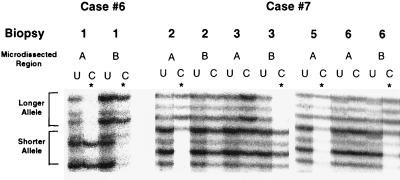
The lesions from four HIV seropositive women (cases 4–7) showed retention of a single X-chromosome allele upon digestion with the methylation-sensitive enzyme, indicating clonality (Table 2). In two women (cases 6 and 7), a different X-inactivation pattern was observed in separate microdissection regions from the same patient, either different regions from the same tumor biopsy (case 6) or different tumor biopsies (case 7), indicating that the KS growth was not clonally related (Fig. 2). One woman (case 7) showed a polyclonal inactivation pattern in 15 different microdissection regions from nine different biopsies. Representative microscopic pictures of two KS regions from this case are shown in Fig. 3. The single specimen available from each of the remaining six HIV seropositive patients (cases 3 and 8–12) showed a polyclonal AR inactivation pattern.
Figure 2.
Clonal analysis of representative KS lesions. The biopsy number and microdissected region of the KS lesions are given above their respective autoradiograph. U, uncut control; C, digested (or cut) with methylation-sensitive restriction enzyme HhaI prior to PCR amplification of the AR gene. The presence of a single AR gene in the digested (C) lane of the KS lesion indicates clonality (e.g., biopsy 2, region A of case 7). The presence of both AR alleles in the digested (C) lane (e.g., biopsy 2, region B of case 7) indicates a polyclonal pattern. Different AR alleles inactivated (e.g., region A and B of case 6; biopsy 2, region A; biopsy 3, region B of case 7) indicate these KS lesions arose independently and are not clonally related either in the same biopsy (case 6) or different biopsies (case 7).
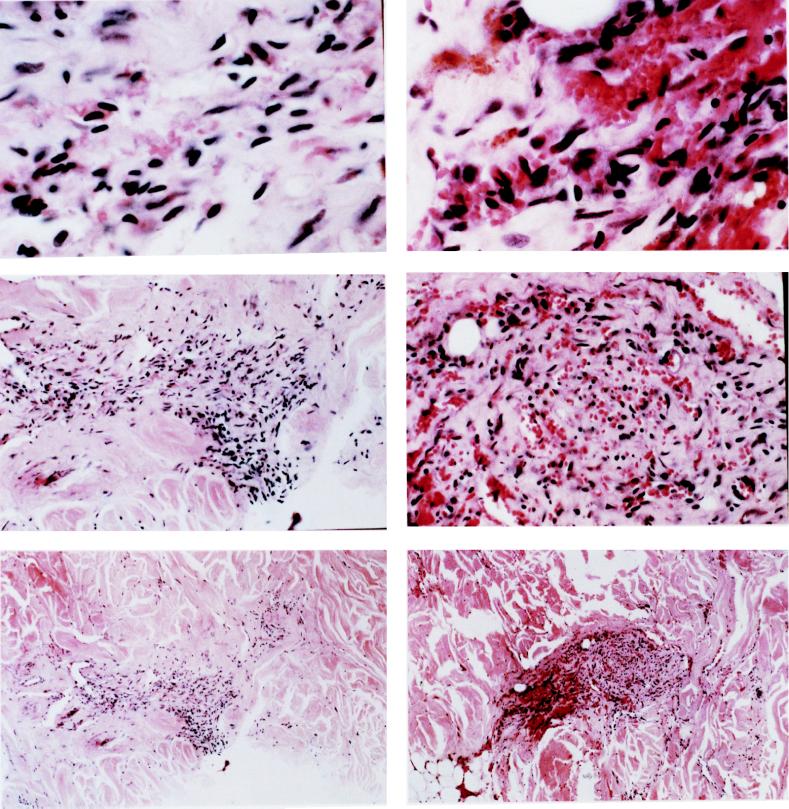
Figure 3.
(A and B) Histologic examination of two KS regions from case 7 that are shown for their X-chromosome inactivation pattern in Fig. 2. The left column represents biopsy 2, and the right column represents biopsy 3. Histologic examinations at low power (×20 magnification, Top), intermediate power (×100 magnification, Middle), and high power (×400 magnification, Bottom) are shown. Discrete tumor regions with minimal cellularity surrounded by normal tissue are noted. The lesions display characteristic features of KS with proliferation of spindle cells and slit-like vascular structures with extravasated red blood cells.
No significant differences were observed between the clonal and nonclonal samples with respect to any of the histopathological characteristics (extravasation, hemosiderin deposition, and lymphocyte infiltration) or overall tumor grade.
DISCUSSION
We found that less than half of the KS lesions from HIV-infected and -uninfected individuals contain one AR methylated allele, indicating clonally derived tumors. This finding confirms that of Rabkin et al. (24), who showed evidence of clonality in two of three KS biopsies. A recent report by this group concluded that multicentric KS lesions from eight different patients showed the same AR methylation pattern, suggesting that multiple separate lesions on the same person are likely to have descended and spread from the single neoplastic cell (24). We disagree with these conclusions based on our findings of more than one neoplastic clone within different biopsies of the same patient (20).
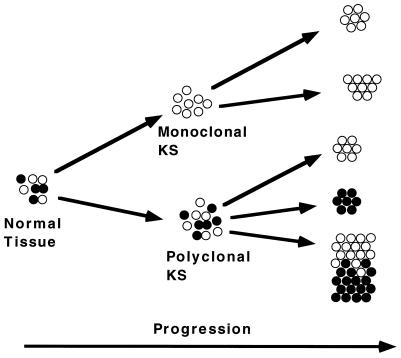
We have recently studied the clonal origin of bilateral breast carcinoma by the same assay and found a different AR methylation pattern in the right and left breast tumors in 3 of 12 patients, indicating that the tumors arose from distinct transformed cells (25). It is important to note that the only cases which give unequivocal polyclonal information from microdissected lesions are those in which different inactivation patterns are observed. This result is because regions of a lesion with the same X-inactivation pattern suggest clonality but multiple individual clones could still be present but contain, by chance, the same inactivated X-chromosome (Fig. 4).
Figure 4.
Model for KS progression. Normal tissue is a mosaic of cells with different methylated AR alleles (represented by open and filled circles) caused by random X-chromosome inactivation. Neoplasms typically develop from clonal outgrowth of a single cell into a monoclonal tumor with subsequent metastasis of the same clone (monoclonal KS pathway). Alternatively, as evidence from our data and shown in the polyclonal KS pathway, individual lesions may have a polyclonal/hyperplastic stage, followed by the outgrowth of distinct clonal neoplasm(s) (seen as groups of either open or closed circles, e.g., case 7). This process may also occur at distinct sites within the same lesion (shown as defined areas of both open and closed circles in the same group, e.g., case 6).
In seven cases (one HIV negative, six HIV positive), all microdissected biopsy regions examined exhibited retention of both AR alleles. In two cases, regions showed a clonal methylation pattern, whereas other regions of the same biopsy (case 7) or different biopsy (cases 2 and 7) showed a polyclonal methylation pattern. It is true that the microdissected regions may appear to be a polyclonal expansion caused by the abundance of normal stroma, infiltration of mononuclear cells within the lesions, or incomplete DNA digestion. The former explanation seems unlikely because the specimens were microdissected to enrich for putative neoplastic cells. Moreover, the degree of lymphocyte infiltration was similar in the clonal and nonclonal tumors. Alternatively, early KS lesions may represent a benign proliferation that subsequently evolves to a clonal lesion by preferential growth (Fig. 4). Rabkin et al. (24) also observed evidence of a polyclonal AR methylation pattern in 4 of 27 tumors, but for unknown reasons discounted this possibility in favor of contamination with stroma cells. Additional examination of multiple KS lesions early or later in the development of the disease may serve to clarity this issue. It is also possible that, analogous to Hodgkin’s disease, only rare cells within the tumor lesion are neoplastic, whereas the majority represent normal immune cells or stroma. In such as case, the X-chromosome inactivation assay, which requires the majority of cells in a given tumor to be neoplastic, may not be sensitive enough. Further analysis of other genetic or biochemical markers, such as alterations in oncogenes or tumor suppressor genes, will be necessary to determine whether specific cells in a polyclonal population ultimately give rise to a monoclonal KS lesion.
One can speculate that an infectious agent or other proliferative stimuli could lead to hyperplasia in KS, after which genetic events could lead to the evolution of independent clones. Such a sequence may be more plausible in HIV-infected individuals, where the proliferative signal is more pronounced because of immune suppression or local production of growth promoting factors/cytokines. Additional studies of a larger series will be needed to determine if the evolution of independent clones is restricted to HIV-related KS alone.
The following interpretations can be made from the available evidence (Fig. 4). (i) KS lesions are clonal in at least some of the cases. (ii) Independent clones may develop in a given patient, either spontaneously or after undergoing a premalignant/hyperplastic stage. (iii) KS may begin as a polyclonal disease with subsequent evolution to a clonal process. (iv) The polyclonal pattern is a false negative result caused by stromal cells in the tumor tissue producing a polyclonal pattern. To better understand the last two possibilities, a much larger study than ours and that of Rabkin et al. has to be done, preferably with examination of early as well as late KS and visceral as well as cutaneous tumors.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: KS, Kaposi’s sarcoma; AR, androgen receptor.
References
- 1.Geddes M, Francheschi S, Barchielli A, Falcini F, Carli S, Cocconi G, Conti E, Crosignani P, Gata L, Giarelli L. Br J Cancer. 1994;69:333–336. doi: 10.1038/bjc.1994.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wahman A, Melnick S, Rhame F S, Potter J D. Epidemiol Rev. 1991;13:178–199. doi: 10.1093/oxfordjournals.epirev.a036068. [DOI] [PubMed] [Google Scholar]
- 3.Penn I. Transplantation. 1979;27:8–11. doi: 10.1097/00007890-197901000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Lifson A R, Darrow W W, Hessol N A, O’Malley P M, Barnhart L, Jaffe H J, Rutherford G W. Am J Epidemiol. 1990;131:221–231. doi: 10.1093/oxfordjournals.aje.a115492. [DOI] [PubMed] [Google Scholar]
- 5.Gill P S, Hamilton A, Naidu Y. AIDS Updates. 1994;7:1–11. [Google Scholar]
- 6.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 7.Cai J, Zhang T, Lotz M, Zhang Y, Masood R, Gill P S. Blood. 1997;89:1491–1500. [PubMed] [Google Scholar]
- 8.Harwood A R, Osoba D, Hofstader S L, Goldstein M B, Cardella C J, Holecek M J, Kunynetz R, Gianmmarco R A. Am J Med. 1979;67:759–765. doi: 10.1016/0002-9343(79)90731-9. [DOI] [PubMed] [Google Scholar]
- 9.Schottstaedt M W, Hurd E R, Stone M J. Am J Med. 1987;82:1021–1026. doi: 10.1016/0002-9343(87)90168-9. [DOI] [PubMed] [Google Scholar]
- 10.Klepp O, Dahl O, Stenwig J T. Cancer. 1978;42:2626–2630. doi: 10.1002/1097-0142(197812)42:6<2626::aid-cncr2820420618>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 11.Gill P S, Loureiro C, Bernstein-Singer M, Rarick M U, Sattler F, Levine A M. Ann Intern Med. 1989;110:937–940. doi: 10.7326/0003-4819-110-11-937. [DOI] [PubMed] [Google Scholar]
- 12.Real F X, Krown S E, Koziner B. Am J Med. 1986;80:119–122. doi: 10.1016/0002-9343(86)90060-4. [DOI] [PubMed] [Google Scholar]
- 13.Delli-Bovi P, Donti E, Knowles D M, II, Friedman-Kien A, Luciw P A, Dina D, Dalla-Favera R, Basilico C. Cancer Res. 1986;46:6333–6338. [PubMed] [Google Scholar]
- 14.Lunardi-Iskandar Y, Gill P S, Lam V H, Zeman R A, Michaels F, Mann D L, Reitz M S, Jr, Kaplan M, Berneman Z N, Carter D, et al. J Natl Cancer Inst. 1995;87:974–981. doi: 10.1093/jnci/87.13.974. [DOI] [PubMed] [Google Scholar]
- 15.Siegal B, Levinton-Kriss S, Schiffer A, Sayer J, Engelberg I, Vonsover A, Ramon Y, Rubinstein E. Cancer. 1990;65:492–498. doi: 10.1002/1097-0142(19900201)65:3<492::aid-cncr2820650320>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 16.Sidransky D, Frost P, Von Escehnbach A, Oyasu R, Preisinger A C, Vogelstein B. N Engl J Med. 1992;326:737–740. doi: 10.1056/NEJM199203123261104. [DOI] [PubMed] [Google Scholar]
- 17.Waldmann T A, Goldman C K, Bongiovanni K F, Sharrow S O, Davey M P, Cease K B, Greenberg S J, Longo D L. Blood. 1988;72:1805–1816. [PubMed] [Google Scholar]
- 18.Vogelstein B, Fearon E R, Hamilton S R, Preisinger A C, Willard H F, Michelson A M, Riggs A D, Orkin S H. Cancer Res. 1987;47:4806–4813. [PubMed] [Google Scholar]
- 19.Allen R C, Zoghbi H Y, Moseley A B, Rosenblatt H M, Belmont J W M. Am J Hum Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- 20.Gill P S, Tsai Y C, Rao A P, Jones P A. N Engl J Med. 1997;337:670–671. doi: 10.1056/NEJM199708213370813. [DOI] [PubMed] [Google Scholar]
- 21.Spruck C H, III, Rideout W M, III, Olumi A F, Ohneseit P F, Yang A S, Tsai T C, Nichols P W, Horn T, Hermann G G, Steven K, et al. Cancer Res. 1993;53:1162–1166. [PubMed] [Google Scholar]
- 22.Tsai Y C, Simoneau A R, Spruck C H, Nichols P W, Steven K, Buckley J D, Jones P A. J Urol. 1995;153:1697–1700. doi: 10.1016/s0022-5347(01)67507-4. [DOI] [PubMed] [Google Scholar]
- 23.Hollandar M, Wolfe D A. Nonparametric Statistical Methods. New York: Wiley; 1973. [Google Scholar]
- 24.Rabkin C S, Janz S, Lash A, Coleman A E, Musaba E, Liotta L, Biggar J, Zhuang A. N Engl J Med. 1997;336:988–993. doi: 10.1056/NEJM199704033361403. [DOI] [PubMed] [Google Scholar]
- 25.Shibata A, Tsai Y C, Press M C, Henderson B E, Jones P A, Ross R K. Clin Cancer Res. 1996;2:743–748. [PubMed] [Google Scholar]