Abstract
RNA interference provides a potent and specific method for controlling gene expression in human cells. To translate this potential into a broad new family of therapeutics, it is necessary to optimize the efficacy of the RNA-based drugs. As discussed in this Review, it might be possible to achieve this optimization using chemical modifications that improve their in vivo stability, cellular delivery, biodistribution, pharmacokinetics, potency, and specificity.
RNA, a remarkable molecule
RNA is a molecule that has been full of surprises, and its potential for controlling biology has been underestimated with startling regularity. First came the unexpected observation from Sharp (1) and Roberts et al. (2) that RNA was made in a long version and then spliced to create a shorter version that encodes protein. A few years later Altman (3) and Cech (4) observed that RNA, presumed to be a passive molecule that existed as a binding partner for proteins, could also act as a catalyst.
Then, in 1998, Fire and Mello made the observation that RNA could be introduced into cells in C. elegans and block gene expression (5–7). This discovery, termed RNA interference (RNAi), overturned dogma by suggesting that RNA was able to play an active role in regulating gene expression, in addition to its traditional role as an agent that transfers genetic information. Once again, RNA was exhibiting its irritating ability to perform a function it was not supposed to be able to.
The significance of Fire and Mello’s discovery was threefold. First, it unveiled a largely unsuspected natural regulatory mechanism that had the potential to exploit the versatility of Watson-Crick base-pairing for the control of gene expression. Second, it provided an important basic research strategy for studying gene function (blocking gene expression is often an excellent approach to investigating the function of a gene). Last, it raised the glimmering possibility that it might be possible to use this remarkably convenient approach to manipulate expression of disease genes in human cells and develop a new class of therapeutics.
RNAi: impact and mechanism
Few discoveries have had the immediate impact of RNAi. Within just a few years of the pioneering work of Fire, Mello, and colleagues (5) and the subsequent demonstration that double-stranded small interfering RNAs (siRNAs) could trigger RNAi in mammalian cells (8), RNAi has become a routine tool for silencing gene expression in the laboratory, with over 12,000 articles on PubMed citing the keyword “siRNA” since 2001. The success of duplex RNA in the laboratory has led to the hope that it might also revolutionize gene silencing in animals and lead to new drugs for the treatment of many classes of human disease.
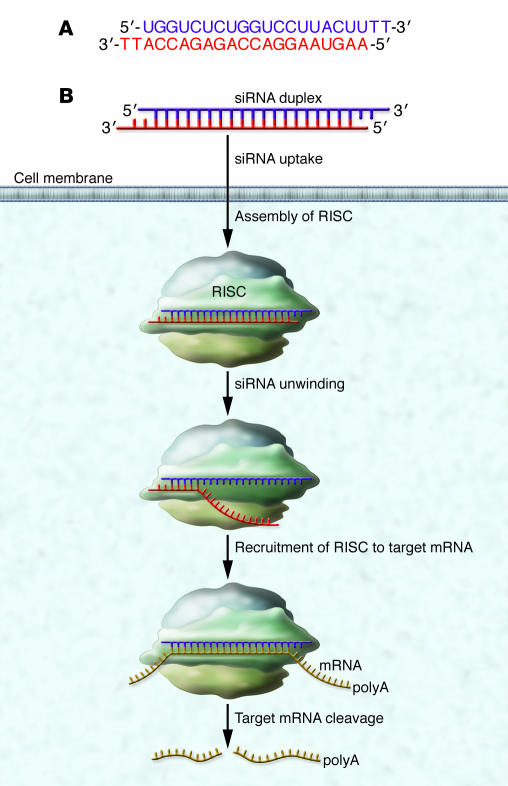
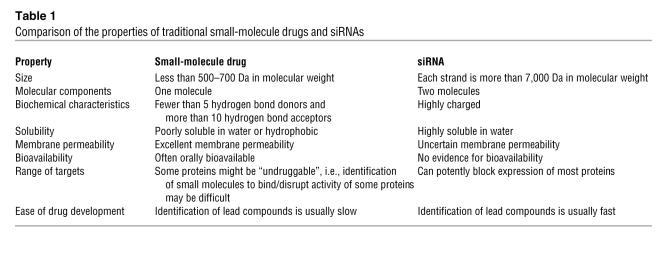
siRNAs are typically approximately 20 base pairs in length and are complementary to mRNA (Figure 1A). Working with cells in culture, laboratories found that they could conveniently block gene expression by either transfecting the siRNA into cells (a process in which the siRNA is complexed with lipids or other molecules to facilitate passage through cell membranes) (9) or by introducing a vector that could express the siRNA within the cells (10). Once inside the cell, the siRNA is bound by the proteins of the RNA-induced silencing complex (RISC; Figure 1B) (11–13).
Figure 1. (A) siRNAs are double-stranded RNAs approximately 20 base-pairs in length.
The duplex shown has staggered ends capped with two thymidines to improve resistance to digestion by nucleases. This is a traditional design, but it is possible to have blunt-ended duplexes, and the thymidines can be omitted. (B) Duplex siRNAs enter cells and bind to the RISC. The RISC proteins unravel the duplex and facilitate the search for mRNA sequences that are complementary to one of the RNA strands. Upon recognition of a complementary mRNA, RISC cleaves the mRNA and prevents translation.
The RISC proteins facilitate searching through the genome for RNA sequences that are complementary to one of the two strands of the siRNA duplex. One strand of the siRNA (the sense or passenger strand) is lost from the complex, while the other strand (the antisense or guide strand) is matched with its complementary RNA target. Recognition of mRNA by the antisense strand of the siRNA can cause destruction of the mRNA, prevent synthesis of protein, and thereby reduce the level of protein inside cells (Figure 1B).
siRNA drugs have already reached phase I clinical trials (14, 15). In spite of this rapid progress, however, it is not clear that siRNA will provide a broadly useful class of drugs. Substantial challenges must be overcome to move from the bench to the clinic, and overcoming these challenges is likely to require that the RNA portion of the RNA-based drug be chemically modified to improve its properties (14). This Review discusses some of these challenges and how chemical modifications might contribute to the clinical development of siRNAs.
siRNAs differ from traditional drug candidates
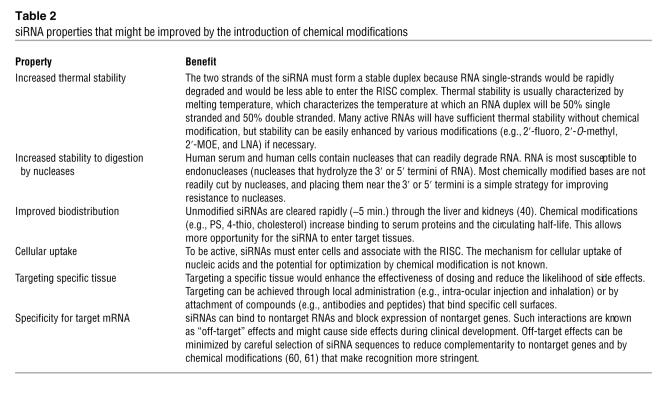
Most approved drugs share important features in common (Table 1). They are relatively small (with a molecular weight of less than 500 Da), apolar, and function by binding to proteins and altering protein function (16, 17). Much is known about designing traditional drugs and how to optimize their likelihood of efficacy in vivo, but even with this vast experience the development of small molecules is often marked by failure in preclinical and clinical trials (18).
Table 1 .
Comparison of the properties of traditional small-molecule drugs and siRNAs
Enter siRNA onto the scene. siRNA drugs are not at all similar to traditional small molecule drugs (Table 1), and the suggestion that they could be developed into an important class of drugs would provoke skepticism among many medicinal chemists. Translating the potential shown by siRNAs in cultured cells to patients requires adjusting our concepts about the molecular requirements for successful drugs.
Complications abound. siRNAs contain two strands, which must be synthesized separately and then hybridized before use. The need for hybridization complicates obtaining and characterizing a drug for clinical use by adding an additional step to the process required to prepare the drug. Each strand has a molecular weight of approximately 7,000 Da and has a negatively charged phosphate backbone. The large size and charged backbone discourage easy passage of RNA duplexes through cell membranes. Large size also raises the cost of the molecule by increasing the complexity of the synthesis and the need for chemical building blocks. In addition, both strands have the potential to trigger unwanted side effects. Finally, the targets for siRNAs are specific sequences within RNA, rather than proteins, which are the target of almost all traditional drugs.
siRNA drugs benefit from an older sibling
The theoretical obstacles to the development and use of siRNA drugs are substantial. Some challenges, however, are not new and have already been addressed by a previous generation of research on the properties of antisense oligonucleotides (ASOs) (see What ASOs have given siRNA) (15). ASOs are single-stranded nucleic acids that are designed to bind mRNA and reduce protein synthesis. Unlike siRNAs, gene inhibition by ASOs does not require binding to RISC proteins. Instead, ASOs bind mRNA and either act as a roadblock for translation by the ribosome or form a DNA-RNA hybrid that recruits RNAse H to cleave the mRNA (19).
For over 20 years, biotechnology and pharmaceutical companies and academic laboratories have worked to develop ASOs as drugs. One ASO has been approved by the FDA for treatment of CMV retinitis (20). A second oligonucleotide drug, Macugen, has been approved for macular degeneration (21), but it functions by recognizing a protein target rather than by binding to a nucleic acid. Many clinical trials with ASOs are ongoing, with some yielding promising physiologic effects in phase II testing (22–25).
Researchers who perform gene-silencing experiments in cell culture almost always need to complex ASOs and siRNAs with cationic lipid or some other delivery agent to facilitate entry into cells (9). It is essential to realize that many animal studies and most clinical trials with ASOs have not required the use of complexing agents — sufficient quantities of oligomer are taken up by tissue, with liver and fat being the tissues in which oligomers most easily accumulate. The requirement for complexing agents in cell culture rather than in animals is a warning that experience in cell culture does not directly extrapolate to the situation in animals.
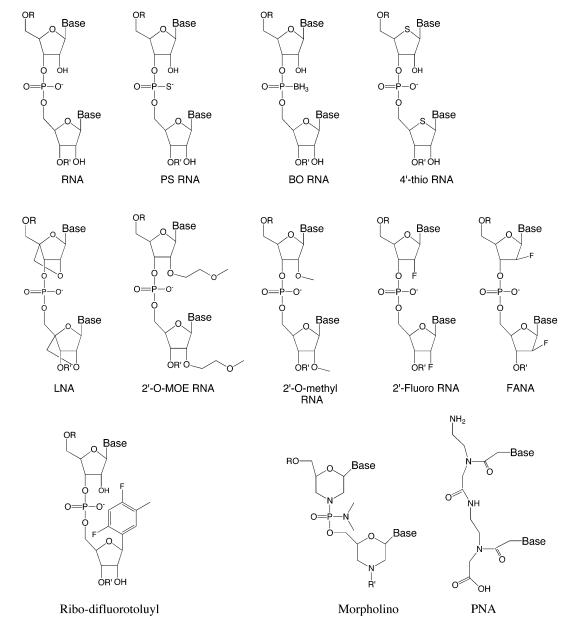
Like siRNAs, ASOs are intended to bind to mRNA by Watson-Crick base-pairing and are large, negatively charged molecules (26, 27). Because of work with ASOs, there exist good protocols for the large-scale synthesis of nucleic acid drugs in amounts needed for clinical trials. Experience with ASOs in animals and in clinical trials also provides substantial information about the biodistribution, toxicology, and pharmacology of nucleic acids in vivo that is likely to apply to siRNAs. Finally, clinical development of ASOs has provided insights into how to choose target genes and diseases. For example, it makes sense to pursue gene targets in tissues where there is proven biodistribution of oligomers and where markers exist that allow inhibition of gene expression to be evaluated during phase I and phase II clinical trials (evidence that a silencing agent is reducing expression of the target gene would help support more costly phase III clinical trials). Chemical modifications have been developed to optimize the properties of ASOs, and these modifications might also be used to improve the properties and efficacy of siRNAs (Table 2).
Table 2 .
siRNA properties that might be improved by the introduction of chemical modifications
Why siRNA?
Given that ASOs are making progress in the clinic and are half the molecular weight of siRNAs, it is fair to ask “What new potential are siRNAs bringing to drug development?” and “Why not focus on the more established ASO technology, especially since it now seems to be generating positive clinical results?”
There is no definitive answer to these questions at the moment. In cell culture, the widespread and rapid adoption of siRNA supports the conclusion that siRNAs are a more “user friendly” approach to gene silencing for basic research than are ASOs (this has certainly been the experience in my laboratory). One explanation for the increased efficacy of siRNAs compared with ASOs in the laboratory is that siRNAs co-opt a natural silencing pathway and the RISC proteins facilitate efficient recognition of target sequences by siRNAs, whereas ASOs must find their targets unassisted. It is reasonable to hypothesize that outstanding results in cell culture will translate into superior potential in the clinic. It is not clear, however, whether experience in cell culture will translate into results in animals.
Chemical modifications for improving siRNA drugs
Many different types of chemical modification have been developed to improve the in vivo properties of nucleic acids (14, 26, 27). Each modification offers a different potential for tailoring the properties of siRNAs (Table 2). They can be used alone or in combination and the number of modified nucleotides can vary relative to the number that remain as unmodified RNA. The modifications also differ in how they are tolerated by the RISC complex. Some modifications can be introduced at most or all bases of both RNA strands, whereas other modifications cannot be placed at some positions. Potential for activity of a given modified RNA is first determined by cell culture studies that examine silencing well-characterized target genes. This section describes the properties of some of the siRNA modifications that are the most promising in terms of improving the properties of siRNAs to render them more suitable clinical modalities (Figure 2).
Figure 2. RNA and the nucleic acid analogs and chemical modifications mentioned in this Review.
Phosphorothioate linkages.
Replacing one nonbridging oxygen atom on the backbone phosphate between two ribonucleotides with a sulphur atom creates a phosphorothioate (PS) linkage (Figure 2) (28). PS linkages within DNA substantially increase the resistance of the DNA to serum nucleases. PS linkages also improve the half-life of ASOs in the circulation by increasing their binding to serum proteins (29, 30). The one FDA-approved ASO drug is a PS-linked DNA oligonucleotide (20), and most ongoing clinical trials use oligonucleotides that contain PS linkages in combination with DNA or 2′-modified RNA bases (15). These clinical trials have shown that ASOs containing PS linkages are well tolerated in patients (22–25).
Several studies have shown that siRNAs with introduced PS linkages retain the ability to silence target sequences (31–39). However, cytotoxic effects were observed when PS linkages were placed at alternating positions throughout either strand of the siRNA (32), suggesting that the number of PS linkages should be limited. Animal experiments using siRNAs containing just one or two terminal PS modifications have been successful (36–38). Relative to unmodified RNA, complete substitution with PS linkages does not substantially improve the resistance of siRNAs to digestion by nucleases (31) or alter their biodistribution in mice (40).
Boranophosphate linkages.
Another option for modifying the phosphate backbone of an oligonucleotide is the introduction of a boron atom in place of one of the nonbridging oxygen atoms to create a boron-phosphorous linkage (Figure 2). This modification has attracted relatively little study, but a preliminary investigation reports improved gene silencing activity at lower concentrations relative to either PS siRNAs or native siRNAs and enhanced resistance to degradation by nucleases (41). There is also a report that single-stranded boranophosphate (BO) siRNAs can engage the RISC machinery and silence target genes (42); if true, this might substantially simplify some of the drug development problems for siRNA by requiring just one rather than two strands for silencing. One problem with using BO siRNAs is that chemical methods for introducing the boron modification have not been optimized and will need to improve prior to large-scale testing.
Locked nucleic acid.
Locked nucleic acid (LNA) nucleotides contain a methylene bridge between the 2′ and 4′ carbons of the ribose ring (Figure 2) (43–45). This linkage constrains the ribose ring, “locking” it into a conformation close to that formed by RNA after hybridization. This constraint preorganizes the LNA base for hybridization to mRNA or DNA, reduces the entropic penalty paid upon binding, and enhances the affinity of LNA oligonucleotides for complementary sequences. For example, a single LNA nucleotide enhances the melting temperature of an RNA-RNA or RNA-DNA duplex by as much as 5°–10° C. As a result, the introduction of LNA nucleotides into an oligonucleotide allows the affinity of binding of the oligonucleotide to be fine-tuned for an application with a minimum of chemical modifications to the parent oligonucleotides. The potential for LNA-modified oligomers to translate their high-affinity binding into enhanced efficacy in vivo is unknown.
The potential of LNA for improving the potency of gene silencing by nucleic acids inside cells has been widely recognized, and one ASO that contains a mix of LNA and DNA bases is being tested in clinical trials (46). The LNA modification increases the thermal stability of RNA duplexes and stabilizes them to digestion by nucleases (31). LNA-containing RNA duplexes are active silencing agents inside cells (31, 47) and animals (48). Gene silencing is sensitive to the position of the LNA nucleotides within the duplex, but well-placed LNA modification within the sense strand can enhance potency (47). ASOs that contain LNA bases have been reported to be highly potent but to also cause liver toxicity in mouse models (49). It is important to understand the origins of this toxicity, whether it can be minimized, whether it applies to humans, and whether it will also be observed with LNA-modified siRNAs.
2′-modified RNA.
Many RNA analogs have been developed that include substitutions for the hydroxyl group on the 2′ carbon atom of the ribose ring. These RNA analogs include 2′-O-methyl RNA (50), 2′-O-methoxyethyl (2′-MOE) RNA (51, 52), and 2′-fluoro RNA (Figure 2) (53). These modifications enhance the affinity of the RNA oligomer for complementary RNA or DNA sequences and increase its resistance to digestion by nucleases. The 2′-modified nucleotides 2′-O-methyl and 2′-MOE have been used to improve the properties of ASOs that have been tested in clinical trials (15, 27).
Several reports have described the introduction of 2′-modified ribonucleotides into RNA duplexes (31–39, 54–62). The extent of substitution varies, from just one modified nucleotide to complete substitution of both strands. In all reports, it was possible to retain silencing activity after replacing some of the ribonucleotides in RNA duplexes with 2′-modified nucleotides. Remarkably, RNA oligonucleotides can be completely substituted with alternating 2′ modifications (2′-O-methyl and 2′-fluoro) and retain the ability to silence gene expression in human cells, even exhibiting up to a 500-fold increase in potency relative to native siRNA (55). Introduction of 2′-O-methyl nucleotides near the termini of the RNA strand that is complementary to the mRNA target can reduce off-target effects (i.e., alteration of the expression of nontarget genes) (60, 61), a finding that might provide a strategy to reduce the potential for side effects and prove critically important for therapeutic development. Several studies have shown that siRNAs containing 2′ modifications can silence gene expression in animal models (35–38, 62).
Most recently, siRNAs targeting the hepatocyte-specific genes apolipoprotein B and factor VII were demonstrated to achieve efficient gene silencing without altering levels of key microRNAs (62). This is a significant result because it suggests that therapeutic dosages of siRNAs will not overload endogenous microRNA pathways, which can cause complicating off-target effects. The siRNAs used in these studies had either 2′-fluoro or 2′-O-methyl bases and contained PS linkages between the two thymidines capping the 3′-end of each strand.
Another class of 2′ modification is 2′-deoxy-2′-fluoro-β-d-arabinonucleic acid (FANA). FANA nucleotides are based on the sugar arabinose rather than ribose and have the stereochemistry at the 2′-position inverted relative to that found in 2′-O-methyl RNA and 2′-fluoro RNA. siRNAs that have completely FANA-substituted sense strands are 4-fold more potent than analogous unmodified RNAs and have a longer half-life in serum (63, 64). FANA substitutions can also be tolerated for gene silencing when placed at the 3′ and 5′ positions of the antisense strand or throughout the sense strand.
4′-thio modified RNA.
4′-Thio modified nucleotides contain a sulphur atom in place of oxygen attached to the 4′ carbon of the ribose ring (Figure 2). This modification enhances the resistance of the RNA oligonucleotide to nuclease digestion when introduced at various positions within both the sense and antisense strands of siRNAs (57, 58). Inclusion of 4′-thio nucleotides yielded active siRNAs with increased resistance to digestion by nucleases in cell culture assays and improved potencies (57). Combining 4′-thio bases with 2′ modifications such as 2′-O-methyl or 2′-MOE further improved potencies (58).
Sulfur atoms improve the biodistribution of ASOs that contain PS linkages by increasing binding to serum proteins (29). It is possible that 4′-thio substitutions might also improve the in vivo properties of siRNAs by the same mechanism, but there have been no published reports on the subject. It is also possible to combine 4′-thio substitution with FANA nucleotides, and 4′-S-FANA–modified RNA duplexes have been shown to possess potencies for silencing in cell culture that were similar to unmodified siRNAs (64).
Ribo-difluorotoluyl nucleotide.
The modifications described thus far in this section involve alterations to the ribonucleotide ribose ring or the oligonucleotide phosphate backbone. However, it is also possible to modify the ribonucleotide base, and one modification that has been tested is the introduction of ribo-difluorotoluyl (rF) nucleotides (65). This modification is especially interesting because rF nucleotides cannot form normal Watson-Crick base-pairs. siRNA duplexes with a single rF nucleotide were shown to be active silencing agents and to exhibit enhanced resistance to degradation compared with the parental native siRNA.
Uncharged nucleic acid mimics.
Single-stranded uncharged nucleic acid mimics, such as peptide nucleic acids (PNAs) (66, 67) and morpholino oligomers (68), have attracted substantial attention from basic researchers and drug developers. PNAs and morpholinos are interesting candidate modifications because they have dramatically different chemical properties relative to modified DNA or RNA. The PNA or morpholino monomers could be chemically attached to RNA strands, creating modified oligomers with novel properties. The chemical features of PNA or morpholino monomers may confer opportunities for optimizing the biodistribution and efficacy of gene-silencing agents that go beyond those conferred by less radical alterations. In spite of their theoretical promise, the application of PNAs or morpholinos to RNAi is likely to be slow because obtaining siRNAs modified with PNA or morpholino to test is not straightforward, as standard methods for RNA synthesis are not compatible with coupling to PNA or morpholino monomers to RNA.
siRNA conjugates
Another strategy for improving the function of siRNA duplexes in vivo is to make more dramatic changes to conjugate the siRNA duplex to small molecules or peptides designed to increase binding to proteins or cellular uptake (69–72). Enhanced binding to serum proteins might improve biological half-life and tissue distribution, while compounds that specifically bind to cell surface proteins might allow targeting of specific diseased tissues. Improving cell uptake would enhance the potency of siRNAs. These strategies have been extensively explored for improving ASOs, although not with any great success (73), and are now being applied to duplex RNA (74). It is possible that better results might be achieved with siRNAs because of their potential for greater potency.
The concept behind siRNA conjugates is simple. One part of the conjugate is duplex RNA, which provides specificity for the target mRNA sequence. The other part of the conjugate is a molecule optimized for improving biodistribution, cellular uptake, or other in vivo properties. A major strength of the approach is that the two portions of the conjugate can be developed as separate modules and then coupled to create hybrid molecules that combine the strengths of the two parts. A weakness is that the synthesis of novel conjugates is complicated by the need to couple a molecule to the siRNA, making a large and complex siRNA even larger and more complex.
A conjugate of duplex RNA and cholesterol has been tested for its ability to inhibit the expression of APOB in mice (36). The RNA portion of the conjugate was resistant to nuclease digestion because it contained PS linkages and 2′-O-methyl RNA modifications. Cholesterol was coupled to one of the RNA strands of the siRNA to create a cholesterol–RNA duplex conjugate. Attachment of the RNA duplex to cholesterol increased binding to human serum albumin and increased biodistribution to various organs including the liver, heart, kidney, adipose tissue, and lung. Evaluation of gene expression in treated mice showed reduced levels of ApoB mRNA in the liver and jejunum and reduced amounts of APOB protein in plasma.
Several other types of conjugate have also been tested. siRNA conjugated to peptides (69) and polyethylene glycol (70, 71) have been reported to yield gene silencing in cultured cells. Conjugates to aptamers (small structured nucleic acids selected to bind target proteins) have been reported to block expression in cell culture and animals (75). At this stage, it is not possible to evaluate whether improved properties justify the added synthetic complexity inherent in generating conjugates, but this is an active area for future research.
It might also be possible to combine the strengths of siRNA with the specificity of antibodies. siRNAs have been mixed with antibodies engineered to display positively charged peptides on their surfaces (76). The duplex RNA is intended to be the silencing agent and is linked to the antibody by noncovalent electrostatic interactions with the positively charged peptide. The antibody used could, in theory, bind to any cell surface protein and in this study (76) was targeted to cells expressing HIV-1 envelope protein. The strength of this concept is its potential versatility and the fact that there is no need to synthesize the conjugate — it forms spontaneously upon mixing. The conjugate yielded inhibition of target gene expression inside cells, but once again data in animals was less conclusive. Work on RNA conjugates is in its early stages, and more research will be necessary before the potential of the approach becomes apparent.
Chemical modifications and local delivery
Another approach to the problem of biodistribution is to simplify the problem by focusing on local rather than systemic delivery. Local delivery involves administration of the siRNA drug to a smaller and potentially more protected area within the body. The first clinical trials of siRNAs have used this approach, with intraocular injection of a VEGF-specific siRNA being assessed as a possible treatment for macular degeneration and inhalation of siRNA being assessed as a possible treatment for infection with respiratory syncytial virus (RSV) (14, 15). Local administration is a good strategy for early groundbreaking siRNA trials because it simplifies distribution, lowers the likelihood of adverse side effects, and reduces the amount and cost of the siRNA dose.
Local delivery has important implications for the role of chemical modifications to siRNA. There might be less need for modification than if the siRNA is systemically administered because the obstacles to biodistribution are much less and the requirements for stability might be less. Indeed, the VEGF-specific siRNA now being tested in clinical trials using intraocular administration is an unmodified RNA (77–79), and unmodified siRNAs have provided relief from coronavirus infection in rhesus macaques (78). Nevertheless, although chemical modifications might be less essential for local delivery than systemic delivery, it is still probable that they can significantly improve the properties of drug candidates, and this possibility should be explored during development.
Chemical modifications and formulation strategies
Another strategy for clinical development of siRNAs is to improve their efficacy by mixing RNA with compounds that can facilitate uptake by cells and tissues that normally resist entry of siRNAs. These agents, which include peptides, nanoparticles, and lipids, can also increase protection of the RNA against nucleases by interacting with the RNA surface and restricting access to enzymes in the serum. Several studies have reported gene silencing in animals using complexes that contain siRNA (81–84).
A full description of these results is beyond the scope of this Review, but several points are worth noting. The first is that formulations require introduction of an additional chemical agent into patients, complicating the procedure and increasing the potential for unexpected toxicities. The benefit of formulations therefore needs to be substantial to justify the added complexity of the approach. The second is that formulations may lessen the need for chemical modifications because the RNA complex might be more protected from nucleases and more likely to enter the target tissue. However, as with local delivery, it is still probable that chemical modifications will play a role in identifying optimal agents for clinical trials.
Summary
The development of duplex RNA as a general class of therapeutic agents faces substantial challenges. To fulfill the potential of RNAi for therapy, it is necessary to develop strategies for in vivo delivery of large, highly charged macromolecules that would give the designers of traditional small-molecule drugs nightmares. An arsenal of chemical modifications is available for improving biological properties such as stability, biodistribution, cellular uptake, and potency. Such modifications are making the development of therapeutic RNA more feasible for some indications. More breakthroughs in the area of delivery are necessary, however, before duplex RNA has the same impact in the clinic as it has had in the research laboratory.
Acknowledgments
This work was supported by grants from the NIH (NIGMS 60642 and 73042) and the Robert A. Welch Foundation (I-1244).
Footnotes
Nonstandard abbreviations used: ASO, antisense oligonucleotide; BO, boranophosphate; FANA, 2′-deoxy-2′-Fluoro-β-d-arabinonucleic acid; LNA, locked nucleic acid; 2′-MOE, 2′-methoxyethyl; PNA, peptide nucleic acid; PS, phosphorothioate; rF, ribo-difluorotoluyl; RISC, RNA-induced silencing complex; RNAi, RNA interference.
Conflict of interest: The author has received research support from Applied Biosystems during the previous fiscal year.
Citation for this article: J. Clin. Invest. 117:3615–3622 (2007). doi:10.1172/JCI33483.
References
- 1.Sharp P.A. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 2.Chow L.T., Gelinas R.E., Broker T.R., Roberts R.J. An amazing sequence rearrangement at the 5′ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- 3.Altman S. Nobel lecture. Enzymatic cleavage of RNA by RNA. Biosci. Rep. 1990;10:317–337. doi: 10.1007/BF01117232. [DOI] [PubMed] [Google Scholar]
- 4.Cech T.R. Nobel lecture. Self-splicing and enzymatic activity of an intervening sequence RNA from tetrahymena. Biosci. Rep. 1990;10:239–261. doi: 10.1007/BF01117241. [DOI] [PubMed] [Google Scholar]
- 5.Fire A., et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Fire A. Gene silencing by double-stranded RNA (Nobel lecture). Angew. Chem. Int. Ed. Engl. 2007;46:6966–6984. doi: 10.1002/anie.200701979. [DOI] [PubMed] [Google Scholar]
- 7.Mello C.C. Return to the RNAi world: Rethinking gene expression and evolution (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 2007;46:6985–6994. doi: 10.1002/anie.200701713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elbashir S.M., et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in mammalian cell culture. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 9.Janowski B.A., Hu J., Corey D.R. Silencing gene expression by targeting chromosomal DNA with antigene peptide nucleic acids and duplex RNAs. Nat. Protoc. 2006;1:436–443. doi: 10.1038/nprot.2006.64. [DOI] [PubMed] [Google Scholar]
- 10.Tiscornia G., Singer O., Verma I.M. Design and cloning of lentiviral vectors expressing small interfering RNAs. Nat. Protoc. 2006;1:234–240. doi: 10.1038/nprot.2006.36. [DOI] [PubMed] [Google Scholar]
- 11.Sen G.L., Blau H.M. A brief history of RNAi: the silence of the genes. FASEB J. 2006;20:1293–1299. doi: 10.1096/fj.06-6014rev. [DOI] [PubMed] [Google Scholar]
- 12.Tomari Y., Zamore P.D. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- 13.Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 14.Bumcrot D., Manoharan M., Koteliansky V., Sah D.W.Y. RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat. Chem. Biol. . 2006;2:711–719. doi: 10.1038/nchembio839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corey D.R. RNAi learns from antisense. Nat. Chem. Biol. 2007;3:8–11. doi: 10.1038/nchembio0107-8. [DOI] [PubMed] [Google Scholar]
- 16.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 17.Juliano R.L., Astriab-Fisher A., Falke D. Macromolecular therapeutics: Emerging strategies for drug discovery in the postgenome era. Mol. Interv. 2001;1:40–53. [PubMed] [Google Scholar]
- 18.Hurko O. Understanding the strategic importance of biomarkers for the discovery and early development phases. Drug Discovery World. 2006;7:63–74. [Google Scholar]
- 19.Wu H., et al. Determination of the role of the human RNase H1 in the pharmacology of DNA-like antisense drugs. J. Biol. Chem. 2004;279:17181–17189. doi: 10.1074/jbc.M311683200. [DOI] [PubMed] [Google Scholar]
- 20.Grillone L.R., Lanz R. Fomivirsen. Drugs Today. 2001;37:245–255. doi: 10.1358/dot.2001.37.4.620590. [DOI] [PubMed] [Google Scholar]
- 21.Ng E.W., et al. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug. Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 22.Akdim F., Stroes E.S., Kastelein J.J. Antisense apolipoprotein B therapy: where do we stand? Curr. Opin. Lipidol. 2007;18:397–400. doi: 10.1097/MOL.0b013e328248b4ad. [DOI] [PubMed] [Google Scholar]
- 23.Miyake H., Hara I., Fujisawa M., Gleave M.E. The potential of clusterin inhibiting antisense oligodeoxynucleotide therapy for prostate cancer. Expert Opin. Investig. Drugs. 2006;15:507–517. doi: 10.1517/13543784.15.5.507. [DOI] [PubMed] [Google Scholar]
- 24.Liu G. Technology evaluation: ISIS-113715, ISIS. Curr. Opin. Mol. Ther. 2004;6:331–336. [PubMed] [Google Scholar]
- 25.Geary R.S., et al. Lack of pharmacokinetic interaction for ISIS 113715, a 2′-O-methoxyethyl modified antisense oligonucleotide targeting protein tyrosine phosphatase 1B messenger RNA, with oral antidiabetic compounds metformin, glipizide or rosiglitazone. Clin. Pharmacokinet. . 2006;45:789–801. doi: 10.2165/00003088-200645080-00003. [DOI] [PubMed] [Google Scholar]
- 26.Braasch D.A., Corey D.R. Novel antisense strategies for controlling gene expression. Biochemistry. 2002;41:4503–4510. doi: 10.1021/bi0122112. [DOI] [PubMed] [Google Scholar]
- 27.Gleave M.E., Monia B.P. Antisense therapy for cancer. Nat. Rev. Cancer. 2005;5:468–479. doi: 10.1038/nrc1631. [DOI] [PubMed] [Google Scholar]
- 28.Eckstein F. Developments in RNA chemistry, a personal view. Biochemie. 2002;84:841–848. doi: 10.1016/s0300-9084(02)01459-1. [DOI] [PubMed] [Google Scholar]
- 29.Geary R.S., Yu R.Z., Levin A.A. Pharmacokinetics of phosphorothioate antisense oligonucleotides. Curr. Opin. Investig. Drugs. 2001;2:562–573. [PubMed] [Google Scholar]
- 30.Watanabe T.A., Geary R.S., Levin A.A. Plasma protein binding of an antisense oligonucleotide targeting human ICAM-1 (ISIS 2302). Oligonucleotides. 2006;16:169–180. doi: 10.1089/oli.2006.16.169. [DOI] [PubMed] [Google Scholar]
- 31.Braasch D.A., et al. RNA interference in mammalian cells by chemically modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 32.Harborth J., et al. Sequence, chemical, and structural variation of small interfering RNAs and short hairpin RNAs and the effect on mammalian gene silencing. Antisense Nucleic Acid Drug Dev. 2003;13:83–105. doi: 10.1089/108729003321629638. [DOI] [PubMed] [Google Scholar]
- 33.Chiu Y., Rana T.M. siRNA function in RNAi: A chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amarzguioui M., Holen T., Babaie E., Prydz H. Tolerance for mutations and chemical modifications in an siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Layzer J.M., et al. In vivo activity of nuclease resistant siRNAs. RNA. . 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soutschek J., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 37.Morrissey D.V., et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- 38.Morrissey D.V., et al. Potent and persistent in vivo anti-HBV activity of chemical modified RNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 39.Kraynack B.A., Baker B.F. Small interfering RNAs containing full 2’-O-methylribonucleotide-modified sense strands display Argonaute2/eIF2C2-dependent activity. RNA. 2006;12:163–173. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Braasch D.A., et al. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorg. Med. Chem. Lett. 2004;14:1139–1143. doi: 10.1016/j.bmcl.2003.12.074. [DOI] [PubMed] [Google Scholar]
- 41.Hall A.H.S., et al. RNA interference using boranophosphate siRNAs: structure-activity relationships. Nucleic Acids Res. . 2004;32:5991–6000. doi: 10.1093/nar/gkh936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hall A.H.S., et al. High potency silencing by single-stranded boranophosphate siRNA. Nucleic Acids Res. 2006;34:2773–2781. doi: 10.1093/nar/gkl339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koshkin A.A., et al. LNA (locked nucleic acids): Synthesis of the adenine, cytosine, guanine, 5-methylcytosine, thymine, and uracil bicyclonucleoside monomers, oligomerisation, and unprecedented nucleic acid recognition. Tetrahedon. 1998;54:3607–3630. [Google Scholar]
- 44.Obika S., et al. Stability and structural features of the duplexes containing nucleoside analogues with a fixed N-type conformation 2′-O,4′-C-methyleneribonucleotides. Tetrahedron Lett. 1998;39:5401–5404. [Google Scholar]
- 45.Braasch D.A., Corey D.R. Locked nucleic acid (LNA): fine-tuning the recognition of DNA and RNA. Chem. Biol. . 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- 46.Frieden M., Orum H. The application of locked nucleic acids in the treatment of cancer. IDrugs. . 2006;9:706–711. [PubMed] [Google Scholar]
- 47.Elmen J., et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mook O.R., Baas F., de Wissel M.B , Fluiter K. Evaluation of locked nucleic acid-modified small interfering RNA in vitro and in vivo. Mol. Cancer Ther. 2007;6:833–843. doi: 10.1158/1535-7163.MCT-06-0195. [DOI] [PubMed] [Google Scholar]
- 49.Swayze E.E., et al. Antisense oligonucleotides containing locked nucleic acid improve potency but cause significant hepatoxicity. Nucleic Acids Res. 2006;35:687–700. doi: 10.1093/nar/gkl1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rusckowski M., Ou T., Roskey A., Agrawal S. Biodistribution and metabolism of a mixed backbone oligonucleotide (GEM 231) following single and multiple dose adminstration in mice. Antisense Nucleic Acid Drug Dev. 2000;10:333–345. doi: 10.1089/oli.1.2000.10.333. [DOI] [PubMed] [Google Scholar]
- 51.Zhang H., et al. Reduction of liver Fad expression by an antisense oligonucleotide protects mice from fulminant hepatitis. Nat. Biotechnol. 2000;18:862–867. doi: 10.1038/78475. [DOI] [PubMed] [Google Scholar]
- 52.Crooke R.M., et al. An apolipoprotein B antisense oligonucleotide lowers LDL cholesterol in hyperlipidemic mice without causing hepatic steatosis. J. Lipid Res. 2005;46:872–884. doi: 10.1194/jlr.M400492-JLR200. [DOI] [PubMed] [Google Scholar]
- 53.Kawasaki A.M., et al. Uniformly modified 2′-deoxy-2′-fluoro phosphorothioate oligonucleotides as nuclease resistant antisense compounds with high affinity and specificity for RNA targets. J. Med. Chem. 1993;36:831–841. doi: 10.1021/jm00059a007. [DOI] [PubMed] [Google Scholar]
- 54.Czauderna F., et al. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Allerson C.R., et al. Fully 2’-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 56.Prakash T.P., et al. Positional effect of chemical modifications on short interference RNA activity in mammalian cells. J. Med. Chem. 2005;48:4247–4253. doi: 10.1021/jm050044o. [DOI] [PubMed] [Google Scholar]
- 57.Hoshika S., et al. RNA interference induced by siRNAs modified with 4′-thioriobnucleosides in cultured mammalian cells. FEBS Lett. 2005;579:3115–3118. doi: 10.1016/j.febslet.2005.04.073. [DOI] [PubMed] [Google Scholar]
- 58.Dande P., et al. Improving RNA interference in mammalian cells by 4’-Thio-modified small interfering RNA (siRNA): Effect on siRNA activity and nuclease stability when used in combination with 2′-O-alkyl modifications. J. Med. Chem. 2006;49:1624–1634. doi: 10.1021/jm050822c. [DOI] [PubMed] [Google Scholar]
- 59.Koller E., et al. Competition for RISC binding predicts in vitro potency of siRNA. Nucleic Acids Res. 2006;34:4467–4476. doi: 10.1093/nar/gkl589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jackson A.L., et al. Position-specific chemical modifications of siRNAs reduces “off-target” transcription silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Snove O., Rossi J.J. Chemical modifications rescue off-target effects. ACS Chem. Biol. 2006;1:274–276. doi: 10.1021/cb6002256. [DOI] [PubMed] [Google Scholar]
- 62.Matthias J., et al. Effective RNAi-mediated gene silencing without interruption of the endogenous microRNAs pathway. Nature. 2007 doi: 10.1038/nature06179. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dowler T., et al. Improvements in siRNA properties mediated by 2′-deoxy-2′-fluoro-S-D-arabinonucleic acid (FANA). Nucleic Acids Res. 2006;34:1669–1675. doi: 10.1093/nar/gkl033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Watts J.K., et al. 2′-Fluoro-4′-thioarabino-modified oligonucleotides: conformational switches linked to siRNA activity. Nucleic Acids Res. 2007;35:1441–1451. doi: 10.1093/nar/gkl1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xia J., et al. Gene silencing activity of siRNAs with ribo-difluorotoluyl nucleotide. ACS Chem. Biol. 2006;1:176–183. doi: 10.1021/cb600063p. [DOI] [PubMed] [Google Scholar]
- 66.Nielsen P.G., Egholm M., Berg R.H., Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 67.Kaihatsu K., Huffman K.E., Corey D.R. Intracellular uptake and inhibition of gene expression by PNAs and PNA-peptide conjugates. Biochemistry. 2004;43:14340–14347. doi: 10.1021/bi048519l. [DOI] [PubMed] [Google Scholar]
- 68.Summerton J. Morpholino antisense oligomers: the case for an RNAse H-independent structural type. Biochim. Biophys. Acta. 1999;1489:141–158. doi: 10.1016/s0167-4781(99)00150-5. [DOI] [PubMed] [Google Scholar]
- 69.Muratovska A., Eccles M.R. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558:63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 70.Oishi M., et al. Lactosylated poly(ethylene glycol)-siRNA conjugate through acid-labile β-thiopropionate linkage to construct pH-sensitive polyion complex micelles achieving enhanced gene silencing in hepatoma cells. J. Am. Chem. Soc. 2005;127:1624–1625. doi: 10.1021/ja044941d. [DOI] [PubMed] [Google Scholar]
- 71.Kim S.H., et al. PEG conjugated VEGF siRNA for anti-angiogenic gene therapy. J. Control. Release. 2006;116:123–129. doi: 10.1016/j.jconrel.2006.05.023. [DOI] [PubMed] [Google Scholar]
- 72.Kim W.J., et al. Cholesteryl oligoarginine delivering vascular endothelial growth factor siRNA effectively inhibits tumor growth in colon adenocarcinoma. Mol. Ther. 2006;14:343–350. doi: 10.1016/j.ymthe.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 73.Manoharan M. Oligonucleotide conjugates as potential antisense drugs with improved uptake, biodistribution, targeted delivery, and mechanism of action. Antisense Nucleic Acid Drug Dev. 2002;12:103–128. doi: 10.1089/108729002760070849. [DOI] [PubMed] [Google Scholar]
- 74.De Paula D., Bentley M.V.L.B., Mahato R.I. Hydrophobization and bioconjugation for enhanced siRNA delivery and targeting. RNA. 2007;13:1–26. doi: 10.1261/rna.459807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McNamara J.O., et al. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 76.Song E., et al. Antibody mediated in vivo delivery of small interfering RNAs via cell surface receptors. Nat. Biotechnol. 2005;23:709–714. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 77.Whelan J. First clinical data on RNAi. Drug Discov. Today. 2005;15:1014–1015. doi: 10.1016/S1359-6446(05)03547-6. [DOI] [PubMed] [Google Scholar]
- 78.Fattal E., Bochot A. Ocular delivery of nucleic acids: antisense oligonucleotides, aptamers, and siRNA. Adv. Drug Deliv. Rev. 2006;58:1203–1223. doi: 10.1016/j.addr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 79.Hadj-Slimane R., Leppelletier Y., Lopez N., Garbay C., Raynaud F. Short interfering RNA (siRNA), a novel therapeutic tool acting on angiogenesis. Biochemie. 2007;89:1234–1244. doi: 10.1016/j.biochi.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 80.Li B., et al. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 2005;9:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar P., et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- 82.Zimmerman T.S., et al. Nature RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 83.Hu-Lieskovan S., et al. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral, delivery of small interfering RNA inhibits tumor growth in a murine model of metatstic Ewing’s sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 84.Landen C.N., et al. Therapeutic EphA2 gene targeting in vivo using neutral liposomal small interfering RNA delivery. Cancer Res. 2005;65:6910–6918. doi: 10.1158/0008-5472.CAN-05-0530. [DOI] [PubMed] [Google Scholar]