Abstract
The association found between breast cancer development and prolonged exposure to estrogens suggests that this hormone is of etiologic importance in the causation of the disease. Studies on estrogen metabolism, formation of DNA adducts, carcinogenicity, cell transformation and mutagenicity have led to the hypothesis that reaction of certain estrogen metabolites, predominantly catechol estrogen-3,4-quinones, with DNA forms depurinating adducts [4-OHE1(E2)-1-N3Ade and 4-OHE1(E2)-1-N7Gua]. These adducts cause mutations leading to the initiation of breast cancer. Catechol-O-methyltransferase (COMT) is considered an important enzyme that protects cells from the genotoxicity and cytotoxicity of catechol estrogens, by preventing their conversion to quinones. The goal of the present study was to investigate the effect of COMT inhibition on the formation of depurinating estrogen-DNA adducts. Immortalized human breast epithelial MCF-10F cells were treated with 4-OHE2 (0.2 or 0.5 μM) for 24 h at 120, 168, 216, and 264 h post-plating or one time at 1–30 μM 4-OHE2 with or without the presence of COMT inhibitor (Ro41-0960). The culture media were collected at each point, extracted by solid-phase extraction and analyzed by HPLC connected with a multichannel electrochemical detector. The results demonstrate that MCF-10F cells oxidize 4-OHE2 to E1(E2)-3,4-Q, which react with DNA to form the depurinating N3Ade and N7Gua adducts. The COMT inhibitor Ro41-0960 blocked the methoxylation of catechol estrogens, with concomitant 3–4 fold increases in the levels of the depurinating adducts. Thus, low activity of COMT leads to higher levels of depurinating estrogen-DNA adducts that can induce mutations and initiate cancer.
Keywords: estrogen metabolism, estrogen protective enzymes, COMT inhibition, depurinating estrogen-DNA adducts
Introduction
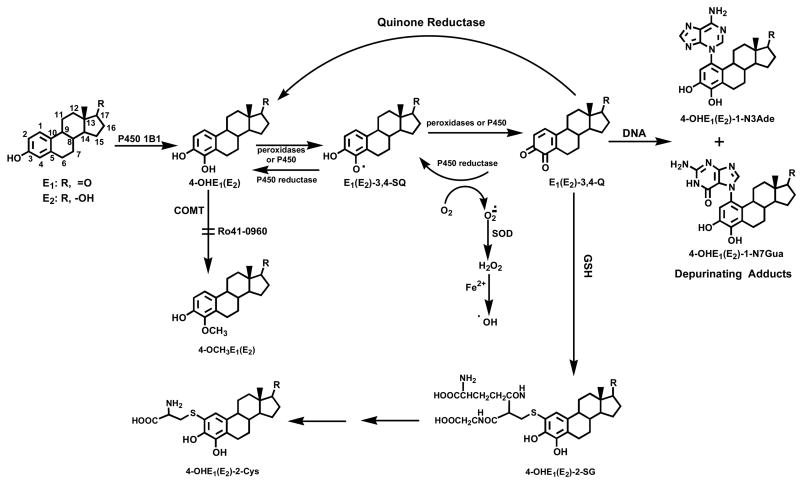
Prolonged exposure of women to high estrogen levels is associated with an elevated incidence of breast cancer [1–5]. Experiments on estrogen metabolism [6–10], formation of DNA adducts [11–17], mutagenicity [17–21], cell transformation [22–24] and carcinogenicity [25–28] have led to the hypothesis that certain estrogen metabolites, predominantly catechol estrogen-3,4-quinones, react with DNA to cause the mutations leading to the initiation of cancer (Fig. 1) [17]. The reaction of estrone(estradiol)-3,4-quinones [E1(E2)-3,4-Q], derived from 4-OHE1(E2), with DNA produces predominantly depurinating adducts and very small amounts of stable adducts [11, 13, 14, 29].
Fig. 1.
Major pathway of estrogen activation in the formation of depurinating estrogen-DNA adducts.
In extrahepatic tissues, cytochrome P450 (CYP)1A1 and CYP1B1 predominantly metabolize the natural estrogens E1 and E2 to 2- and 4- catechol estrogens (CE), respectively [30–32], which can be competitively oxidized to their respective semiquinones and quinones. In general, the CE are inactivated by conjugating reactions, such as glucuronidation and sulfation. A common pathway of inactivation in extrahepatic tissues, however, occurs by O-methylation catalyzed by the ubiquitous catechol-O-methyltransferase (COMT) [33]. If the formation of E1 and E2 is excessive, due to overexpression of aromatase and/or the presence of excess sulfatase that converts stored E1 sulfate to E1, increased formation of CE is expected (Fig. 1). In particular, the presence and/or induction of CYP1B1 and other 4-hydroxylases could dramatically increase the formation of 4-OHE1(E2). Thus, conjugation of 4-OHE1(E2) via methylation can become insufficient, and competitive oxidation of 4-OHE1(E2) to E1(E2)-3,4-Q could be more abundant [34]. The increased level of quinones would generate more reaction with DNA at the N-3 of adenine (Ade) and N-7 of guanine (Gua) to form depurinating adducts (Fig. 1) [17, 29, 34]. These adducts are lost from DNA by destabilization of the glycosyl bond. The apurinic sites generated in the DNA can produce mutations by error-prone repair [18–20], which in turn can lead to initiation of cancer.
The Phase II enzyme COMT is considered to be a key enzyme in decreasing the effects of 4-OHE1(E2) by converting the catechol estrogens into the corresponding methoxy derivatives [33]. COMT is an intracellular enzyme that is present as both soluble and membrane-bound forms encoded by the same gene with different transcription start sites [35]; the soluble form is the major one in most organs [33]. COMT activity can be altered by either endogenous factors, such as genetic polymorphisms and levels of expression, or exogenous factors such as inhibition by environmental compounds. Genetic epidemiology studies have proposed a possible correlation between the low activity allele (COMTLL) and increased breast cancer risk [36–38].
COMT activity can be inhibited by many natural and synthetic compounds [33, 39, 40]. Ro41-0960 is a nitrocatechol-type inhibitor of COMT that inhibits methylation of catechol estrogens. It is a poor substrate for COMT, but binds tightly to catalytic sites of the enzyme, thus inhibiting methylation of other substrates without depleting cofactors [41, 42]. We hypothesize that COMT inhibition decreases inactivation of CE, which may in turn lead to increased formation of CE-Q and DNA damage that initiates cancer.
To fully understand how estrogens can become carcinogenic in the human breast through metabolic activation to CE-Q an experimental system is required in which estrogens or their metabolites (e.g., 4-OHE2) would induce transformation phenotypes in human breast epithelial cells in vitro that are indicative of neoplasia. Data from a recent study showed that successive treatment of MCF-10F cells with 4-OHE2 induced mutations, cell transformation and cancer [21, 24]. The ERα-negative MCF-10F cell line is a good experimental model for researching the carcinogenicity and mutagenic potential of 4-OHE2. To investigate the implications of possible COMT inhibition by Ro41-0960 and increased formation of depurinating adducts, the cells were preincubated with 3 μM Ro41-0960 and then treated with 4-OHE2 (0.2–30 μM) for 24 h. The profile of 4-OHE2 metabolites, conjugates and depurinating DNA adducts was determined in cell culture medium by HPLC equipped with a multichannel electrochemical detector (ECD) and validated by ultraperformance liquid chromatography (UPLC)-MS/MS techniques. This is the first report on the metabolic profile of 4-OHE2 in MCF-10F cells treated in a dose-response manner
Materials and Methods
Chemicals and Reagents
4-OHE2 and all standards were synthesized in our laboratory, as previously described [13, 43–45]. Ro41-0960 and all other chemicals were purchased from Sigma (St. Louis, MO). MCF-10F cells were obtained from the ATCC (Rockville, MD).
Cell culture and treatment
MCF-10F cells were cultured in phenol red DMEM/F12 (1:1) medium containing 20 ng/ml epidermal growth factor, 0.01 mg/ml insulin, 500 ng/ml hydrocortisone, 5 % horse serum and 100 μg/ml penicillin/streptomycin mixture and maintained in a humidified incubator at 37 ºC and 5% CO2. Estrogen-free medium was prepared in phenol red-free DMEM/F12 medium with charcoal-stripped fetal bovine serum (FBS). To keep the concentration of DMSO the same (0.001%) in all experiments, different stock solutions of 4-OHE2 (0.2–30 mM) were prepared. A stock of 9 mM Ro41-0960 was prepared in ethanol.
The MCF-10F cells (2.5 × 105 cells) were seeded for 48 h in estrogen-containing medium. The medium was changed to estrogen-free medium and the cells were grown for another 72 h. To investigate the direct relationship of COMT inhibition on the formation of depurinating adducts, the cells were first treated with 3 μM Ro41-0960 for 1 h and then treated once with various concentrations of 4-OHE2 (0–30 μM) for 24 h.
For multiple treatment experiments, 1.0 × 105 MCF-10F cells were seeded and treated with 0.2 or 0.5 μM 4-OHE2 for 24 h at 120, 168, 216 and 264 h post-seeding. Cell cultures were or were not pre-incubated with Ro41-0960 for 1 h prior to the addition of 4-OHE2. After each treatment, the medium was removed, ascorbic acid was added (2 mM final concentration) to prevent further oxidation of desired compounds, and the mixture was processed immediately. Media from four T-150 flasks of MCF-10F cells treated with 10 μl DMSO and 3.3 μl of ethanol were used as controls.
Sample preparation and analysis by HPLC-ECD and by UPLC-MS/MS
i. Sample Preparation
Culture media from four flasks (40 mL) were processed by passing through Varian C8 Certify II cartridges (Varian, Harbor City, CA). The cartridges were pre-equilibrated by sequentially passing 1 ml of methanol, distilled water, and potassium phosphate buffer (100 mM, pH 8) through them. Culture media were adjusted with 1 ml of 1 M potassium phosphate buffer to pH 8.0 and passed through the cartridges. After washing with 200 μl of the phosphate buffer, the analytes were eluted with 1 ml of elution buffer [methanol:acetonitrile:water: trifluoroacetic acid (8:1:1:0.1)] and evaporated by using a Jouan concentrator (Thermo Scientific, Waltham, MA). The residue was resuspended in 150 μl of methanol/water (1:1) and filtered through a 5000-MW cut-off filter (Millipore, Bedford, MA).
ii. HPLC
Analyses of all samples were conducted on an HPLC system equipped with dual ESA Model 580 solvent delivery modules, an ESA Model 540 auto-sampler and a 12-channel CoulArray electrochemical detector (ESA, Chelmsford, MA). The two mobile phases used were A: acetonitrile:methanol:buffer:water (15:5:10:70) and B: acetonitrile:methanol: buffer:water (50:20:10:20). The buffer was a mixture of 0.25 M citric acid and 0.5 M ammonium acetate in triple-distilled water, and the pH was adjusted to 3.6 with acetic acid. The 95-μl injections were carried out on a Phenomenex Luna-2 C-18 column (250 × 4.6 mm, 5 μm; Phenomenex, Torrance, CA), initially eluted isocratically at 90% A/10% B for 15 min, followed by a linear gradient to 90% B in the next 40 min, and held there for 5 min (total 50 min gradient) at a flow rate of 1 ml/min and a temperature of 30 °C. The serial array of 12 coulometric electrodes was set at potentials of -35, 10, 70, 140, 210, 280, 350, 420, 490, 550, 620 and 690 mV. The system was controlled and the data were acquired and processed using the CoulArray software package (ESA). Peaks were identified by both retention time and peak height ratios between the dominant peak and the peaks in the two adjacent channels. The metabolites, conjugates and depurinating adducts were quantified by comparison of peak response ratios with known amounts of standards.
iii. UPLC-MS/MS
Some of the results from HPLC-ECD determinations were validated by analyzing the same samples on a Waters Acquity UPLC equipped with a MicroMass QuattroMicro triple stage quadrupole mass spectrometer (Waters, Milford, MA). The 10-μl injections were carried out on a Waters Acquity UPLC BEHC18 column (1.7 μm, 1 × 100 mm). The instrument was operated in the positive electrospray ionization mode. All aspects of system operation, data acquisition and processing were controlled using QuanLynx v4.0 software (Waters). The column was eluted starting with 5% acetonitrile in water (0.1% formic acid) for 1 min at a flow rate of 150 μl/min; then a gradient to 55% acetonitrile in 10 min was run. Ionization was achieved using the following settings: capillary voltage 3 kV; cone voltage 15–40 V; source block temperature 100 °C; desolvation temperature 200 °C, with a nitrogen flow of 400 l/h. Nitrogen was used as both the desolvation and auxiliary gas. Argon was used as the collision gas. Three-point calibration curves were run for each standard. Triplicate samples were analyzed for each data point.
Results and Discussion
To examine the profile of estrogen metabolism in MCF-10F cells, an HPLC method with a CoulArray ECD [10] was used to quantify the relative concentrations of estrogen metabolites, conjugates and depurinating DNA adducts. Standard solutions of each compound were combined to generate equimolar mixtures containing varying concentrations of each estrogen standard and injected onto the column. These standard solutions were then used to generate calibration curves. Standard curves were linear between 125 and 500 pg/μl. The limit of detection under the conditions of analysis was ~10 pg/μl on column (for 95-μl injection volume).
To determine the effect of COMT inhibition on 4-OHE2 metabolism, MCF-10F cells were pretreated with 3 μM Ro41-0960 for 1 h and then exposed to different concentrations of 4-OHE2. This concentration of the inhibitor reduced COMT activity by 90–95% at 24 h, as assessed by methylation of 4-OHE2 (data not shown).
To mimic intermittent exposure of immortalized MCF-10F cells to estrogen [22, 23], the cells were successively treated with 0.2 or 0.5 μM 4-OHE2 twice a week for two weeks (Tables 1 and 2). The cell culture medium was removed 24 h after each treatment and processed for analysis of estrogen metabolites, conjugates and depurinating DNA adducts. Treatment with the COMT inhibitor decreased the concentration of methoxylated conjugates formed from 0.2 μM 4-OHE2 to undetectable levels and increased the recovery of 4-OHE1(E2) 6–12-fold (Table 1).
Table 1.
Intermittent incubation of MCF-10F cells with 0.2 μM 4-OHE2 with or without 3 μM Ro41-09601
| 4-OHE2 treatment | 4-OHE2+ Ro41-0960 treatment | |||||||
|---|---|---|---|---|---|---|---|---|
| Detected Compounds2 | 1st | 2nd | 3rd | 4th | 1st | 2nd | 3rd | 4th |
| pmol/106cells | ||||||||
| 4-OHE1(E2) | 0.08 ± 0.02 | 0.22 ± 0.02 | 0.27 ± 0.08 | 0.31 ± 0.04 | 0.98 ± 0.01 | 1.64 ± 0.03 | 1.81 ± 0.11 | 1.91 ± 0.20 |
| 4-OCH3E1(E2) | 0.10 ± 0.01 | 0.11 ± 0.04 | 0.39 ± 0.21 | 0.76 ± 0.12 | nd3 | nd | nd | nd |
| 4-OH E1(E2)-2-SG | nd | nd | nd | 0.01 ± 0.01 | nd | nd | 0.09 ± 0.03 | 0.36 ± 0.09 |
| 4-OH E1(E2)-2-Cys | nd | nd | nd | 0.18 ± 0.03 | 0.12 ± 0.03 | 0.16 ± 0.03 | 1.66 ± 0.35 | 2.35 ± 0.96 |
| 4-OH E1(E2)-1-N3Ade | nd | nd | loq4 | 0.30 ± 0.07 | nd | loq | 0.55 ± 0.13 | 0.59 ± 0.07 |
| 4-OH E1(E2)-1-N7Gua | nd | nd | loq | 0.81 ± 0.12 | nd | loq | 0.44 ± 0.11 | 0.85 ± 0.12 |
4-OHE2 was incubated with MCF-10F cells at 37 °C for 24 h in the presence or absence of Ro41-0960 (COMT inhibitor) twice weekly for two weeks.
The compounds were identified and quantified by HPLC-ECD, and values are an average of three replicates.
Not detected.
Although the compound was identified, it could not be quantified due to the limit of quantification.
Table 2.
Intermittent incubation of MCF-10F cells with 0.5 μM 4-OHE2 with or without 3 μM Ro41-09601
| 4-OHE2 treatment | 4-OHE2 + Ro41-0960 treatment | |||||||
|---|---|---|---|---|---|---|---|---|
| Detected Compounds2 | 1st | 2nd | 3rd | 4th | 1st | 2nd | 3rd | 4th |
| pmol/106cells | ||||||||
| 4-OHE1(E2) | 0.14 ± 0.02 | 0.46 ± 0.02 | 0.49 ± 0.04 | 0.47 ± 0.04 | 1.14 ± 0.01 | 2.30 ± 0.03 | 2.49 ± 0.98 | 2.58 ± 0.96 |
| 4-OCH3 E1(E2) | 0.25 ± 0.01 | 2.58 ± 0.54 | 7.64 ± 0.40 | 5.18 ± 0.11 | nd3 | nd | nd | nd |
| 4-OH E1(E2)-2-SG | nd | nd | 0.27 ± 0.01 | 0.62 ± 0.04 | 0.04 ± 0.03 | 0.34 ± 0.03 | 0.38 ± 0.02 | 0.60 ± 0.06 |
| 4-OH E1(E2)-2-Cys | nd | 0.32 ± 0.03 | 0.75 ± 0.42 | 1.38 ± 0.04 | 0.75 ± 0.01 | 1.18 ± 0.03 | 1.16 ± 0.05 | 1.49 ± 0.01 |
| 4-OH E1(E2)-1-N3Ade | nd | nd | loq4 | 0.25 ± 0.12 | nd | 0.21 ± 0.01 | 0.55 ± 0.01 | 0.83 ± 0.09 |
| 4-OH E1(E2)-1-N7Gua | nd | nd | loq | loq | nd | loq | 0.81 ± 0.12 | 0.70 ± 0.19 |
4-OHE2 was incubated with MCF-10F cells at 37 °C for 24 h in the presence or absence of Ro41-0960 (COMT inhibitor) twice weekly for two weeks.
The compounds were identified and quantified by HPLC-ECD, and values are an average of three replicates.
Not detected.
Although the compound was identified, it could not be quantified due to the limit of quantification.
In the absence of the inhibitor, 4-OCH3E1(E2) was the major product of metabolism from 0.2 μM 4-OHE2. Small, but increasing amounts of the GSH and Cys conjugates were detected after successive treatments with 4-OHE2. The 4-OHE1(E2)-2-Cys conjugates are presumably obtained by mercapturic acid biosynthesis from the 4-OHE1(E2)-2-SG [46] and by conjugation of E1(E2)-3,4-Q with the 0.4 mM Cys present in the culture medium. The presence of Cys in the medium diminishes the significance of these results. The depurinating adducts were also detected in small amounts after the 3rd and 4th treatments (Table 1). In the presence of the COMT inhibitor, the methoxy conjugates were undetectable, whereas the GSH and Cys conjugates, as well as the two depurinating DNA adducts were increased (Table 1). In the presence of the COMT inhibitor, low levels of both adducts were detected after the 2nd treatment. After the 3rd treatment, the adducts could be quantified if the inhibitor was present. After the 4th treatment, the presence of the inhibitor resulted in approximately twice as much N3Ade, although the level of the N7Gua adduct was about the same as without the inhibitor.
When the cells were treated with 0.5 μM 4-OHE2, inclusion of the COMT inhibitor increased the level of 4-OHE1(E2) in the medium 5–8 fold and the 4-OCH3E1(E2) was undetectable. The levels of the GSH and Cys conjugates increased a little in the presence of the COMT inhibitor, especially at the early time points. With the inhibitor, the level of N3Ade adduct was increased after the 2nd, 3rd and 4th treatments with 4-OHE2, and the N7Gua adduct was quantifiable after the 3rd and 4th treatments. Statistical comparison of adduct levels could be carried out only for N3Ade after the 4th treatment; the presence of the COMT inhibitor led to a significant increase (p < 0.01).
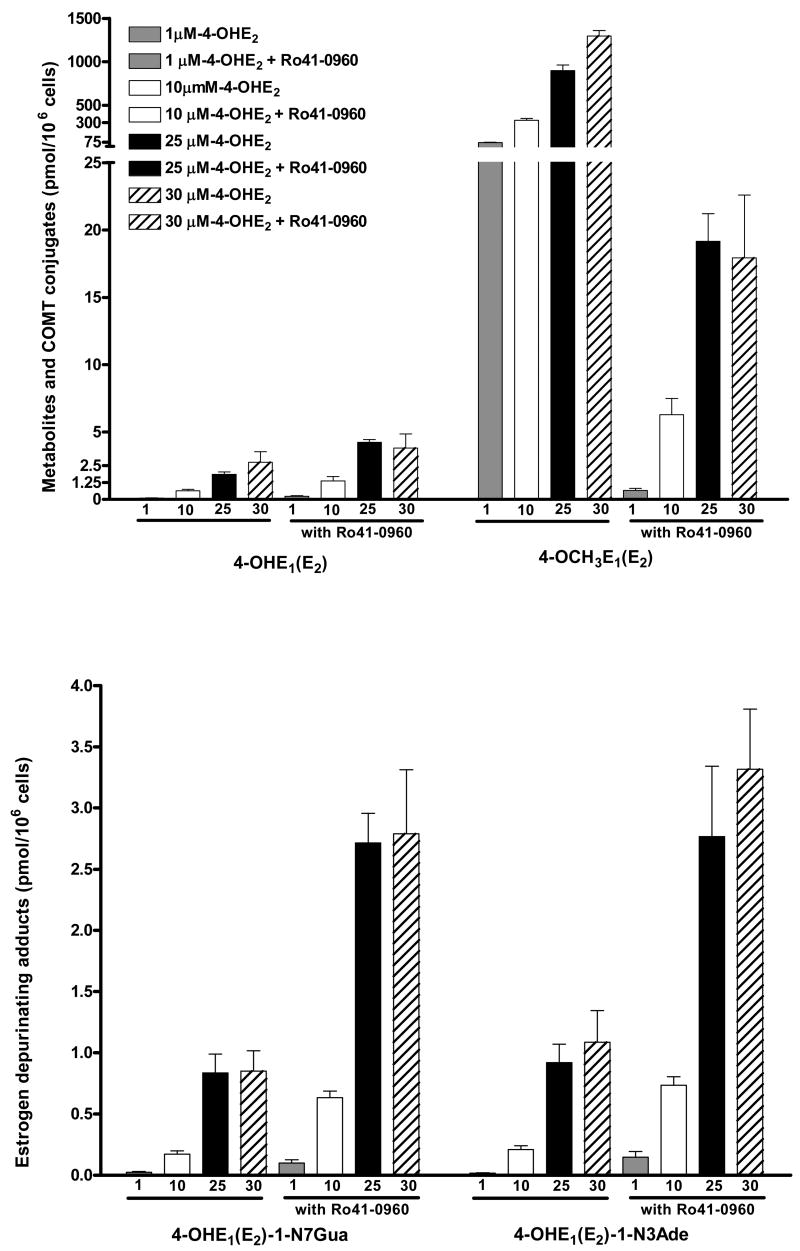
When MCF-10F cells were incubated with 1–30 μM 4-OHE2 for 24 h, a dose-response was observed (Table 3, Fig. 2). Increased concentrations of 4-OHE2 produced a large increase in 4-OCH3E1(E2) formed. When the COMT inhibitor was present, however, formation of 4-OCH3E1(E2) decreased 98–99% (p < 0.003). In parallel, the higher concentrations of 4-OHE2 yielded an increase in the depurinating N3Ade and N7Gua adducts (Table 3, Fig. 2). A more dramatic 3 to 4-fold increase occurred when Ro41-0960 was present. In the presence of the inhibitor the levels of both adducts were significantly different, p < 0.05. The level of 4-OHE1(E2)-2-SG conjugate was low and was unchanged by the presence of the inhibitor. The Cys conjugate, in contrast, had a dose-response based on the concentration of 4-OHE2 and was 4-fold higher in the presence of the COMT inhibitor (Table 3, Fig. 2). In the presence of the COMT inhibitor, we speculate that the overall recovery of estrogen compounds is much lower because the catechol estrogens are oxidized to E1(E2)-3,4-Q, which readily react with the SH groups of Cys and amino group of lysine in proteins.
Table 3.
Incubation of MCF-10F cells with 1–30 μM of 4-OHE2 with or without 3 μM Ro41-0960 for 24 hrs at 37 °C
| 4-OHE2 treatment | 4-OHE2 + Ro41-0960 treatment | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Compounds Detected | 1 μM | 10 μM | 25 μM | 30 μM | 1 μM | 10 μM | 25 μM | 30 μM | |
| pmol/106 cells | |||||||||
| 4-OHE1(E2) | 0.10 ± 0.03 | 0.63 ± 0.21 | 1.86 ± 0.3 | 2.75 ± 1.36 | 0.60 ± 0.05 | 3.64 ± 0.57 | 4.91 ± 0.85 | 5.81 ± 1.02 | |
| 4-OCH3E1(E2) | 70 ± 5 | 340 ± 34 | 899 ± 112 | 1297 ± 110 | 0.66 ± 0.26 | 6.37 ± 2.17 | 19.2 ± 3.5 | 17.9 ± 2.3 | |
| 4-OHE1(E2)-2-SG | 0.08 ± 0.03 | 0.35 ± 0.04 | 0.54 ± 0.23 | 1.1 ± 0.36 | 0.14 ± 0.02 | 0.47 ± 0.13 | 0.59 ± 0.18 | 0.87 ± 0.32 | |
| 4-OHE1(E2)-2-Cys | 0.25 ± 0.03 | 0.62 ± 0.12 | 3.89 ± 0.49 | 7.9 ± 1.93 | 0.3 ± 0.21 | 1.51 ± 0.56 | 16.1 ± 4.7 | 33.3 ± 6.4 | |
| 4-OHE1(E2)-1-N7Gua | 0.02 ± 0.01 | 0.17 ± 0.05 | 0.84 ± 0.27 | 0.85 ± 0.29 | 0.10 ± 0.05 | 0.63 ± 0.09 | 2.71 ± 0.42 | 2.79 ± 0.90 | |
| 4-OHE1(E2)-1-N3Ade | 0.02 ± 0.01 | 0.21 ± 0.05 | 0.92 ± 0.26 | 1.09 ± 0.45 | 0.15 ± 0.08 | 0.74 ± 0.12 | 2.77 ± 1.00 | 3.32 ± 0.85 | |
Fig. 2.
Incubation of MCF-10F cells with 1–30 μM 4-OHE2 with or without 3 μM Ro41-0960 (COMT inhibitor) for 24 h.
As shown in Figure 1, a balanced estrogen metabolism involves conversion of 4-OHE2 to its methoxy derivative. This process is catalyzed by the enzyme COMT. A competing pathway for 4-OHE2 is its oxidation to E2-3,4-semiquinone and then to E2-3,4-Q. Two other pathways can inhibit the formation of depurinating DNA adducts formed by reaction of E1(E2)-3,4-Q with DNA. One is the reduction of the quinone to the CE catalyzed by quinone reductase (Fig. 1) [47,48]. The second pathway is the reaction of the quinone with GSH.
In the present study, we have found that in MCF-10F cells increasing concentrations of 4-OHE2 afford higher levels of the depurinating adducts (Fig. 2, Table 3). At the same time, very large amounts of 4-OCH3E1(E2) are observed, indicating that large amounts of COMT are present in the cells. When the COMT inhibitor was present, inhibition of CE methylation was almost total and the levels of the N3Ade and N7Gua adducts increased four-fold (Fig. 2, Table 3). From these studies inhibition of COMT activity clearly unbalances estrogen metabolism toward excessive formation of E1(E2)-3,4-Q. As part of this imbalance, greater formation of hydroxyl radicals occurs. This is demonstrated by formation of 8-hydroxy-2′-deoxyguanosine, which is derived from the increased redox cycling between the estrogen semiquinones and quinones (Fig. 1) [49].
Another important factor is excessive formation of 4-OHE1(E2) as a major metabolite of E1(E2), catalyzed by CYP1B1 [30–32]. Minimization of estrogen-DNA adduct formation occurs when COMT is present at high levels because methoxylation of 4-OHE1(E2) is one of the key elements in reducing adduct formation. This important role of COMT in protecting cells from cancer initiation by estrogens suggests that COMT polymorphisms could have a significant effect on cancer incidence. For example, the common val108met polymorphism decreases COMT activity 3–4-fold [50,51]. Thus, persons homozygous for this polymorphism could be at increased risk for estrogen-induced cancers. The critical events described above are extremely useful in determining the agents that can minimize the formation of estrogen-DNA adducts, thereby inhibiting the initiation of breast and other human cancers.
Acknowledgments
Grant support: This research was supported by U.S. Public Health Service grant P01 CA49210 from the National Cancer Institute and the U.S. Army Breast Cancer Research Program grant DAMD 17-03-1-0229. Core support at the Eppley Institute was provided by grant P30 CA36727 from the National Cancer Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dorgan JF, Longcope C, Stephenson HE, Jr, Falk RT, Miller R, Franz C, Kahle L, Campbell WS, Tangrea JA, Schatzkin A. Relation of prediagnostic serum estrogen and antrogen levels to breast cancer risk. Cancer Epidemiol Biomarkers Prev. 1996;5:533–539. [PubMed] [Google Scholar]
- 2.Thomas HV, Key TJ, Allen DS, Moore JW, Dowsett M, Fentiman IS, Wang DY. A prospective study of endogenous serum hormone concentrations and breast cancer risk in postmenopausal women on the island of Guernsey. Br J Cancer. 1997;76:401–405. doi: 10.1038/bjc.1997.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hankinson SE, Willett WC, Manson JE, Colditz GA, Hunter DJ, Spiegelman D, Barbieri RL, Speizer FE. Plasma sex steroid hormone levels and risk of breast cancer in postmenopausal women. J Natl Cancer Inst. 1998;90:1292–1299. doi: 10.1093/jnci/90.17.1292. [DOI] [PubMed] [Google Scholar]
- 4.Kabuto M, Akiba S, Stevens RG, Neriishi K, Land CE. A prospective study of estradiol and breast cancer in Japanese women. Cancer Epidemiol Biomarkers Prev. 2000;9:575–579. [PubMed] [Google Scholar]
- 5.Endogenous Hormones and Breast Cancer Collaborative Group. Endogenous sex hormones and breast cancer in postmenopausal women: reanalysis of nine prospective studies. J Natl Cancer Inst. 2002;94:606–616. doi: 10.1093/jnci/94.8.606. [DOI] [PubMed] [Google Scholar]
- 6.Zhu BT, Conney AH. Functional role of estrogen metabolism in target cells: Review and perspectives. Carcinogenesis. 1998;19:1–27. doi: 10.1093/carcin/19.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Cavalieri EL, Kumar S, Todorovic R, Higginbotham S, Badawi AF, Rogan EG. Imbalance of estrogen homeostasis in kidney and liver of hamsters treated with estradiol: Implications for estrogen-induced initiation of renal tumors. Chem Res Toxicol. 2001;14:1041–1050. doi: 10.1021/tx010042g. [DOI] [PubMed] [Google Scholar]
- 8.Devanesan P, Santen RJ, Bocchinfuso WP, Korach KS, Rogan EG, Cavalieri EL. Catechol estrogen metabolites and conjugates in mammary tumors with hyperplastic tissue from estrogen receptor-α knock-out (ERKO)/Wnt-1 Mice: Implications for initiation of mammary tumors. Carcinogenesis. 2001;22:1573–1576. doi: 10.1093/carcin/22.9.1573. [DOI] [PubMed] [Google Scholar]
- 9.Cavalieri EL, Devanesan P, Bosland MC, Badawi AF, Rogan EG. Catechol estrogen metabolites and conjugates in different regions of the prostate of Noble rats treated with 4-hydroxyestradiol: implications for estrogen-induced initiation of prostate cancer. Carcinogenesis. 2002;23:329–333. doi: 10.1093/carcin/23.2.329. [DOI] [PubMed] [Google Scholar]
- 10.Rogan EG, Badawi AF, Devanesan PD, Meza JL, Edney JA, West WW, Higginbotham SM, Cavalieri EL. Relative imbalances in estrogen metabolism and conjugation in breast tissue of women with carcinoma: Potential biomarkers of susceptibility to cancer. Carcinogenesis. 2003;24:697–702. doi: 10.1093/carcin/bgg004. [DOI] [PubMed] [Google Scholar]
- 11.Cavalieri EL, Stack DE, Devanesan PD, Todorovic R, Dwivedy I, Higginbotham S, Johansson SL, Patil KD, Gross ML, Gooden JK, Ramanathan R, Cerny RL, Rogan EG. Molecular origin of cancer: Catechol estrogen-3,4-quinones as endogenous tumor initiators. Proc Natl Acad Sci USA. 1997;94:10937–10942. doi: 10.1073/pnas.94.20.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markushin Y, Zhong W, Cavalieri EL, Rogan EG, Small GJ, Yeung ES, Jankowiak R. Spectral characterization of catechol estrogen quinone (CEQ)-derived DNA adducts and their identification in human breast tissue extract. Chem Res Toxicol. 2003;16:1107–1117. doi: 10.1021/tx0340854. [DOI] [PubMed] [Google Scholar]
- 13.Li K-M, Todorovic R, Devanesan P, Higginbotham S, Kofeler H, Ramanathan R, Gross ML, Rogan EG, Cavalieri EL. Metabolism and DNA binding studies of 4-hydroxyestradiol and estradiol-3,4-quinone in vitro and in female ACI rat mammary gland in vivo. Carcinogenesis. 2004;25:289–297. doi: 10.1093/carcin/bgg191. [DOI] [PubMed] [Google Scholar]
- 14.Zahid M, Kohli E, Saeed M, Rogan E, Cavalieri E. The greater reactivity of estradiol-3,4-quinone versus estradiol-2,3-quinone with DNA in the formation of depurinating DNA adducts. Implications for tumor–initiating activity. Chem Res Toxicol. 2006;19:164–172. doi: 10.1021/tx050229y. [DOI] [PubMed] [Google Scholar]
- 15.Markushin Y, Gaikwad N, Zhang H, Kapke P, Rogan EG, Cavalieri EL, Trock BJ, Pavlovich C, Jankowiak R. Potential biomarker for early risk assessment of prostate cancer. Prostate. 2006;66:1565–1571. doi: 10.1002/pros.20484. [DOI] [PubMed] [Google Scholar]
- 16.Saeed M, Rogan E, Fernandez SV, Sheriff F, Russo J, Cavalieri E. Formation of depurinating N3Adenine and N7Guanine adducts by MCF-10F cells cultured in the presence of 4-hydroxyestradiol. Int J Cancer. 2007;120:1821–1824. doi: 10.1002/ijc.22399. [DOI] [PubMed] [Google Scholar]
- 17.Cavalieri E, Chakravarti D, Guttenplan J, Hart E, Ingle J, Jankowiak R, Muti P, Rogan E, Russo J, Santen R, Sutter T. Catechol estrogen quinones as initiators of breast and other human cancers: implications for biomarkers of susceptibility and cancer prevention. BBA Reviews on Cancer. 2006;1766:63–78. doi: 10.1016/j.bbcan.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Chakravarti D, Mailander P, Li K-M, Higginbotham S, Zhang HL, Gross ML, Meza JL, Cavalieri EL, Rogan EG. Evidence that a burst of DNA depurination in SENCAR mouse skin induces error-prone repair and form mutations in the H-ras gene. Oncogene. 2001;20:7945–7953. doi: 10.1038/sj.onc.1204969. [DOI] [PubMed] [Google Scholar]
- 19.Mailander PC, Meza JL, Higginbotham S, Chakravarti D. Induction of A.T to G.C mutations by erroneous repair of depurinated DNA following estrogen treatment of the mammary gland of ACI rats. J Steroid Biochem Mol Biol. 2006;101:204–215. doi: 10.1016/j.jsbmb.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Z, Kosinska W, Khmelnitsky M, Cavalieri EL, Rogan EG, Chakravarti D, Sacks PG, Guttenplan JB. Mutagenic activity of 4-hydroxyestradiol, but not 2-hydroxyestradiol, in BB rat2 embryonic cells, and the mutational spectrum of 4-hydroxyestradiol. Chem Res Toxicol. 2006;19:475–479. doi: 10.1021/tx0502645. [DOI] [PubMed] [Google Scholar]
- 21.Fernandez SV, Russo IH, Russo J. Estradiol and its metabolites 4-hydroxyestradiol and 2-hydroxyestradiol induce mutations in human breast epithelial cells. Int J Cancer. 2006;118:1862–1868. doi: 10.1002/ijc.21590. [DOI] [PubMed] [Google Scholar]
- 22.Russo J, Lareef MH, Balogh G, Guo S, Russo IH. Estrogen and its metabolites are carcinogenic agents in human breast epithelial cells. J Steroid Biochem Mol Biol. 2003;87:1–25. doi: 10.1016/s0960-0760(03)00390-x. [DOI] [PubMed] [Google Scholar]
- 23.Lareef MH, Garber J, Russo PA, Russo IH, Heulings R, Russo J. The estrogen antagonist ICI-182-780 does not inhibit the transformation phenotypes induced by 17-beta-estradiol and 4-OH estradiol in human breast epithelial cells. Int J Oncol. 2005;26:423–429. [PubMed] [Google Scholar]
- 24.Russo J, Fernandez SV, Russo PA, Fernbaugh R, Sheriff FS, Lareef HM, Garber J, Russo IH. 17-Beta-estradiol induces transformation and tumorigenesis in human breast epithelial cells. Faseb J. 2006;20:1622–1634. doi: 10.1096/fj.05-5399com. [DOI] [PubMed] [Google Scholar]
- 25.Liehr JG, Fang WF, Sirbasku DA, Ari-Ulubelen A. Carcinogenicity of catecholestrogens in Syrian hamsters. J Steroid Biochem. 1986;24:353–356. doi: 10.1016/0022-4731(86)90080-4. [DOI] [PubMed] [Google Scholar]
- 26.Li JJ, Li SA. Estrogen carcinogenesis in Syrian hamster tissue: Role of metabolism. Fed Proc. 1987;46:1858–1863. [PubMed] [Google Scholar]
- 27.Newbold RR, Liehr JG. Induction of uterine adenocarcinoma in CD-1 mice by catechol estrogens. Cancer Res. 2000;60:235–237. [PubMed] [Google Scholar]
- 28.Yue W, Santen RJ, Wang JP, Li Y, Verderame MF, Bocchinfuso WP, Korach KS, Devanesan P, Todorovic R, Rogan EG, Cavalieri EL. Genotoxic metabolites of estradiol in breast: potential mechanism of estradiol induced carcinogenesis. J Steroid Biochem Mol Biol. 2003;86:477–486. doi: 10.1016/s0960-0760(03)00377-7. [DOI] [PubMed] [Google Scholar]
- 29.Cavalieri E, Rogan E, Chakravarti D. Methods in enzymology. Vol. 382. Duesseldorf, Germany: Elsevier; 2004. The role of endogenous catechol quinones in the initiation of cancer and neurodegenerative diseases: In: Sies, H., Packer, L., eds. Quinones and quinone enzymes, part B; pp. 293–319. [DOI] [PubMed] [Google Scholar]
- 30.Spink DC, Hayes CL, Young NR, Christou M, Sutter TR, Jefcoate CR, Gierthy JF. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin on estrogen metabolism in MCF-7 breast cancer cells: evidence for induction of a novel 17 beta-estradiol 4-hydroxylase. J Steroid Biochem Mol Biol. 1994;51:251–258. doi: 10.1016/0960-0760(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 31.Hayes CL, Spink DC, Spink BC, Cao JQ, Walker NJ, Sutter TR. 17 Beta-estradiol hydroxylation catalyzed by human cytochrome P450 1B1. Proc Natl Acad Sci USA. 1996;93:9776–9781. doi: 10.1073/pnas.93.18.9776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spink DC, Spink BC, Cao JQ, DePasquale JA, Pentecost BT, Fasco MJ, Li Y, Sutter TR. Differential expression of CYP1A1 and CYP1B1 in human breast epithelial cells and breast tumor cells. Carcinogenesis. 1998;19:291–298. doi: 10.1093/carcin/19.2.291. [DOI] [PubMed] [Google Scholar]
- 33.Mannisto PT, Kaakkola S. Catechol-O-methyltransferase (COMT): biochemistry, molecular biology, pharmacology, and clinical efficacy of the new selective COMT inhibitors. Pharmacol Rev. 1999;51:592–628. [PubMed] [Google Scholar]
- 34.Lu F, Zahid M, Saeed M, Cavalieri EL, Rogan EG. Estrogen metabolism and formation of estrogen DNA-adducts in estradiol-treated MCF-10F cells. The effects of 2,3,7,8-tetrachlorodibenzo-p-dioxin induction and catechol-O-methyltransferase inhibition. J Steroid Biochem Mol Biol. 2007 doi: 10.1016/j.jsbmb.2006.12.102. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hersey RM, Williams KI, Weisz J. Catechol estrogen formation by brain tissue characterized of a direct product isolation assay for estrogen 2- and 4-hydroxylase activity and its application to studies of 2- and 4-hydroxy estradiol formation by rabbit hypothalamus. Endocrinology. 1981;109:1912–1920. doi: 10.1210/endo-109-6-1912. [DOI] [PubMed] [Google Scholar]
- 36.Yim DS, Park SK, Yoo KY, Yoon KS, Chung HH, Kang HL, Ahn SH, Noh DY, Choe KJ, Jang IJ, Shin SG, Strickland PT, Hirvonen A, Kang D. Relationship between the vall58met polymorphism of catechol-O-methyl transferase and breast cancer. Pharmacogenetics. 2001;11:279–286. doi: 10.1097/00008571-200106000-00001. [DOI] [PubMed] [Google Scholar]
- 37.Huang CS, Chern HD, Chang KJ, Cheng CW, Hsu SM, Shen CY. Breast cancer risk associated with genotype polymorphism of the estrogen metabolizing genes CYP17, CYP1A1 and COMT: a mutagenic study on cancer susceptibility. Cancer Res. 1999;59:4870–4875. [PubMed] [Google Scholar]
- 38.Lavigne JA, Helzlsouer KJ, Huang HY, Strickland PT, Bell DA, Selmin O, Watson MA, Hoffman S, Comstock GW, Yager JD. An association between the allele coding for low activity of catechol-O-methytransferase and the risk for breast cancer. Cancer Res. 1997;57:5493–5497. [PubMed] [Google Scholar]
- 39.Garner CE, Burka LT, Etheridge AE, Matthews HB. Catechols metabolites of polychlorinated biphenyls inhibit the catechol-O-methyltransferase mediated metabolism of catechol estrogen. Toxcol Appl Pharmacol. 2000;162:115–123. doi: 10.1006/taap.1999.8823. [DOI] [PubMed] [Google Scholar]
- 40.Hollman PC, Katan MB. Dietry flavanoids: Intake, health effects and bioavailability. Food Chem Toxicol. 1999;37:937–942. doi: 10.1016/s0278-6915(99)00079-4. [DOI] [PubMed] [Google Scholar]
- 41.Backstrom R, Honkanen E, Pippuri A, Kairisalo P, Pystynen J, Heinola K, Nissinen E, Linden IB, Mannisto PT, Kaakkola S, Pohto P. Synthesis of some novel potent and selective catechol-O-methyltransferase inhibitors. J Med Chem. 1989;32:841–846. doi: 10.1021/jm00124a017. [DOI] [PubMed] [Google Scholar]
- 42.Ding YS, Gately SJ, Fowler JS, Chen R, Volkow ND, Logan J, Shea CE, Sugano Y, Koomen J. Mapping catechol-O-methytransferase in vivo: Initial studies with [18F] Ro41-0960. Life Sci. 1996;58:195–208. doi: 10.1016/0024-3205(95)02277-5. [DOI] [PubMed] [Google Scholar]
- 43.Saeed M, Zahid M, Rogan E, Cavalieri E. Synthesis of the catechols of natural and synthetic estrogens by using 2′-iodoxybenzoic acid (IBX) as the oxidizing agent. Steroids. 2005;70:173–178. doi: 10.1016/j.steroids.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 44.Stack DE, Byun J, Gross ML, Rogan EG, Cavalieri EL. Molecular characteristics of catechol estrogen quinones in reactions with deoxyribonucleosides. Chem Res Toxicol. 1996;9:851–859. doi: 10.1021/tx960002q. [DOI] [PubMed] [Google Scholar]
- 45.Cao K, Stack DE, Ramanathan R, Gross ML, Rogan EG, Cavalieri EL. Synthesis and structure elucidation of estrogen quinones conjugated with cysteine, N-acetylcysteine, and glutathione. Chem Res Toxicol. 1998;11:909–916. doi: 10.1021/tx9702291. [DOI] [PubMed] [Google Scholar]
- 46.Boyland E, Chasseaud LF. The role of glutathione and glutathione S-transferases in mercapturic acid biosynthesis. Adv Enzymol Relat Areas Mol Biol. 1969;32:173–219. doi: 10.1002/9780470122778.ch5. [DOI] [PubMed] [Google Scholar]
- 47.Montano MM, Chaplin LJ, Deng H, Mesia-Vela S, Gaikwad N, Zahid M, Rogan E. Protective roles of quinone reductase and tamoxifen against estrogen-induced mammary tumorigenesis. Oncogene. 2007;26:3587–3590. doi: 10.1038/sj.onc.1210144. [DOI] [PubMed] [Google Scholar]
- 48.Gaikwad NW, Rogan EG, Cavalieri EL. Evidence by ESI-MS for NQO1-catalyzed reduction of estrogen ortho-quinones. Free Radic Biol Med. 2007 doi: 10.1016/j.freeradbiomed.2007.07.021. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lavigne JA, Goodman JE, Fonong T, Odwin S, He P, Roberts DW, Yager JD. The effects of catechol-O-methyltransferase inhibition on estrogen metabolite and oxidative DNA damage levels in estradiol-treated MCF-7 cells. Cancer Res. 2001;61:7488–7494. [PubMed] [Google Scholar]
- 50.Scanlon PD, Raymond FA, Weinshilboum RM. Catechol-O-methyltransferase: thermolabile enzyme in erythrocytes of subjects homozygous for allele for low activity. Science. 1979;203:63–65. doi: 10.1126/science.758679. [DOI] [PubMed] [Google Scholar]
- 51.Lachman HM, Papolos DF, Saito T, Yu YM, Szumlanski CL, Weinshilboum RM. Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996;6:243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]