Abstract
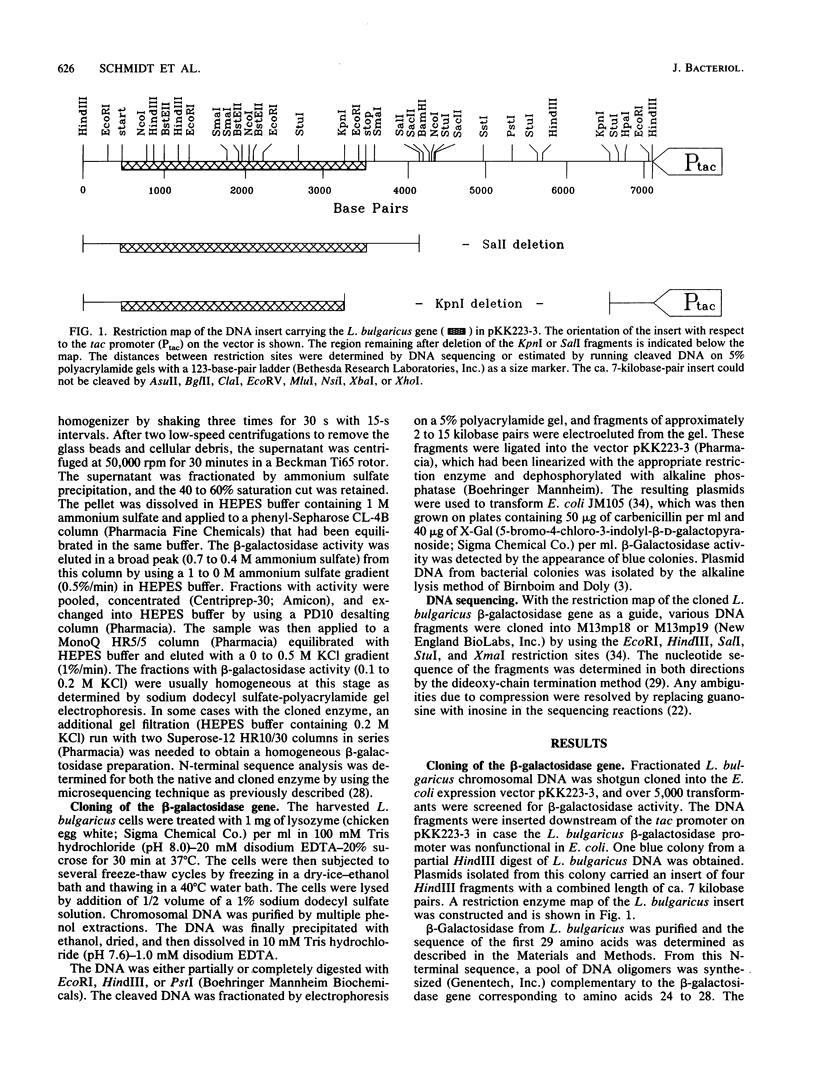
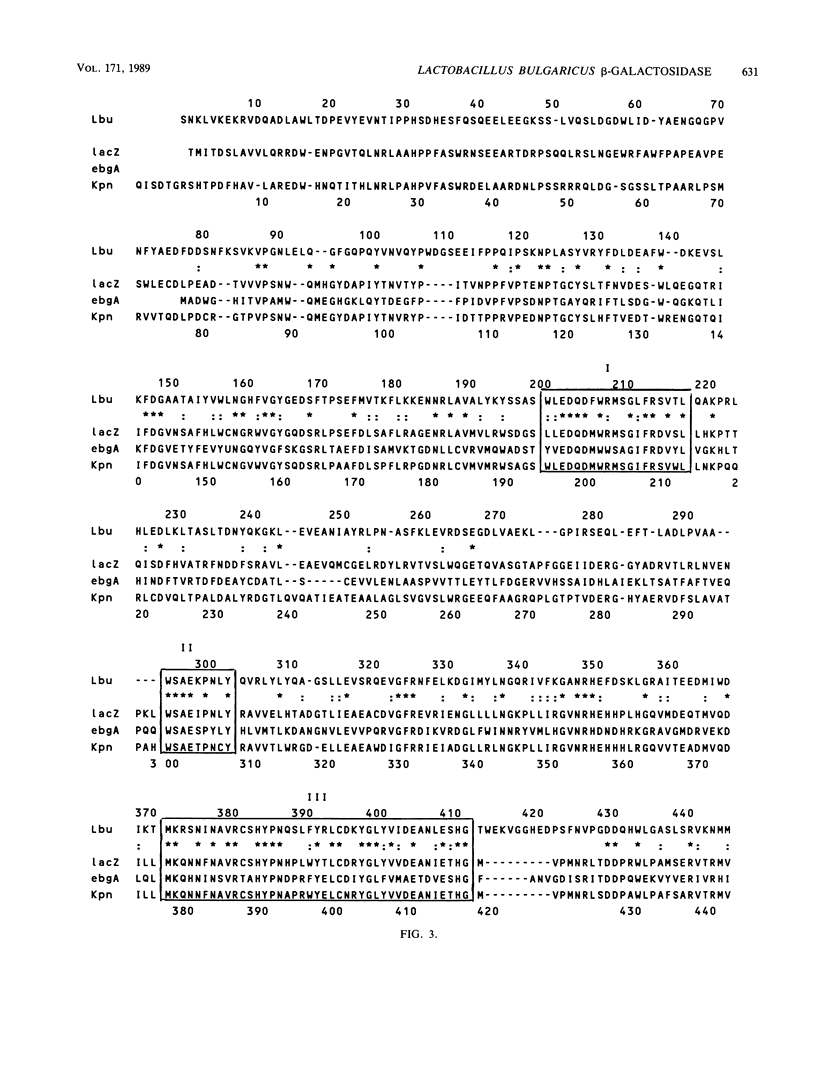
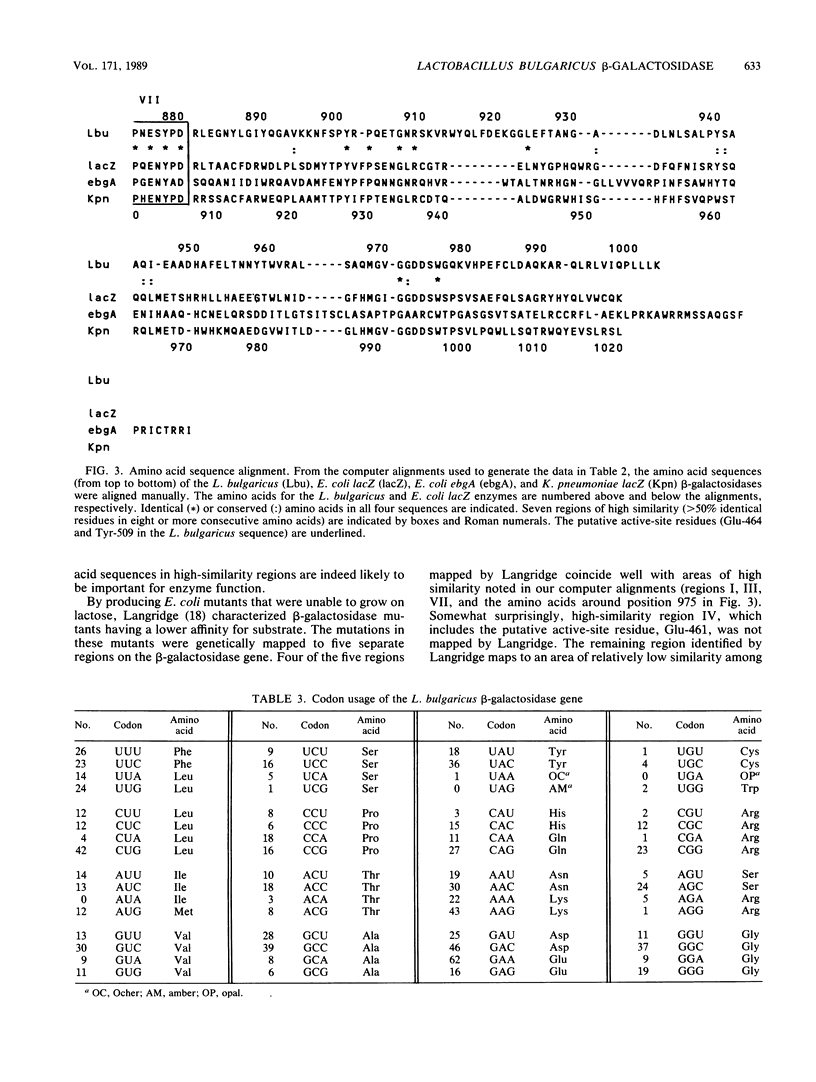
The Lactobacillus bulgaricus beta-galactosidase gene was cloned on a ca. 7-kilobase-pair HindIII fragment in the vector pKK223-3 and expressed in Escherichia coli by using its own promoter. The nucleotide sequence of the gene and approximately 400 bases of 3'- and 5'-flanking sequences was determined. The amino acid sequence of the beta-galactosidase, deduced from the nucleotide sequence of the gene, yielded a monomeric molecular mass of ca. 114 kilodaltons, slightly smaller than the E. coli lacZ and Klebsiella pneumoniae lacZ enzymes but larger than the E. coli evolved (ebgA) beta-galactosidase. The cloned beta-galactosidase was found to be indistinguishable from the native enzyme by several criteria. From amino acid sequence alignments, the L. bulgaricus beta-galactosidase has a 30 to 34% similarity to the E. coli lacZ, E. coli ebgA, and K. pneumoniae lacZ enzymes. There are seven regions of high similarity common to all four of these beta-galactosidases. Also, the putative active-site residues (Glu-461 and Tyr-503 in the E. coli lacZ beta-galactosidase) are conserved in the L. bulgaricus enzyme as well as in the other two beta-galactosidases mentioned above. The conservation of active-site amino acids and the large regions of similarity suggest that all four of these beta-galactosidases evolved from a common ancestral gene. However, these enzymes are quite different from the thermophilic beta-galactosidase encoded by the Bacillus stearothermophilus bgaB gene.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrews J., Clore G. M., Davies R. W., Gronenborn A. M., Gronenborn B., Kalderon D., Papadopoulos P. C., Schäfer S., Sims P. F., Stancombe R. Nucleotide sequence of the dihydrofolate reductase gene of methotrexate-resistant Lactobacillus casei. Gene. 1985;35(1-2):217–222. doi: 10.1016/0378-1119(85)90174-x. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breunig K. D., Dahlems U., Das S., Hollenberg C. P. Analysis of a eukaryotic beta-galactosidase gene: the N-terminal end of the yeast Kluyveromyces lactis protein shows homology to the Escherichia coli lacZ gene product. Nucleic Acids Res. 1984 Mar 12;12(5):2327–2341. doi: 10.1093/nar/12.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvinger W. E., Riley M. Nucleotide sequence of Klebsiella pneumoniae lac genes. J Bacteriol. 1985 Sep;163(3):850–857. doi: 10.1128/jb.163.3.850-857.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchel D. E., Gronenborn B., Müller-Hill B. Sequence of the lactose permease gene. Nature. 1980 Feb 7;283(5747):541–545. doi: 10.1038/283541a0. [DOI] [PubMed] [Google Scholar]
- Eckhardt T., Strickler J., Gorniak L., Burnett W. V., Fare L. R. Characterization of the promoter, signal sequence, and amino terminus of a secreted beta-galactosidase from "Streptomyces lividans". J Bacteriol. 1987 Sep;169(9):4249–4256. doi: 10.1128/jb.169.9.4249-4256.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards L. A., Huber R. E. A detailed examination of the iodination of beta-galactosidase: stoichiometric inactivation by nonspecific iodination. Biochem Cell Biol. 1986 Jun;64(6):523–527. doi: 10.1139/o86-073. [DOI] [PubMed] [Google Scholar]
- Edwards L. A., Tian M. R., Huber R. E., Fowler A. V. The use of limited proteolysis to probe interdomain and active site regions of beta-galactosidase (Escherichia coli). J Biol Chem. 1988 Feb 5;263(4):1848–1854. [PubMed] [Google Scholar]
- Fitch W. M., Smith T. F. Optimal sequence alignments. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1382–1386. doi: 10.1073/pnas.80.5.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler A. V., Smith P. J. The active site regions of lacZ and ebg beta-galactosidases are homologous. J Biol Chem. 1983 Sep 10;258(17):10204–10207. [PubMed] [Google Scholar]
- Fowler A. V., Zabin I., Sinnott M. L., Zabin I. Methionine 500, the site of covalent attachment of an active site-directed reagent of beta-galactosidase. J Biol Chem. 1978 Aug 10;253(15):5283–5285. [PubMed] [Google Scholar]
- Herrchen M., Legler G. Identification of an essential carboxylate group at the active site of lacZ beta-galactosidase from Escherichia coli. Eur J Biochem. 1984 Feb 1;138(3):527–531. doi: 10.1111/j.1432-1033.1984.tb07947.x. [DOI] [PubMed] [Google Scholar]
- Hirata H., Fukazawa T., Negoro S., Okada H. Structure of a beta-galactosidase gene of Bacillus stearothermophilus. J Bacteriol. 1986 Jun;166(3):722–727. doi: 10.1128/jb.166.3.722-727.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber R. E., Fowler A. V., Zabin I. Inactivation of beta-Galactosidase by iodination of tyrosine-253. Biochemistry. 1982 Sep 28;21(20):5052–5055. doi: 10.1021/bi00263a031. [DOI] [PubMed] [Google Scholar]
- Kalnins A., Otto K., Rüther U., Müller-Hill B. Sequence of the lacZ gene of Escherichia coli. EMBO J. 1983;2(4):593–597. doi: 10.1002/j.1460-2075.1983.tb01468.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge J. Genetic and enzymatic experiments relating to the quaternary structure of beta-galactosidase. Aust J Biol Sci. 1974 Jun;27(3):321–330. doi: 10.1071/bi9740321. [DOI] [PubMed] [Google Scholar]
- Langridge J. Genetic and enzymatic experiments relating to the tertiary structure of beta-galactosidase. J Bacteriol. 1968 Nov;96(5):1711–1717. doi: 10.1128/jb.96.5.1711-1717.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge J. Genetic evidence for the disposition of the substrate binding site of beta-galactosidase. Proc Natl Acad Sci U S A. 1968 Aug;60(4):1260–1267. doi: 10.1073/pnas.60.4.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- London J. The ecology and taxonomic status of the lactobacilli. Annu Rev Microbiol. 1976;30:279–301. doi: 10.1146/annurev.mi.30.100176.001431. [DOI] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci U S A. 1979 May;76(5):2232–2235. doi: 10.1073/pnas.76.5.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naider F., Bohak Z., Yariv J. Reversible alkylation of a methionyl residue near the active site of -galactosidase. Biochemistry. 1972 Aug 15;11(17):3202–3208. doi: 10.1021/bi00767a010. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Poolman B., Royer T. J., Mainzer S. E., Schmidt B. F. Lactose transport system of Streptococcus thermophilus: a hybrid protein with homology to the melibiose carrier and enzyme III of phosphoenolpyruvate-dependent phosphotransferase systems. J Bacteriol. 1989 Jan;171(1):244–253. doi: 10.1128/jb.171.1.244-253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring M., Armitage I. M., Huber R. E. m-Fluorotyrosine substitution in beta-galactosidase; evidence for the existence of a catalytically active tyrosine. Biochem Biophys Res Commun. 1985 Sep 16;131(2):675–680. doi: 10.1016/0006-291x(85)91290-2. [DOI] [PubMed] [Google Scholar]
- Rodriguez H., Kohr W. J., Harkins R. N. Design and operation of a completely automated Beckman microsequencer. Anal Biochem. 1984 Aug 1;140(2):538–547. doi: 10.1016/0003-2697(84)90205-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stokes H. W., Betts P. W., Hall B. G. Sequence of the ebgA gene of Escherichia coli: comparison with the lacZ gene. Mol Biol Evol. 1985 Nov;2(6):469–477. doi: 10.1093/oxfordjournals.molbev.a040372. [DOI] [PubMed] [Google Scholar]
- Vanderslice P., Copeland W. C., Robertus J. D. Cloning and nucleotide sequence of wild type and a mutant histidine decarboxylase from Lactobacillus 30a. J Biol Chem. 1986 Nov 15;261(32):15186–15191. [PubMed] [Google Scholar]
- Varenne S., Buc J., Lloubes R., Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J Mol Biol. 1984 Dec 15;180(3):549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



