Abstract
Condensed Abstact
The functional polymorphic deletion of the androgen metabolizing UGT2B17 gene was examined for involvement in incident prostate cancer risk. In this study of 411 Caucasian cases and 397 Caucasian controls, the UGT2B17 deletion is not associated with prostate cancer risk.
Background
UDP-glucuronosyltransferase (UGT) 2B17 is a phase II metabolizing enzyme that mediates the glucuronidation of C19 steroids. A deletion polymorphism in the UGT2B17 gene is associated with a substantial reduction in glucuronidation activity in vitro.
Methods
We examined the association between the UGT2B17 deletion polymorphism and the risk of incident prostate cancer in a population-based study from central Arkansas that included 411 Caucasian cases and 397 Caucasian controls. We developed a novel high-throughput procedure that uses real-time PCR and allelic discrimination for genotyping analysis.
Results
The prevalence of the UGT2B17 deletion [(0/0)] was 12% in the controls, which was consistent with previous population estimates and with Hardy Weinberg equilibrium. There was no association between the UGT2B17 deletion polymorphism and prostate cancer risk in unconditional logistic regression analysis. Compared to the wild type group (+/+), the adjusted odds ratio (OR) was 0.89 (95% CI = 0.55–1.45) for the homozygous deletion (0/0), and the OR was 0.99 (95% CI = 0.73–1.35) for the heterozygote group (+/0).
Conclusion
These findings show that the UGT2B17 deletion polymorphism is not associated with prostate cancer risk in Caucasians.
Keywords: prostate, cancer, UGT, deletion, polymorphism
Introduction
UDP-glucuronosyltransferases (UGTs) are a superfamily of phase II detoxification enzymes that catalyze the glucuronidation of a variety of compounds including steroid hormones, bilirubin, pharmaceuticals, and carcinogens [1–3]. Several UGTs exhibit activity against C19 steroids; of these UGT2B15 and UGT2B17 are the only isoforms that are expressed in human prostate [4]. Elevated serum androgen levels have been implicated as a risk factor for prostate cancer, but study results have not been consistent [5–9]. Polymorphic variants in androgen metabolizing enzymes may alter androgen levels and therefore affect risk for prostate cancer. Data from two recent studies indicate the presence of a prevalent polymorphic deletion of the entire UGT2B17 gene [10, 11]. This deletion polymorphism is associated with significantly reduced glucuronidation rates of several compounds [12]. In a recent study, Park et al [13] found that the homozygous UGT2B17 gene deletion significantly increased the risk of prostate cancer in Caucasians. They observed no association in African-American subjects. In the current study, we examined the association of this deletion polymorphism with risk for prostate cancer in a large Arkansas-based Caucasian population using a novel real-time PCR approach that distinguishes between the 3 genotypes of the UGT2B17 deletion polymorphism.
Materials and Methods
Study Population
Subjects with incident prostate cancer, diagnosed from 1998–2003 in central Arkansas, were recruited into the study within six months of their diagnosis. The patients were recruited from the University of Arkansas for Medical Sciences, the University Hospital in Little Rock, the Central Arkansas Veteran’s Health Care System in Little Rock, and the Jefferson Regional Medical Center in Pine Bluff, Arkansas. These institutions treat the vast majority of cancer patients in the Little Rock metropolitan area. The medical records were reviewed to obtain information on tumor pathology and post-diagnostic PSA testing results. All subjects had histologically-confirmed prostate cancer. Community controls were recruited primarily from a mass-mailing database that covers 80% of Arkansas residents. This database was supplemented by records from the Arkansas State Driver’s License records. Elderly controls were also identified and recruited from the Centers for Medicare and Medicaid Services records. The controls were frequency matched to cases on age (± 5 years) and race.
Exclusion criteria for the case-control study included a history of cancer (except nonmelanoma skin cancer), uncontrolled cardiovascular disease, and hepatic dysfunction. All subjects were interviewed by a trained interviewer using a structured lifestyle questionnaire, and were asked to provide a blood sample for DNA analysis and prostate-specific antigen (PSA) measurements. PSA levels were collected for 99% of the cases and 52% of the controls.
The appropriate institutional review board approvals were obtained for the study protocol, and further details of the methods have been previously published [14]. The study population consists of 411 Caucasian cases and 397 Caucasian controls which provided >95% power to detect a statistically significant OR of 2.0 at an exposure rate of 10%, which has been observed for the UGT2B17 homozygous deletion.
UGT2B17 deletion polymorphism genotyping methodology
Details describing the UGT2B17 gene deletion polymorphism have been reported elsewhere [10, 11]. For large sample screening, we developed a high-throughput genotyping assay using real-time PCR with allelic discrimination. Although the use of two primers and two probes has become common practice for SNP genotyping and has recently been used in an analysis of a small deletion polymorphism [15], the amplification of both the UGT2B17 wild-type and deletion alleles could not be performed using the same set of PCR primers due to the large size (~120Kb) of the UGT2B17 gene deletion polymorphism. Each reaction included two primers and one 6-FAM-labeled probe to amplify exon 1 of UGT2B17, and two primers and one JOE-NHS-Ester-labeled probe (that spans the deletion cut site) that amplify only if the deletion is present (Figure 1). Due to high sequence homology between UGT2B17 other UGT genes and pseudogenes, primers were designed using Blast 2.2.14 (http://www.ncbi.nlm.nih.gov/BLAST/) maximizing for 3′ sequence mismatches with other homologous genes (Table 1), with primers obtained from Integrated DNA Technologies (Coralville, IA). Reactions (20 μl) were performed in 384-well plates using the ABI 7900 HT Sequence Detection System, with incubations performed at 50°C for 2 min; 95°C for 15 min; and 40 cycles of 94°C for 1 min, 60°C for 1 min 30 sec. Reactions included QuantiTect Multiplex PCR Master Mix (1x final concentration; Qiagen, Valencia, CA), 0.4 μM for each primer, 0.2 μM for each probe, and 20–100 ng of DNA. Negative controls (no DNA template) were run on every plate and genotypes were assigned by the automatic calling feature of the allelic discrimination option in SDS 2.2.2 software (Applied Biosystems, Foster City, CA). Individuals homozygous for the wild-type allele (intact UGT2B17 gene) are represented as (+/+), heterozygous individuals are represented as (+/0), and individuals who were homozygous for the UGT2B17 gene deletion allele (UGT2B17 null) are represented as (0/0).
Figure 1.
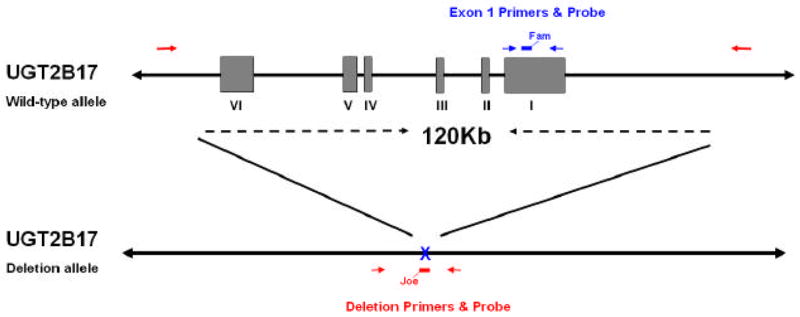
Gene structure and primer and probe locations for the UGT2B17 multiplex real-time PCR assay. The roman numerals I–VI indicate the exons of the UGT2B17 gene on the wild-type allele. The deletion allele is shown below the wild-type allele indicating the 120 kb that are deleted, including the entire UGT2B17 gene. Red arrows indicate the primers that will amplify the deletion allele and the red bar (with fluorescent label Joe) indicates the deletion probe. Blue arrows indicate the primers that will amplify exon 1 of UGT2B17 from the wild-type allele and the blue bar (with fluorescent label Fam) indicates the deletion probe.
Table 1.
Primer and probe sequences for the UGT2B17 genotype assay.
| primer name | primer composition (5′ to 3′) |
|---|---|
| Exon 1 – forward | TGAAAATGTTCGATAGATGGACATATAGTA |
| Exon 1 – reverse | GACATCAAATTTTGACTCTTGTAGTTTTC |
| Exon 1 – probe | 6-FAM-TACATTTTGGTCATATTTTTCACAACTACAAGAATTGT-BHQ1 |
| Deletion – forward | TTTAATGTTTTCTGCCTTATGCCAC |
| Deletion – reverse | AGCCTATGCAATTTTCATTCAACATAG |
| Deletion – probe | JOE-ACTACACTGAGATTTACAAAAGAATTCTGTCAGGATATAG-BHQ1 |
To validate the new genotyping methodology, we used 63 normal human liver genomic DNA specimens that has been previously described [16]. The methodology was also validated using 96 oral buccal cell genomic DNA samples from cancer-free community residents. Details of this study have also been reported previously [17]. DNA from both of these studies was extracted using a commercial kit (Qiagen Inc., Valencia, CA).
Statistical Analysis
Deviation from Hardy-Weinberg equilibrium in the controls was tested using ?2 analysis. Unconditional logistic regression was used to estimate the odds ratios (OR) and 95% confidence intervals (CI) associated with prostate cancer risk. Potential confounders were identified by Spearman rank correlation analyses and multivariate regression models, including stepwise regression models. Age (continuous), pack-years of smoking (continuous), and family history of prostate cancer (categorical) were found to be significant predictors of prostate cancer risk, and were entered into the final models. All statistical analyses were conducted using SPSS 15.0 statistical software (SPSS 15.0, SPSS Inc., Chicago IL).
Results
The genotyping method was initially tested in a blinded study of 63 normal liver genomic DNA samples by comparing the findings from the real-time method to that using a previously-validated methodology [12]. This previous methodology differentiated between PCR amplifications of the two highly homologous genes UGT2B15 and UGT2B17 (95% homology in nucleotide sequence). Briefly, the previous method of genotyping included 3 PCR amplification reactions: exon 1 of UGT2B17 or the highly-homologous UGT2B15 was amplified using sense and antisense primers that contained 1 or 2 bp mismatches between UGT2B15 or UGT2B17 sequences. This was followed by gene-specific restriction fragment length polymorphism (RFLP) to confirm the presence of UGT2B17 or UGT2B15. A third PCR reaction amplified the UGT2B17 deletion allele with primers spanning the deletion cut-site, as previously described [11, 12]. We found 100% concordance when comparing the high-throughput real-time method versus our previously validated RFLP methodology. In a second blinded quality-control study that included the 96 oral buccal cell genomic DNA samples, samples were genotyped in duplicate by real-time PCR, and again there was 100% concordance for the UGT2B17 genotype.
The basic demographic characteristics of the cases and controls are shown in Table 2. The mean age of the cases was higher than the controls (66 vs. 62), and the cases had a higher mean amount of smoking pack-years than the controls (30 vs. 22). More cases than controls were ever smokers (72% vs. 59%) and heavy smokers (39% vs. 28%). In subset analysis by smoking level, subjects were classified into light and heavy smoking categories (stratified at the median for all smokers in the study population). More cases than controls (19% vs. 8%) had a family history of prostate cancer. The mean PSA level was 42 ng/ml in cases and 1.6 ng/ml in controls. There were no controls with PSA levels above 10 ng/ml. Among the 302 case subjects with a Gleason score, 59% had a score of 6 or less.
Table 2.
Demographics of the prostate cancer cases and controls.
| Demographic | Cases (n=411) | Controls (n=397) |
|---|---|---|
| Age (years)a | 66 ± 8 | 62 ± 11 |
| Smoking (pack-years)a | 30 ± 35 | 22 ± 32 |
| Non-smokers | 28% | 42% |
| Light-smokers | 33% | 31% |
| Heavy-smokers | 39% | 28% |
| Family history of prostate cancer | 19% | 8% |
| PSA level (ng/ml)a,b | 42 ± 437 | 1.6 ± 1.7 |
| Gleason score = 6c | 59% | NAd |
| Gleason score > 6c | 41% | NA |
Mean ± Standard Deviation shown for continuous variables.
PSA levels are shown when available (405 cases and 205 controls).
Gleason scores were available for 302 cases.
NA, not applicable
The UGT2B17 genotype frequencies, OR, and 95% CI are shown in Table 3. The frequency of the UGT2B17 deletion (0/0) genotype in controls was 12%, which was similar to that observed in other Caucasian study populations [11–13, 18]. The UGT2B17 gene deletion polymorphism was determined to be consistent with Hardy-Weinberg equilibrium for the control population.
Table 3.
Genotype frequencies and risk of prostate cancer.
| UGT2B17 genotypea | Cases (%) | Controls (%) | OR (95% CI)b |
|---|---|---|---|
| (+/+) | 201 (49) | 190 (48) | 1.00 (Ref.) |
| (+/0) | 168 (41) | 159 (40) | 0.99 (0.73–1.35) |
| (0/0) | 42 (10) | 48 (12) | 0.89 (0.55–1.45) |
| (+/+) | 201 (49) | 190 (48) | 1.00 (Ref.) |
| (+/0) + (0/0) | 210 (51) | 207 (52) | 0.97 (0.73–1.30) |
| (+/+) + (+/0) | 369 (90) | 349 (88) | 1.00 (Ref.) |
| (0/0) | 42 (10) | 48 (12) | 0.89 (0.56–1.43) |
The three genotypes of the UGT2B17 deletion polymorphism are shown as: (+/+), two alleles of the UGT2B17 gene; (+/0), one allele of the UGT2B17 gene and one gene deletion allele; (0/0), UGT2B17 null, two copies of the deletion allele.
Adjusted for age, smoking pack-years, and family history of prostate cancer.
There was no significant difference in the prevalence of the UGT2B17 deletion genotype between prostate cancer cases (10%) and controls (12%). The OR, adjusted for age, smoking, and family history of prostate cancer, was not significant when comparing deletion homozygote subjects (0/0) (OR=0.89, 95% CI 0.55–1.45) or heterozygote subjects (+/0) (OR=0.99, 95% CI 0.73–1.35) to wild type subjects (+/+) (Table 3). There was also no association with prostate cancer risk when collapsing genotype categories. The risk when comparing all other subjects to the wild-type group was 0.97 (95% CI 0.73–1.30). Alternatively, when comparing deletion homozygotes to the combined heterozygotes and wild-type group, the OR was 0.89 (95% CI 0.56–1.43).
When stratified by smoking level (non-smokers, light-smokers, and heavy-smokers), there was also no association between UGT2B17 genotype and prostate cancer risk. The adjusted OR was not elevated when comparing deletion homozygote subjects (0/0) or heterozygote subjects (+/0) to wild type subjects (+/+) in any of the three smoking categories (Table 4). There was also no association with prostate cancer risk in any of the three smoking categories when collapsing genotype categories.
Table 4.
Genotype frequencies and risk of prostate cancer in non-, light-, and heavy-smoking categories.
| UGT2B17 genotypea | Cases (%) | Controls (%) | OR (95% CI)b |
|---|---|---|---|
| Non-smokers | |||
| (+/+) | 61 (53) | 76 (48) | 1.00 (Ref.) |
| (+/0) | 44 (38) | 63 (39) | 0.84 (0.49–1.44) |
| (0/0) | 10 (9) | 21 (13) | 0.62 (0.26–1.49) |
| (+/+) | 61 (53) | 76 (48) | 1.00 (Ref.) |
| (+/0) + (0/0) | 54 (47) | 84 (52) | 0.79 (0.47–1.31) |
| (+/+) + (+/0) | 105 (91) | 139 (87) | 1.00 (Ref.) |
| (0/0) | 10 (9) | 21 (13) | 0.67 (0.29–1.56) |
| Light-smokersc | |||
| (+/+) | 65 (49) | 61 (50) | 1.00 (Ref.) |
| (+/0) | 50 (37) | 42 (35) | 1.25 (0.70–2.21) |
| (0/0) | 19 (14) | 18 (15) | 1.10 (0.49–2.46) |
| (+/+) | 65 (49) | 61 (50) | 1.00 (Ref.) |
| (+/0) + (0/0) | 69 (51) | 60 (50) | 1.20 (0.71–2.04) |
| (+/+) + (+/0) | 115 (86) | 103 (85) | 1.00 (Ref.) |
| (0/0) | 19 (14) | 18 (15) | 1.00 (0.46–2.16) |
| Heavy-smokersc | |||
| (+/+) | 75 (46) | 53 (46) | 1.00 (Ref.) |
| (+/0) | 74 (46) | 54 (46) | 0.94 (0.57–1.56) |
| (0/0) | 13 (8) | 9 (8) | 0.97 (0.38–2.47) |
| (+/+) | 75 (46) | 53 (46) | 1.00 (Ref.) |
| (+/0) + (0/0) | 87 (54) | 63 (54) | 0.95 (0.58–1.54) |
| (+/+) + (+/0) | 149 (92) | 107 (92) | 1.00 (Ref.) |
| (0/0) | 13 (8) | 9 (8) | 1.00 (0.40–2.45) |
The three genotypes of the UGT2B17 deletion polymorphism are shown as: (+/+), two alleles of the UGT2B17 gene; (+/0), one allele of the UGT2B17 gene and one gene deletion allele; (0/0), UGT2B17 null, two copies of the deletion allele.
Adjusted for age, smoking pack-years (except non-smokers), and family history of prostate cancer.
Light- versus heavy-smoker categories, subjects stratified at the median for all smokers in the study population.
Analysis was also conducted in the subset of individuals who have no family history of prostate cancer to avoid potential bias if other, more penetrant, genetic factors are involved in prostate cancer risk. This subset of individuals without family history of prostate cancer had a similar frequency of the UGT2B17 deletion (11% in the cases and 12% in the controls) as the overall population. In addition, logistic regression analysis indicated no association of the UGT2B17 genotype with prostate cancer in this subset of individuals. The OR, adjusted for age and smoking, was not significant when comparing deletion homozygote subjects (0/0) (OR=1.01, 95% CI 0.61–1.69), or heterozygote subjects (+/0) (OR=1.03, 95% CI 0.74–1.43), to wild type subjects (+/+) when individuals with a family history of prostate cancer were excluded (Supplementary Table 1).
Discussion
This study did not detect an association between the UGT2B17 gene deletion polymorphism and risk for prostate cancer in Caucasians. It was previously reported that the UGT2B17 deletion polymorphism was not associated with an increased risk of prostate cancer in African-Americans from Arkansas [13], but was associated with an increased risk (OR=1.9) in Caucasian subjects from Florida that included 293 cases and a similar number of controls [13]. One difference between our study and the previous finding is that the real-time PCR genotyping technique employed in the present study is a high-throughput method that facilitates genotyping for large sample sets with the automated assignment of genotypes, whereas the traditional PCR techniques used in the previous study require gel electrophoresis and the manual calling of genotypes [12, 13]. The current method also distinguishes between heterozygous (+/0) and homozygous (+/+) UGT2B17 genotypes, a distinction not performed in the previous study. However, the prevalence of the UGT2B17 deletion (0/0) in the control groups of both studies was similar (11–12%), which is consistent with other reports in samples of healthy individuals [11, 12, 18]. It is possible that the lack of association in the current study is due in part to undetected prostate cancer in the controls. Approximately half of the control subjects were not screened for PSA; therefore some controls may have had latent prostatic adenocarcinoma. However, none of the controls that were screened for PSA had a value above 10 ng/ml, indicating that there was little or no misclassification of controls. The Gleason score distribution among cases in this study was very similar to that reported in a case series of 54,200 subjects [19], indicating that the case population was representative of the stage at which most prostate tumors are detected in the PSA screening era. Therefore the differences between these two studies might reflect chance, lack of control for unknown confounders, or some other factor that might mediate a potential affect of UGT2B17 on prostate cancer risk.
The lack of association of the UGT2B17 gene deletion polymorphism with prostate cancer may be due in part to the down-regulation of UGT2B17 transcription by androgens in the prostate [20–22]. This may result in low expression of UGT2B17 in the prostate compared to UGT2B15 expression, which is not regulated by androgens [21, 22]. A deletion in the UGT2B17 gene would therefore not result in a dramatic difference in overall androgen glucuronidation rates within the prostate, which would consequently have little overall impact on the risk for prostate cancer. Another potential consideration in understanding the role of glucuronidation in prostate cancer is the presence of functional polymorphisms in UGT2B15. Some studies have examined the association between polymorphisms in UGT2B15 and prostate cancer risk but the results have been conflicting [23–26]. Larger studies enabling us to examine functional polymorphisms in both genes and their interactions will be necessary to better evaluate the role of glucuronidating enzymes and risk for prostate cancer.
Supplementary Material
Acknowledgments
These studies were supported by Public Health Service grants P01-CA68384 (P. Lazarus), R01-DE13158 (P. Lazarus), K07-CA104231 (J. Muscat), and 1R01-AG15722 (N. P. Lang) from the National Institutes of Health and a formula grant under the Pennsylvania Department of Health’s Health Research Formula Funding Program (State of PA, Act 2001–77 – part of the PA Tobacco Settlement Legislation; to P. Lazarus). We thank the Functional Genomics Core Facility at Penn State University College of Medicine for their technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gueraud F, Paris A. Glucuronidation: a dual control. Gen Pharmacol. 1998;31(5):683–688. doi: 10.1016/s0306-3623(98)00114-1. [DOI] [PubMed] [Google Scholar]
- 2.Ren Q, Murphy SE, Zheng Z, Lazarus P. O-Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1- (3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab Dispos. 2000;28(11):1352–1360. [PubMed] [Google Scholar]
- 3.Tephly TR, Burchell B. UDP-glucuronosyltransferases. a family of detoxifying enzymes. Trends Pharmacol Sci. 1990;11(7):276–279. doi: 10.1016/0165-6147(90)90008-v. [DOI] [PubMed] [Google Scholar]
- 4.Beaulieu M, Levesque E, Hum DW, Belanger A. Isolation and characterization of a novel cDNA encoding a human UDP-glucuronosyltransferase active on C19 steroids. J Biol Chem. 1996;271(37):22855–22862. doi: 10.1074/jbc.271.37.22855. [DOI] [PubMed] [Google Scholar]
- 5.Hsing AW, Reichardt JK, Stanczyk FZ. Hormones and prostate cancer. current perspectives and future directions. Prostate. 2002;52(3):213–235. doi: 10.1002/pros.10108. [DOI] [PubMed] [Google Scholar]
- 6.Ross R, Bernstein L, Judd H, Hanisch R, Pike M, Henderson B. Serum testosterone levels in healthy young black and white men. J Natl Cancer Inst. 1986;76(1):45–48. [PubMed] [Google Scholar]
- 7.Eaton NE, Reeves GK, Appleby PN, Key TJ. Endogenous sex hormones and prostate cancer. a quantitative review of prospective. Br J Cancer. 1999;80(7):930–934. doi: 10.1038/sj.bjc.6690445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mohr BA, Feldman HA, Kalish LA, Longcope C, McKinlay JB. Are serum hormones associated with the risk of prostate cancer? Prospective results from the Massachusetts Male Aging Study. Urology. 2001;57(5):930–935. doi: 10.1016/s0090-4295(00)01116-x. [DOI] [PubMed] [Google Scholar]
- 9.Heikkila R, Aho K, Heliovaara M, Hakama M, Marniemi J, Reunanen A, et al. Serum testosterone and sex hormone-binding globulin concentrations and the risk of prostate carcinoma. a longitudinal study. Cancer. 1999;86(2):312–315. [PubMed] [Google Scholar]
- 10.Murata M, Warren EH, Riddell SR. A human minor histocompatibility antigen resulting from differential expression due to a gene deletion. J Exp Med. 2003;197(10):1279–1289. doi: 10.1084/jem.20030044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson W, 3rd, Pardo-Manuel de Villena F, Lyn-Cook BD, Chatterjee PK, Bell TA, Detwiler DA, et al. Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics. 2004;84(4):707–714. doi: 10.1016/j.ygeno.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 12.Lazarus P, Zheng Y, Aaron Runkle E, Muscat JE, Wiener D. Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet Genomics. 2005;15(11):769–778. doi: 10.1097/01.fpc.0000175596.52443.ef. [DOI] [PubMed] [Google Scholar]
- 13.Park J, Chen L, Ratnashinge L, Sellers TA, Tanner JP, Lee JH, et al. Deletion polymorphism of UDP-glucuronosyltransferase 2B17 and risk of prostate cancer in African American and Caucasian men. Cancer Epidemiol Biomarkers Prev. 2006;15(8):1473–1478. doi: 10.1158/1055-9965.EPI-06-0141. [DOI] [PubMed] [Google Scholar]
- 14.Stone A, Ratnasinghe LD, Emerson GL, Modali R, Lehman T, Runnells G, et al. CYP3A43 Pro(340)Ala polymorphism and prostate cancer risk in African Americans and Caucasians. Cancer Epidemiol Biomarkers Prev. 2005;14(5):1257–1261. doi: 10.1158/1055-9965.EPI-04-0534. [DOI] [PubMed] [Google Scholar]
- 15.Robledo R, Beggs W, Bender P. A simple and cost-effective method for rapid genotyping of insertion/deletion polymorphisms. Genomics. 2003;82(5):580–582. doi: 10.1016/s0888-7543(03)00172-1. [DOI] [PubMed] [Google Scholar]
- 16.Wiener D, Doerge DR, Fang JL, Upadhyaya P, Lazarus P. Characterization of N-glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human liver. importance of UDP-glucuronosyltransferase 1A4. Drug Metab Dispos. 2004;32(1):72–79. doi: 10.1124/dmd.32.1.72. [DOI] [PubMed] [Google Scholar]
- 17.Elahi A, Zheng Z, Park J, Eyring K, McCaffrey T, Lazarus P. The human OGG1 DNA repair enzyme and its association with orolaryngeal cancer risk. Carcinogenesis. 2002;23(7):1229–1234. doi: 10.1093/carcin/23.7.1229. [DOI] [PubMed] [Google Scholar]
- 18.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006;38(1):86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 19.O’Dowd G, Veltri R, Miller M, Strum S. The Gleason Score. A significant biologic manifestation of prostate cancer agressiveness on biopsy. PCRI Insights. 2001:4.1. [Google Scholar]
- 20.Beaulieu M, Levesque E, Tchernof A, Beatty BG, Belanger A, Hum DW. Chromosomal localization, structure, and regulation of the UGT2B17 gene, encoding a C19 steroid metabolizing enzyme. DNA Cell Biol. 1997;16(10):1143–1154. doi: 10.1089/dna.1997.16.1143. [DOI] [PubMed] [Google Scholar]
- 21.Guillemette C, Levesque E, Beaulieu M, Turgeon D, Hum DW, Belanger A. Differential regulation of two uridine diphospho-glucuronosyltransferases, UGT2B15 and UGT2B17, in human prostate LNCaP cells. Endocrinology. 1997;138(7):2998–3005. doi: 10.1210/endo.138.7.5226. [DOI] [PubMed] [Google Scholar]
- 22.Hum DW, Belanger A, Levesque E, Barbier O, Beaulieu M, Albert C, et al. Characterization of UDP-glucuronosyltransferases active on steroid hormones. J Steroid Biochem Mol Biol. 1999;69(1–6):413–423. doi: 10.1016/s0960-0760(99)00061-8. [DOI] [PubMed] [Google Scholar]
- 23.Gsur A, Preyer M, Haidinger G, Schatzl G, Madersbacher S, Marberger M, et al. A polymorphism in the UDP-Glucuronosyltransferase 2B15 gene (D85Y) is not associated with prostate cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11(5):497–498. [PubMed] [Google Scholar]
- 24.MacLeod SL, Nowell S, Plaxco J, Lang NP. An allele-specific polymerase chain reaction method for the determination of the D85Y polymorphism in the human UDP-glucuronosyltransferase 2B15 gene in a case-control study of prostate cancer. Ann Surg Oncol. 2000;7(10):777–782. doi: 10.1007/s10434-000-0777-3. [DOI] [PubMed] [Google Scholar]
- 25.Okugi H, Nakazato H, Matsui H, Ohtake N, Nakata S, Suzuki K. Association of the polymorphisms of genes involved in androgen metabolism and signaling pathways with familial prostate cancer risk in a Japanese population. Cancer Detect Prev. 2006;30(3):262–268. doi: 10.1016/j.cdp.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Park J, Chen L, Shade K, Lazarus P, Seigne J, Patterson S, et al. Asp85tyr polymorphism in the udp-glucuronosyltransferase (UGT) 2B15 gene and the risk of prostate cancer. J Urol. 2004;171(6 Pt 1):2484–2488. doi: 10.1097/01.ju.0000117748.44313.43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


