Abstract
Cell polarization is necessary for directed migration and leukocyte recruitment to inflamed tissues. Recent progress has been made in defining the molecular mechanisms that regulate chemoattractant-induced cell polarity during chemotaxis, including the contribution of phosphoinositide 3-kinase (PI3K)-dependent phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3] synthesis at the leading edge. However, less is known about the molecular composition of the cell rear and how the uropod functions during cell motility. Here, we demonstrate that phosphatidylinositol phosphate kinase type Iγ (PIPKIγ661), which generates PtdIns(4,5)P2, is enriched in the uropod during chemotaxis of primary neutrophils and differentiated HL-60 cells (dHL-60). Using time-lapse microscopy, we show that enrichment of PIPKIγ661 at the cell rear occurs early upon chemoattractant stimulation and is persistent during chemotaxis. Accordingly, we were able to detect enrichment of PtdIns(4,5)P2 at the uropod during chemotaxis. Overexpression of kinase-dead PIPKIγ661 compromised uropod formation and rear retraction similar to inhibition of ROCK signaling, suggesting that PtdIns(4,5)P2 synthesis is important to elicit the backness response during chemotaxis. Together, our findings identify a previously unknown function for PIPKIγ661 as a novel component of the backness signal that regulates rear retraction during chemotaxis.
INTRODUCTION
Neutrophils are critical participants in the innate immune response to inflammatory stimuli such as tissue injury and infection. The rapid recruitment of neutrophils to inflammatory sites requires a highly specialized form of directed cell migration in which shallow gradients of chemoattractant are translated into intracellular signals that establish polarized protrusion in the direction of migration (Devreotes and Janetopoulos, 2003; Niggli, 2003; Parent, 2004). A key component of this process is the asymmetric recruitment of phosphatidylinositol (3,4,5)-trisphosphate [PtdIns(3,4,5)P3] to the membrane adjacent to the highest concentration of chemoattractant (Meili et al., 1999; Servant et al., 2000). PtdIns(3,4,5)P3 and the RhoA family GTPases Cdc42 and Rac mediate a positive feedback mechanism that promotes actin polymerization and the formation of a dominant pseudopod at the cell front (Weiner et al., 1999; Servant et al., 2000; Van Keymeulen et al., 2006). In contrast, actin-based protrusions are inhibited at the cell rear, where RhoA-stimulated actomyosin contractility mediates rear retraction (Niggli, 1999; Eddy et al., 2000). Research into the molecular events that regulate chemotaxis has progressed rapidly; however, there is still limited understanding of the molecular components that define the cell rear and mediate retraction during directed cell migration.
Phosphatidylinositol (4,5)-bisphosphate [PtdIns(4,5)P2] is a membrane phospholipid that regulates a variety of cellular functions including actin cytoskeleton and focal adhesion dynamics, cell signaling, and vesicular trafficking (Ling et al., 2006). PtdIns(4,5)P2 binds to and regulates the function of many cytoskeleton-associated proteins including talin, vinculin, α-actinin, WASP, ezrin-radixin-moesin (ERM) proteins, and cofilin (Ling et al., 2006). The localized synthesis of PtdIns(4,5)P2 has also been implicated in modulating cell motility by functioning as a substrate for phosphoinositide 3-kinase (PI3K) and phospholipase C to generate PtdIns(3,4,5)P3 and inositol (1,4,5)-trisphosphate, respectively (Ling et al., 2006). There has been substantial interest in understanding the temporal and spatial regulation of PtdIns(4,5)P2 synthesis during cell migration. However, because of the relative abundance of PtdIns(4,5)P2 and the difficulty in detecting localized changes in its production, the temporal and spatial dynamics of PtdIns(4,5)P2 synthesis during chemotaxis have remained largely undefined.
One mechanism by which PtdIns(4,5)P2 is generated is through type I phosphatidylinositol phosphate kinases (PIPKI), which produce PtdIns(4,5)P2 through the phosphorylation of the fifth hydroxyl of phosphatidylinositol (4)-phosphate (PI4P) (Doughman et al., 2003). The PIPKI family includes three isoforms: α, β, and γ. Furthermore, PIPKIγ mRNA is alternatively spliced to form PIPKIγ635 and PIPKIγ661, which differ by a 26-amino acid C-terminal extension. Previous work has shown that the PIPKI family of lipid kinases display distinct subcellular distributions in fibroblasts (Di Paolo et al., 2002; Ling et al., 2002; Doughman et al., 2003). PIPKIα targets to the leading edge of cells where it regulates membrane ruffling (Doughman et al., 2003), whereas PIPKIγ661 targets to focal adhesions and may regulate integrin-mediated adhesions during cell migration via an interaction with the cytoskeletal protein talin (Di Paolo et al., 2002; Ling et al., 2002). However, PIPKIγ635 does not target to focal adhesions or interact with talin (Ling et al., 2002). Although PIPKI family members have been implicated in cell motility (Ling et al., 2006), the generation of localized PtdIns(4,5)P2 and functions of PIPKI family members during neutrophil chemotaxis have not been explored.
In this report, we demonstrate that the type Iγ phosphatidylinositol phosphate kinase (PIPKIγ661), which generates PtdIns(4,5)P2, is enriched in the uropod during directed migration. Using primary murine neutrophils that express green fluorescent protein (GFP)-tagged PIPKIγ661 and DMSO-differentiated HL-60 cells (dHL-60) retrovirally infected with GFP-tagged PIPKIγ661, we show that enrichment of PIPKIγ661 at the cell rear occurs early during chemoattractant-induced cell polarization and that cells overexpressing PIPKIγ661 form a prominent uropod. Our data indicate that although lipid kinase activity of PIPKIγ661 is not required for targeting to the rear, overexpression of kinase-dead PIPKIγ661 compromised uropod formation, and retraction of the cell rear in dHL-60 cells. Finally, we were able to detect enrichment of the PIPKIγ661 product, PtdIns(4,5)P2, at the uropod during chemotaxis, and this enrichment was impaired by expression of the kinase-dead PIPKIγ661. Taken together, our findings identify PIPKIγ661 as a novel component of the uropod and suggest that PIPKIγ661 may regulate rear release via the localized generation of PtdIns(4,5)P2 and contribute to backness signaling during chemotaxis.
MATERIALS AND METHODS
Isolation of Primary Neutrophils and Nucleofection
C57Bl/6 mice (Harlan Sprague-Dawley, Madison, WI) were killed in accordance with an institution approved protocol, and the long bones were removed and flushed with cold PBS/HSA/hep (Dulbecco's phosphate-buffered saline without Ca2+ or Mg2+ containing 0.15% human serum albumin and 100 U/ml preservative-free heparin) using a 30-gauge needle. Cells were dispersed using an 18-gauge needle, pelleted, and resuspended in PBS/HSA/hep. Subsequently, cells were passed through a 70-μm cell sieve, overlayed onto a discontinuous gradient of Histopaque 1083 and Histopaque 1119 (Sigma Aldrich, St. Louis, MO), and spun for 30 min at 700 × g with no brake. Red blood cells were lysed using ACK buffer (155 mM NH4Cl, 10 mM KHCO3, and 127 μM EDTA) and washed with PBS/HSA/hep. Cells were resuspended in PBS/HSA/hep and held at 4°C until use. Murine bone marrow–derived neutrophils were nucleofected as previously described (Kunisaki et al., 2006) and maintained at 37°C at 5% CO2 in the presence of 25 ng/ml recombinant murine GM-CSF (R&D Systems, Minneapolis, MN) for 24 h. The transfection efficiency was generally between 10 and 15% at 24 h, with good viability after 16–24 h. Primary human neutrophils used in RT-PCR and immunoblot analysis were obtained from healthy donors as described previously (Lokuta et al., 2003).
Cell Culture and Retroviral Infection
HL-60 cells (UCSF tissue culture facility) were cultured and differentiated as previously described (Nuzzi et al., 2007b). Phoenix viral packaging cells were transiently transfected by calcium-phosphate precipitation (Nuzzi et al., 2007a). Viral supernatant was harvested and used to retrovirally infect HL-60 cells as previously described (Nuzzi et al., 2007a). Populations of GFP-positive cells were obtained by fluorescence-activated cell sorting (FACS) and verified for expression by immunoblotting.
Time-Lapse Microscopy of Focal Adhesion Dynamics
HeLa cells (ATCC, Manassas, VA) were maintained according to the manufacturer's instructions, transiently transfected with GFP-PIPKIγ661 and dsRed-paxillin using Lipofectamine (Invitrogen, Carlsbad, CA), plated on 35-mm glass-bottom dishes coated with 10 μg ml−1 fibronectin, and allowed to adhere for 1 h. Focal adhesion dynamics of GFP-PIPKIγ661 and dsRed-paxillin was recorded using a Nikon Eclipse TE300 inverted fluorescence microscope (Melville, NY) with a cooled charge-coupled device video camera (Hamamatsu Photonics, Bridgewater, NJ) using a 60× objective and captured into Metamorph V 7.0r2 (Universal Imaging, West Chester, PA) at 2-min intervals for 60 min. Image processing was performed as described (Franco et al., 2004). The results are representative of three separate experiments.
RNA Isolation and RT-PCR
Total RNA was extracted from primary human neutrophils, undifferentiated and differentiated HL-60 cells, HEK 293 cells, Jurkat T-cells, and D10 T-cells using RNA STAT-60 (Iso-Tex Diagnostics, Friendswood, TX). After treatment with DNase (Promega, Madison, WI), RT-PCR was performed with 1 μg of RNA and 40 U RNAsin (Promega) using the One Step RT-PCR kit (Qiagen, Chatsworth, CA) and gene-specific primers as follows: PIPKIγ forward, 5′-CGCCCAGAGCACCTCAGATG-3′; reverse, 5′-GCTCCACCTGCACTGTAATCTG-3′; PIPKIγ661 forward, 5′- CGCCCAGAGCACCTCAGATG-3′; reverse, 5′-GCGGGGAGTACACCCAGCTCCTCT-3′; GAPDH forward, 5′-GAGTCAACGGATTTGGTCGTAT-3′; and reverse, 5′-AGTCTTCTGGGTGGCAGTGAT-3′. The following thermal cycling parameters were used: 50°C for 30 min, 95°C for 15 min, 94°C for 1 min, 55°C for 30 s, 72°C for 1 min (35 cycles), and a final extension at 72°C for 10 min. Each RT-PCR sample was resolved by electrophoresis on a 4.25% nondenaturing polyacrylamide gel, stained with ethidium bromide, and analyzed using a UVP Bio-dock system (UVP, Upland, CA). Data are representative of RT-PCR from multiple separate RNA preparations.
Protein Extraction, Antibodies, and Immunoblots
Primary human neutrophils, undifferentiated and differentiated HL-60 cells, and HEK 293 cells were lysed (Nuzzi et al., 2007b), samples were clarified by centrifugation, and protein concentration was determined using a bicinchoninic acid (BCA) protein assay (Pierce, Rockford, IL). Equal amounts of total protein were denatured in SDS sample buffer, resolved on 4–20% gradient SDS-PAGE gels, and transferred to nitrocellulose. Immunoblots were performed using standard conditions with polyclonal antibodies against PIPKIγ (Ling et al., 2002) or GFP (BD Biosciences, San Jose, CA). IRDye 800CW goat-α-rabbit IgG (Rockland Immunochemicals, Gilbertsville, PA) and Alexa Fluor 680 goat-α-mouse IgG (Invitrogen) were used as the secondary antibodies. Immunoblots were imaged on an Odyssey Infrared Imaging System with the Odyssey v2.0 software (LI-COR Bioscience, Lincoln, NE). Data are representative of immunoblots from multiple separate lysate preparations.
Immunofluorescence
Primary murine neutrophils were resuspended in Dulbecco's phosphate-buffered saline (DPBS) alone, or DPBS containing 100 nM formylmethionylleucylphenylalanine (fMLP) and were allowed to adhere for 10 min to glass coverslips coated with 10 μg ml−1 fibrinogen. Cells were fixed and permeabilized with 6.6% paraformaldehyde, 0.05% glutaraldehyde, and 0.25 mg ml−1 saponin in PBS, pH 7.2, for 15 min and then quenched with 0.15 M glycine for 15 min. Nonspecific binding was blocked with PBS containing 10% heat-inactivated fetal bovine serum and 0.25 mg/ml saponin at 4°C overnight. Cells were then stained for 30 min with α-PIPKIγ (Ling et al., 2002) and costained with rhodamine-labeled phalloidin (Molecular Probes, Eugene, OR) and DAPI (Molecular Probes). FITC goat α-rabbit IgG (Jackson ImmunoResearch, West Grove, PA) was used as the secondary antibody. Detection of active RhoA was performed as previously described (Berdeaux et al., 2004) using GST-rhotekin (Cytoskeleton, Denver, CO). D10 T-cells expressing GFP-PIPKIγ661 were plated on poly-l-lysine–coated coverslips, stimulated with LTB4, fixed, and stained with α-ERM (Cell Signaling Technology, Beverly, MA). Cells were mounted in mounting media and viewed on a Nikon Eclipse TE300 inverted fluorescence microscope (Melville, NY) using a 100× oil immersion DIC objective. Fluorescent images were digitally acquired using a cooled charge-coupled device video camera (Hamamatsu Photonics) and processed with Metamorph V 7.0r2 (Universal Imaging). Data are representative of a minimum of 50 cells from at least two independent experiments performed in duplicate.
Expression Constructs
Wild-type GFP-PIPKIγ661 was generated by PCR amplification of a murine PIPKIγ661 cDNA (Ling et al., 2002; a generous gift from Dr. Richard Anderson) and subcloning into pEGFP (Clontech, Palo Alto, CA) as an N-terminal GFP fusion, which was then subcloned into the empty pMX retroviral vector (a generous gift from Dr. Clive Svendsen). Kinase-dead GFP-PIPKIγ661 (D253A; Ling et al., 2002) was generated by mutating wild-type pMX-GFP-PIPKIγ661 with the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). GFP-PIPKIγ635 was generated by PCR amplification of the PIPKIγ661cDNA (in which a stop sequence was inserted after amino acid 635) and subcloning into the pMX vector. Wild-type and kinase-dead (D253A) GFP-PIPKIγ661-FLAG were generated by PCR amplification of GFP-PIPKIγ661 and subcloning into pCDNA3.1(+) (Invitrogen) containing a C-terminal FLAG tag. Wild-type and kinase-dead (D253A) mCherry-PIPKIγ661 were generated by PCR amplification of PIPKIγ661 cDNA and subcloning into the pmCherry-C1 vector. pmCherry-C1 was generated by PCR amplification of pRSET-B-mCherry (a generous gift from Dr. Roger Tsien) with primers containing a 5′ NheI site and 3′ BglII site and subcloning into pEGFP-C1 (Clontech) to replace the enhanced GFP (EGFP) sequence. GFP-PH-PLCδ and GFP-PHAKT constructs were a generous gift from Dr. Tamas Balla. GFP-Ezrin was kindly provided by Monique Arpin and Richard Lamb. dsRed-paxillin was a kind gift from Rick Horwitz. The accuracy of all constructs was verified by DNA sequencing.
Chemotaxis Assay
For each experiment, 5 × 105 cells were plated in Gey's media (dHL-60) or EGM-2MV (Cambrex Bioscience, Walkersville, MD; neutrophils) for 10 min on a glass-bottom dishes coated with 2.5 μg ml−1 fibrinogen (Sigma) and 10 μg ml−1 fibronectin and either left untreated or pretreated for 30 min with 10 μM ROCK inhibitor (Y-27632, Sigma). An Eppendorf Femtotip was loaded with 58 μM C5a, and a chemotactic gradient was formed by slow release of the chemoattractant from the tip into the media using an Eppendorf FemtoJet microinjection system (Westbury, NY) as described (Servant et al., 1999; Lokuta et al., 2003). Chemotaxis was recorded using a Nikon Eclipse TE300 inverted fluorescence microscope with a cooled charge-coupled device video camera (Hamamatsu Photonics) using a 60× oil immersion DIC objective and captured into Metamorph v7.0r2 (Universal Imaging) at 10-s (″) intervals for 10 min. Localization studies were performed on at least three independent samples from multiple cell lines or neutrophil preparations.
Kinase Activity Assays
dHL60 cells that express either GFP, wild-type GFP-PIPKIγ661, or kinase-dead GFP-PIPKIγ661 were washed in PBS and resuspended in lysis buffer (0.2% NP-40, 142.5 mM KCL, 5 mM MgCl2, 10 mM HEPES, pH 7.4, supplemented with protease inhibitor cocktail [P-8340; Sigma], phosphatase inhibitor cocktail [P-5726; Sigma], 2 mM phenylmethylsulfonyl fluoride, 100 mM sodium orthovanadate, 900 mM benzamidine, and 1 mM phenanthroline). Lysates were cleared by centrifugation, and protein concentrations were determined by BCA assay (Pierce). α-GFP (Molecular Probes) was used to immunoprecipitate from 200 μg total lysate at 4°C for 3 h with tilting. Immune complexes were washed twice with lysis buffer and once in kinase buffer without ATP or substrate. Reactions were carried out for 15 min at 32°C in 50 μl volume containing 50 mM Tris, pH 7.5, 10 mM MgCl2, 0.25 mM EGTA, 25 μM PtdIns4P (Echelon Biosciences, Salt Lake City, UT; P-4016), 50 μM ATP, and 10 μCi [32P]γATP. PtdIns [32P]4,5-P2 generation was terminated by the addition of 100 μl 1 N HCl and extracted with 200 μl chloroform/methanol (1:1). The organic phase containing radiolabeled lipids was washed one time with 80 μl methanol, 1 N HCl (1:1) and spotted on thin-layer chromatography (TLC) plates as described previously (Zhang et al., 1997). [32P]PIP2 generation was analyzed by PhosphorImager (Molecular Dynamics, Sunnyvale, CA). The results presented are representative of three separate experiments.
Quantification
MetaMorph software version 7.1.2 (Universal Imaging) was used to analyze fluorescence localization and shape. Localization was determined by drawing a line down the center of the cell from the rear of the cell to the front of the cell. The fluorescence intensity along that line was then obtained using the linescan function of MetaMorph. Peak fluorescence intensities for each cell were taken as 1 and minima as 0. These relative fluorescence values were then averaged for 15–45 cells in each condition. The difference in fluorescence shape between the GFP-PIPKIγ661 and GFP-PIPKIγ635 was determined through the integrated morphometry shape factor analysis in Metamorph. This algorithm assigns a value from 0 to 1, depending upon how closely the object represents a circle, with 1 being a perfect circle. Metamorph cell tracking was used to determine the speed of the cell front and the cell rear. Only cells that were actively chemotaxing for at least 2 min and both the front and rear of the cells were fully visible for the entire time were tracked. Speeds were averaged for the trackable time interval for each cell front and cell rear. Based on this exclusion criteria a total of 10 cells per condition from three separate experiments were tracked.
Online Supplementary Material
Dual color time-lapse microscopy of HeLa cells coexpressing GFP-PIPKIγ661 and dsRed-paxillin (see Figure 2D) is shown in Supplementary Video S1 (scale bar, 5 μm). Time-lapse microscopy of primary neutrophils (see Figure 3) that express GFP-PIPKIγ661 (Supplementary Video S2), GFP-ezrin (Supplemementary Video S3), or GFP-PIPKIγ635 (Supplementary Video S4) migrating toward a micropipette tip releasing C5a are shown (scale bars, 10 μm). Time-lapse microscopy of primary neutrophils migrating toward a micropipette tip releasing C5a (see Figure 5) is shown in Supplementary Videos S5 (GFP-PH-PLCδ), and S6 (GFP-PHAKT; scale bars, 10 μm). Time-lapse microscopy of primary neutrophils that express wild-type GFP-PIPKIγ661 (see Figure 6B) left either untreated (Supplementary Video S7) or pretreated with ROCK inhibitor (Supplementary Video S8) migrating toward a micropipette tip are shown. Time-lapse microscopy of dHL-60 cells (see Figure 7) that express either GFP (Supplementary Video S9), wild-type GFP-PIPKIγ661 (Supplementary Video S10), or kinase-dead GFP-PIPKIγ661 (Supplementary Videos S11 and S12) migrating toward a pipette tip are shown. Images for Video S1 were captured at 2-min intervals for 60 min. All other images were captured at 10-s (″) intervals for 10 min. All time-lapse microscopy images were captured using a 60× objective with a Nikon Eclipse TE300 inverted fluorescence microscope with a cooled charge-coupled device video camera (Hamamatsu Photonics) using differential interference contrast microscopy (DIC) and captured into Metamorph v7.0r2 (Universal Imaging). Time-lapse series were converted into QuickTime movie format with CinePak compression and were prepared using MetaMorph v7.Or2 (Universal Imaging).
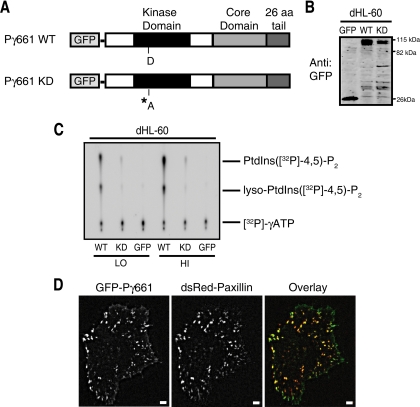
Figure 2.
GFP-PIPKIγ661 is functional and targets to focal adhesion in HeLa cells. (A) Schematic representation of the PIPKIγ661 constructs. The domains and active site residues of PIPKIγ661 are noted. The C-terminal 26-amino acid extension that defines PIPKIγ661 is shaded (dark gray). The kinase-dead construct was generated by mutating the active site aspartate (D253A denoted with an asterisk). (B) Stable HL-60 cell lines were generated that express N-terminal GFP-tagged versions of PIPKIγ661. Expression of GFP-PIPKIγ661 wild-type (WT) and PIPKIγ661 kinase-dead (KD) in dHL-60 cells was detected by immunoblot with α-GFP. (C) Lysates from dHL-60 cells that express either low or high levels of GFP-PIPKIγ661 wild-type (WT) and kinase-dead (KD) were immunoprecipitated with α-GFP antibody and assayed for PI4P kinase activity using TLC. Generation of PtdIns(4,5)P2 was detected by autoradiography. Shown is a representative experiment of more than three replicates. (D) Representative images of HeLa cells that express GFP-PIPKIγ661 demonstrate regions of colocalization of PIPKIγ661 with the focal adhesion protein paxillin. HeLa cells that express GFP-PIPKIγ661 and dsRed-paxillin were plated on 35-mm glass-bottom dishes coated with 10 μg ml−1 fibronectin and analyzed by time-lapse fluorescence microscopy. Shown is a representative of three separate experiments with more than six cells imaged. Scale bar, 5 μm. Corresponding time-lapse microscopy is shown in Supplementary Video S1. Scale bar, 5 μm.
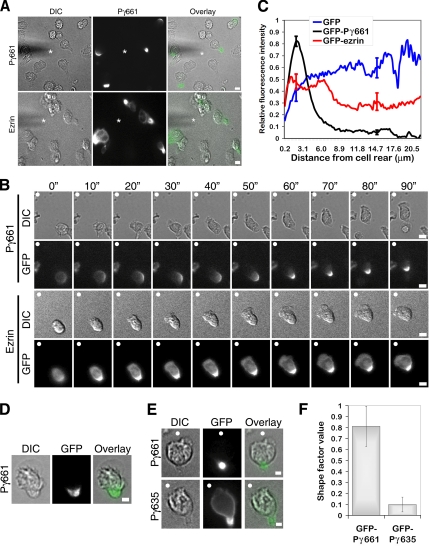
Figure 3.
GFP-PIPKIγ661 targets to the uropod of primary neutrophils. (A) Representative DIC and fluorescent images of neutrophils that express either GFP-PIPKIγ661 or GFP-ezrin. Merged image shows enrichment of GFP-PIPKIγ661 at the cell rear similar to GFP-ezrin in response to a chemotactic gradient. Primary murine neutrophils that express either GFP-PIPKIγ661 or GFP-ezrin were plated onto 2.5 μg ml−1 fibrinogen and 10 μg ml−1 fibronectin and exposed to a chemotactic gradient generated by the slow release of C5a from a Femtotip micropipette. DIC and fluorescent time-lapse images were taken at 100-s (″) intervals. The tip of the micropipette is marked with an asterisk (*). Scale bars, 5 μm. Corresponding time-lapse microscopy is shown in Supplementary Videos S2 (GFP-PIPKIγ661) and S3 (GFP-ezrin). Scale bars, 10 μm. (B) Time-lapse sequence of neutrophils that express either GFP-PIPKIγ661 or GFP-ezrin. Note enrichment of both GFP-PIPKIγ661 and GFP-ezrin in the uropod. The direction of the chemoattractant source is denoted with a filled white circle (○). Scale bars, 5 μm. (C) Fluorescence intensities from the cell rear to the cell front were determined for at least 45 cells for each condition from four different experiments. Shown are the average relative fluorescence intensities for GFP, GFP-PIPKIγ661, and GFP-ezrin. Note the rear localization of GFP-PIPKIγ661 and GFP-ezrin. These differences in localization between GFP, GFP-PIPKIγ661, and GFP-ezrin at 2 μm from the cell rear were confirmed using repeated measures ANOVA and subsequent Bonferroni's multiple comparison test and were found to have a p < 0.05. Similarly, GFP-PIPKIγ661 and GFP-ezrin fluorescence intensities are reduced compared with GFP at the point 15 μm from the cell rear, as indicated by repeated-measures ANOVA and Bonferroni's multiple comparison test with a p < 0.05. Shown are the SEM for 2 and 15 μm from the cell rear. (D) Representative DIC and fluorescent images of neutrophils that express GFP-PIPKIγ661. Magnified merged image (right) shows localization of GFP-PIPKIγ661 (middle) at the cell rear in response to uniform chemoattractant stimulation. Scale bar, 5 μm. (E) Representative DIC and fluorescent images of neutrophils that express GFP-PIPKIγ661 or GFP-PIPKIγ635. Merged image (right) shows enrichment of GFP-PIPKIγ661 (top middle) in the uropod, whereas GFP-PIPKIγ635 (bottom middle) is localized at the cell membrane, at the front of the cell, and the uropod. The direction of the chemoattractant source is denoted with a filled white circle (○). Scale bars, 2 μm. Corresponding time-lapse microscopy of neutrophils expressing GFP-PIPKIγ635 is shown in Supplementary Video S4. Scale bar, 10 μm. (F) The shapes of the fluorescent signals in neutrophils expressing GFP-PIPKIγ661 or GFP-PIPKIγ635 were analyzed using the integrated morphometry shape factor analysis of Metamorph. This algorithm assigns a value from 0 to 1, describing the shape of the fluorescence signal where a perfect circle attains a value of 1 and a line is assigned a 0. At least 15 cells from three different experiments were analyzed, and the SD is shown. Two-tailed Student's t test, p < 0.001.
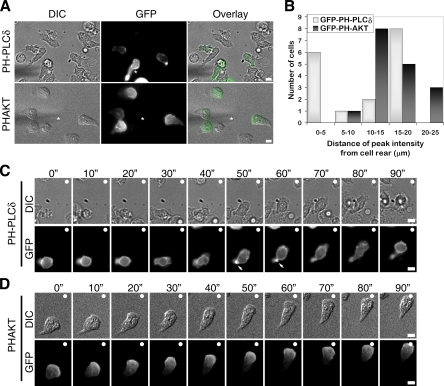
Figure 5.
Asymmetric distribution of GFP-PH-PLCδ and GFP-PHAKT in primary neutrophils exposed to a gradient of chemoattractant. (A) Representative DIC and fluorescent images of neutrophils that express either GFP-PH-PLCδ or GFP-PHAKT. Bone marrow derived murine neutrophils were nucleofected with either GFP-PH-PLCδ or GFP-PHAKT and plated on 35-mm glass-bottom dishes coated with a mixture of 2.5 μg ml−1 fibrinogen and 10 μg ml−1 fibronectin and exposed to a chemotactic gradient generated by the slow release of C5a from a micropipette. DIC and fluorescent time-lapse images were taken at 10-s (″) intervals. The tip of the micropipette is marked with an asterisk (*). Merged image shows enrichment of PHAKT at the leading edge, whereas PH-PLCδ is periodically enriched at both the cell front and cell rear. Scale bars, 5 μm. Corresponding time-lapse microscopy is shown in Supplementary Videos S5 (GFP-PH-PLCδ) and S6 (GFP-PHAKT). Scale bars, 10 μm. (B) Quantification of fluorescence localization in neutrophils expressing either GFP-PH-PLCδ or GFP-PHAKT. Fluorescence intensities from the cell rear to the cell front were determined for 17 cells per condition. Shown are the numbers of cells and the distance from the cell rear of their peak fluorescence intensities for GFP-PH-PLCδ or GFP-PHAKT. (C) Time-lapse sequence of neutrophils that express GFP-PH-PLCδ. Note periodic enrichment of PH-PLCδ at the cell rear (arrows). (D) Time-lapse sequence of neutrophils that express GFP-PHAKT. Note enrichment of PHAKT at the leading edge. The direction of the chemoattractant source is denoted with a filled white circle (○). Scale bars, 5 μm.
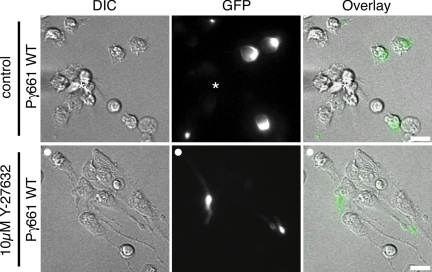
Figure 6.
PIPKIγ661 retains uropod localization upon ROCK inhibition. Representative DIC and fluorescent images of neutrophils pretreated with ROCK Y-27632 inhibitor that express wild-type GFP-PIPKIγ661 (Pγ661 WT). Bone marrow–derived murine neutrophils were nucleofected with Pγ661 wild type (WT), plated on 35-mm glass-bottom dishes coated with a mixture of 2.5 μg ml−1 fibrinogen and 10 μg ml−1 fibronectin, pretreated for 30 min with Y-27632 or vehicle control, and subsequently exposed to a chemotactic gradient generated by the slow release of C5a from a micropipette. DIC and fluorescent time-lapse images were taken at 10-s (″) intervals as described in Materials and Methods. The tip of the micropipette is marked with an asterisk (*) or the direction of chemoattractant source is denoted with a filled white circle (○). Merged image shows enrichment of PIPKIγ661 in the cell rear despite elongated morphology observed upon inhibition of ROCK. Scale bars, 10 μm. Corresponding time-lapse microscopy for cells that express wild-type GFP-PIPKIγ661 are shown in Supplementary Videos S7 (control) and S8 (Y-27632). Scale bars, 10 μm.
Figure 7.
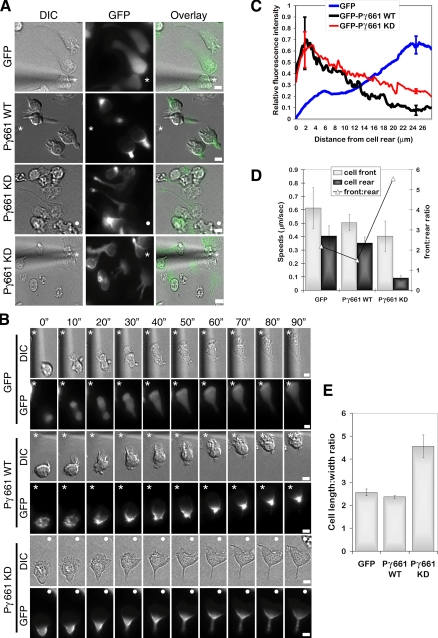
PIPKIγ expression modulates cell morphology and rear retraction. (A) Representative DIC and fluorescent images of dHL-60 cells that express GFP control, GFP-PIPKIγ661 wild-type (WT), or GFP-PIPKIγ661 kinase-dead (KD)show enrichment of GFP-PIPKIγ661 WT in the uropod and GFP-PIPKIγ661 KD at the cell rear in elongated tails. Differentiated HL-60 cells (dHL-60) that express GFP-PIPKIγ661 WT or KD were plated onto 2.5 μg ml−1 fibrinogen and 10 μg ml−1 fibronectin andexposed to a chemotactic gradient generated by the slow release of C5a from a Femtotip micropipette. DIC and fluorescent time-lapse images were taken at 10-s (″) intervals. The tip of the micropipette is marked with an asterisk (*) or the direction of the chemoattractant source is denoted with a filled white circle (○). Scale bars, 5 μm. Corresponding time-lapse microscopy is shown in Supplementary Videos S9 (GFP), S10 (GFP-PIPKIγ661 WT), and S11–12 (GFP-PIPKIγ661 KD). (B) Time-lapse sequence of dHL-60 cells that express control GFP (top), GFP-PIPKIγ661 WT (middle), or GFP-PIPKIγ661 KD (bottom). Scale bars, 10 μm. Note prominent uropod in cells that express GFP-PIPKIγ661 WT compared with the elongated morphology and reduced rear retraction in cells that express GFP-PIPKIγ661 KD. Scale bars, 10 μm. The tip of the micropipette is marked with an asterisk (*) or the direction of the chemoattractant source is denoted with a filled white circle (○). Scale bars, 5 μm. (C) Fluorescence intensities from the cell rear to the cell front were determined for at least 25 cells in each condition from four different experiments. Shown are the average relative fluorescence intensities for GFP, GFP-PIPKIγ661 WT, and GFP-PIPKIγ661 KD. Note the rear localization of both GFP-PIPKIγ661 WT and GFP-PIPKIγ661 KD compared with the GFP control. These differences in localization at 2 μm from the cell rear were confirmed using repeated-measures ANOVA and subsequent Bonferroni's multiple comparison test and were found to have a p < 0.05. Similarly, GFP-PIPKIγ661 WT and GFP-PIPKIγ661 KD fluorescence intensities are reduced at the point 25 μm from the cell rear as compared with GFP as indicated by repeated-measures ANOVA and Bonferroni's multiple comparison test with a p < 0.05. Shown are the standard deviations for 2 and 25 μm from the cell rear. (D) Speeds of the cell front and cell rear for dHL-60 cells expressing GFP, GFP-PIPKIγ661 WT, or GFP-PIPKIγ661 KD were determined for 10 cells in each condition from three different experiments. Shown are the average speeds ± SEM. The ratio of the speed of the cell front to the speed of the cell rear is also shown. Note the reduced speed of the cell rear in dHL-60 cells expressing GFP-PIPKIγ661 KD compared with those expressing GFP. These differences were confirmed using repeated measures ANOVA and subsequent Bonferroni's multiple comparison test and found to have a p < 0.05. (E) Cell length of dHL-60 cells expressing GFP, GFP-PIPKIγ661 WT, or GFP-PIPKIγ661 KD. Cell length was measured for at least 30 cells per condition from four different experiments. Shown are the averages ± the SD. Note the increase in cell length in the dHL-60 cells expressing GFP-PIPKIγ661 KD compared with those expressing GFP. These differences were confirmed using repeated measures ANOVA and subsequent Bonferroni's multiple comparison test and found to have a p < 0.05.
RESULTS
Expression of PIPKIγ661 in Neutrophils and HL-60 Cells
The role of PIPKIγ661 in the regulation of adhesions in fibroblasts makes PIPKIγ661 an attractive candidate to contribute to localized PtdIns(4,5)P2 production during neutrophil motility. To determine expression of the type Iγ PIPKI in neutrophils and the neutrophil-like HL-60 cell line, we used a series of isoform specific reagents for RT-PCR and immunoblot analysis. The type Iγ PIPKI, specifically PIPKIγ661, was readily detected in primary neutrophils by RT-PCR (Figure 1A) and immunoblotting (Figure 1B). PIPKIα and PIPKIβ were also detected by RT-PCR in neutrophils but were not readily detected by immunoblotting (data not shown). Additionally, we observed an increase in PIPKIγ expression upon DMSO-induced differentiation of neutrophil-like HL-60 cells, suggesting that PIPKIγ may be specifically up-regulated in neutrophil-like cells (Figure 1B).
Figure 1.
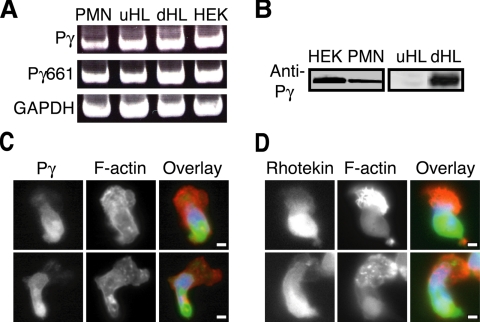
Endogenous PIPKIγ is expressed in neutrophils and localizes toward the cell rear upon chemoattractant stimulation. (A) RT-PCR was used to determine the expression of PIPKIγ in primary neutrophils (PMN), undifferentiated and differentiated HL-60 cells (uHL and dHL), and control HEK-293 cells (HEK) using gene-specific primers against human PIPKIγ (Pγ) and the PIPKIγ661 splice variant (Pγ661). GAPDH loading controls indicate equal sample was loaded. (B) Representative immunoblots show endogenous PIPKIγ (Pγ) expression in primary neutrophils, HEK cells, and HL-60 cells in separate blots (boxes). (C and D) Representative images of neutrophils demonstrate localization of endogenous PIPKIγ (Pγ; C) and active RhoA (rhotekin; D) to the cell rear upon fMLP stimulation. Neutrophils were plated on coverslips coated with 10 μg ml−1 fibrinogen in the presence of 100 nM fMLP for 15 min, fixed, and stained for PIPKIγ or GST-rhotekin (green), rhodamine phalloidin for F-actin (red), and DAPI (blue). Note enrichment of PIPKIγ at the cell rear. Shown are representative images from at least three experiments with more than 50 cells examined. Scale bars, 5 μm.
Asymmetric Distribution of PIPKIγ in Neutrophils
To determine the localization of endogenous PIPKIγ, fMLP-stimulated primary bone-marrow neutrophils were analyzed by immunofluorescence using an antibody that recognizes PIPKIγ (Ling et al., 2002). Our data indicate that endogenous PIPKIγ was enriched toward the rear of the cell (Figure 1C) and did not colocalize with actin at the cell front. Previous studies have shown that the active form of RhoA is enriched in the rear of neutrophil-like HL-60 cells (Xu et al., 2003; Wong et al., 2006). Using GST-rhotekin as a probe for RhoA-GTP, we were able to detect enrichment of active RhoA toward the cell rear of primary neutrophils, similar to the staining observed for PIPKIγ (Figure 1D). Together these data suggest that PIPKIγ is a novel component of the cell rear of polarized neutrophils.
GFP-PIPKIγ661 Retains Lipid Kinase Activity and Focal Adhesion Targeting
To define the dynamics of PIPKIγ localization in live cells, we constructed a GFP-PIPKIγ fusion protein for both wild-type and kinase-dead PIPKIγ661 (Figure 2). Expression of the GFP-fusions in differentiated HL-60 cells (dHL-60) was confirmed by immunoblotting (Figure 2B), and the activity of the GFP-tagged kinase was measured (Figure 2C). We found that wild-type GFP-PIPKIγ661 retained kinase activity when expressed in dHL-60 cells and that the D253A kinase-dead mutant PIPKIγ661 showed markedly reduced kinase activity (Figure 2C). Previous work has shown that PIPKIγ661 targets to focal adhesions (Di Paolo et al., 2002; Ling et al., 2002). To determine if GFP-PIPKIγ661 retained its focal adhesion targeting properties, GFP-PIPKIγ661 was expressed in HeLa cells and analyzed by live fluorescence imaging. We found that GFP-PIPKIγ661 targeted to focal adhesions where it colocalized with paxillin (Figure 2D), displaying dynamic assembly and disassembly at both leading and trailing edge adhesion complexes (Supplementary Video S1). Taken together, we demonstrate that GFP-PIPKIγ661 is functional and shows appropriate targeting to focal adhesions in HeLa cells.
PIPKIγ661 Is a Novel Component of the Uropod in Polarized Leukocytes
To characterize PIPKIγ661 localization in primary neutrophils, GFP-PIPKIγ661 was exogenously expressed in primary mouse neutrophils using nucleofection. This experiment was made possible by recent technical advances using nucleofection to transiently express transgenes in primary neutrophils at relatively high efficiency (see Materials and Methods). When expressed in primary neutrophils, PIPKIγ661 showed specific targeting to the uropod during chemotaxis with exclusion from the leading edge (Figure 3A, Supplementary Video S2), which is in accordance with the localization of endogenous PIPKIγ (Figure 1C), and is similar to GFP-ezrin (Figure 3A, Supplementary Video S3; Lamb et al., 1997). After 20 s of chemoattractant exposure, PIPKIγ was enriched toward the cell rear, which occurred before the acquisition of a polarized morphology (Figure 3B). Additionally, as the cell began to chemotax up the gradient of chemoattractant, PIPKIγ remained localized at the uropod (Figure 3A). Because ERM proteins represent established components of the uropod in myeloid cells (Alonso-Lebrero et al., 2000; Gomez-Mouton et al., 2001; Lee et al., 2004), we examined the localization of GFP-ezrin in primary neutrophils. The enrichment and recruitment of PIPKIγ to the uropod was similar to that of ezrin (Figure 3B). Quantification of fluorescence intensity indicates a striking localization of PIPKIγ specifically at the uropod, whereas GFP-ezrin shows targeting at the rear with a more diffuse distribution (Figure 3C). To determine if PIPKIγ661 targeting to the cell rear required a gradient of chemoattractant, we exposed the cell to a uniform concentration of chemoattractant (Figure 3D). The targeting of PIPKIγ661 to the uropod was also observed in a uniform concentration of chemoattractant, suggesting that this redistribution is a key component of neutrophil polarization. To further characterize the region of PIPKIγ involved in targeting, we expressed GFP-PIPKIγ635, which lacks the 26 amino acids required for talin binding and targeting to focal adhesions, in primary neutrophils. Interestingly, PIPKIγ635 also showed some enrichment toward the cell rear. However, in contrast to PIPKIγ661, PIPKIγ635 was also found along the membrane and at the leading edge of the cell as indicated by morphometric analysis of the shape of the fluorescence in cells expressing GFP-PIPKIγ635 or GFP-PIPKIγ661 (Figure 3, E and F, Supplementary Video S4). This suggests that the C-terminal 26-amino acid extension that defines PIPKIγ661 is not required for membrane targeting but is likely needed to reinforce uropod localization in primary neutrophils (Figure 3D).
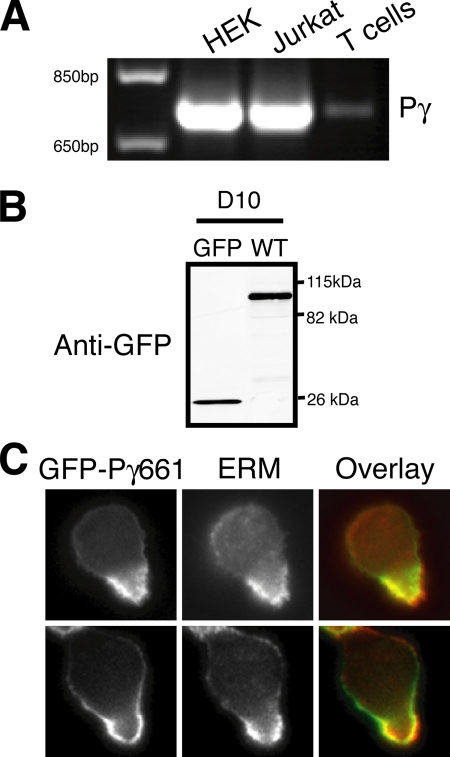
To determine if PIPKIγ is expressed in other leukocytes, we analyzed expression of the type Iγ PIPKI in primary T lymphocytes and Jurkat T-cells by RT-PCR. The type Iγ PIPKI was readily detected in both T lymphocytes and Jurkat cells (see Figure 4A). To determine if PIPKIγ is a component of the uropod in other leukocytes, GFP-PIPKIγ661 was expressed in the D10 T lymphocyte cell line (Figure 4B). In accordance with our findings in primary neutrophils, PIPKIγ661 also targeted to the uropod in T-cells, colocalizing with ERM proteins (Figure 4C). Taken together, our data indicate that PIPKIγ661 specifically targets to the rear of polarized neutrophils and lymphocytes, thereby identifying PIPKIγ661 as a novel component of the leukocyte uropod.
Figure 4.
PIPKIγ is expressed in T-cells and localizes to the uropod. (A) RT-PCR was used to determine the expression of PIPKIγ in primary human T-cells, Jurkat T-cells, and control HEK-293 cells (HEK) using gene-specific primers against human PIPKIγ (Pγ). (B) Stable D10 T-cell lines were generated that express GFP control and wild-type GFP-PIPKIγ661. Expression of GFP-PIPKIγ661 wild-type (WT) in D10 T-cells was detected by immunoblot with α-GFP. (C) Representative images of D10 T-cells demonstrate colocalization of GFP-PIPKIγ661 at the cell rear with ERM proteins upon LTB4 stimulation. D10 T-cells that express GFP-PIPKIγ661 were plated on coverslips coated with poly-l-lysine for 30 min, treated with LTB4, fixed, stained with α-ERM (middle). Merged image (overlay) shows colocalization of GFP-PIPKIγ661 with ERM proteins at the cell rear.
Periodic Enrichment of PtdIns(4,5)P2 in the Uropod During Neutrophil Chemotaxis
To determine if there is localized production of the PIPKIγ661 product PtdIns(4,5)P2 in the uropod during neutrophil chemotaxis, we transiently expressed a probe for PtdIns(4,5)P2 consisting of GFP fused to the pleckstrin homology (PH) domain of PLCδ (GFP-PH-PLCδ; Stauffer et al., 1976; Varnai and Balla, 1998) in primary neutrophils (Figure 5). On exposure to a chemotactic gradient, the localization of GFP-PH-PLCδ was highly dynamic, with a strong periodic enrichment of GFP-PH-PLCδ in the uropod (Figure 5, A and C, Supplementary Video S5). To determine if PtdIns(3,4,5)P3 also showed periodic targeting to the cell rear, we transiently expressed a PtdIns(3,4,5)P3 probe consisting of GFP fused to the PH domain of AKT (Varnai and Balla, 1998; Meili et al., 1999; Servant et al., 2000). In contrast to GFP-PH-PLCδ, GFP-PHAKT showed persistent enrichment at the cell front during neutrophil chemotaxis (Figure 5, A and D, Supplementary Video S6). Quantitative analysis of fluorescence intensity indicated the relative exclusion of the GFP-PHAKT probe from the cell rear compared with the GFP-PH-PLCδ probe, which displayed periodic targeting to the both the uropod and the leading edge of the cell (Figure 5B). Together, these findings suggest that enrichment of both PIPKIγ661 and its PtdIns(4,5)P2 product occurs within the uropod in response to chemoattractant stimulation.
PIPKIγ661 Uropod Localization Is Independent of ROCK Signaling
To understand the signaling pathways involved in recruitment of PIPKIγ661 to the uropod, we examined whether inhibition of ROCK, a downstream effector of RhoA involved in the backness response, would alter the uropod targeting of PIPKIγ661. Primary neutrophils expressing GFP-PIPKIγ661 exhibited a normal polarized morphology and targeting of GFP-PIPKIγ 661 to the uropod (Figure 6, Supplementary Video S7). However, treatment of primary neutrophils expressing GFP-PIPKIγ 661 with the ROCK inhibitor Y-27632 induced an abnormal elongated morphology with defects in rear release during chemotaxis, in agreement with previous reports (Somlyo et al., 2000; Alblas et al., 2001; Worthylake et al., 2001). However, targeting of GFP-PIPKIγ661 to the uropod was not affected by ROCK inhibition (Figure 6, Supplementary Video S8), suggesting that the localization of GFP-PIPKIγ661 to the uropod is independent of ROCK activity.
GFP-PIPKIγ Localizes to the Uropod and Regulates Rear Retraction in Neutrophil-like HL-60 Cells
To extend our work with primary neutrophils and further define the function of PIPKIγ661 during chemotaxis, we have utilized neutrophil-like HL-60 cells (Collins et al., 1977). HL-60 cells represent a widely used model system to study neutrophil chemotaxis (Servant et al., 2000; Weiner et al., 2002; Gomez-Mouton et al., 2004; Van Keymeulen et al., 2006; Wong et al., 2006), because this promyelocytic leukemia cell line differentiates into a neutrophil-like state when cultured with 1.25% DMSO (Collins et al., 1978) and becomes responsive to chemoattractants (Collins et al., 1979; Gallagher et al., 1979; Hauert et al., 2002). Wild-type and kinase-dead GFP-PIPKIγ661 were stably expressed in undifferentiated HL-60 cells. GFP-positive cells were then sorted by flow cytometry, differentiated into a neutrophil-like state with DMSO, and analyzed by time-lapse microscopy. Both wild-type and kinase-dead PIPKIγ661 showed targeting to the rear of polarized dHL-60 cells compared with GFP alone based on quantitative analysis of fluorescence intensity from the rear to the front of the cell (Figure 7, A–C). Time-lapse imaging revealed the dynamic redistribution of GFP-PIPKIγ661 away from the highest concentration of chemoattractant within 20 s after exposure to a gradient of attractant and before the formation of a polarized morphology (Figure 7B), compared with the diffuse distribution of GFP (Figure 7B).
The morphology and chemotactic response of dHL-60 cells varied depending on the expression level of PIPKIγ661. Cell lines that expressed high levels (more than 10-fold overexpression) of wild-type or kinase-dead PIPKIγ661 exhibited reduced polarization and motility in response to chemoattractant (Supplementary Figure S1). Therefore, cells were sorted into high and low expressing populations and phenotypic analysis was performed using the “low” cell lines that had less than fivefold overexpression of the PIPK constructs (Figure 7). Exogenously expressed wild-type GFP-PIPKIγ661 induced an intensely polarized morphology with the formation of a prominent uropod with a distinctive, rounded morphology at the cell rear (Figure 7A, Supplementary Video S10) compared with GFP alone (Figure 7, A–C, Supplementary Video S9). This is in contrast with the phenotype observed with expression of the kinase-dead PIPKIγ661 in which the cells lacked a defined uropod and displayed an elongated morphology with impaired rear retraction (Figure 7, A and B, Supplementary Videos S11 and S12), similar to the phenotype observed with inhibition of ROCK (Figure 6). Quantitative analysis of cell polarity was performed by measuring cell length:width ratio of cells exposed to a gradient of chemoattractant. The analysis revealed an almost twofold increase in elongation of cells that expressed kinase-dead PIPKIγ661 as compared with wild-type PIPKIγ661 or GFP alone (Figure 7E). To further analyze the effects of expression of kinase-dead PIPKIγ661 on motility, cell speeds were measured by tracking the motility of the front and rear of the cell in a gradient of chemoattractant (Figure 7D). Interestingly, the cell speed at the leading edge of the cell was not significantly affected by expression of kinase dead PIPKIγ661, whereas the speed of the uropod was dramatically impaired (Figure 7D). Taken together, our findings identify PIPKIγ661 as a novel uropod component that regulates rear release during neutrophil chemotaxis.
PIPKIγ Regulates the Localized Generation of PtdIns(4,5)P2 at the Uropod during Chemotaxis
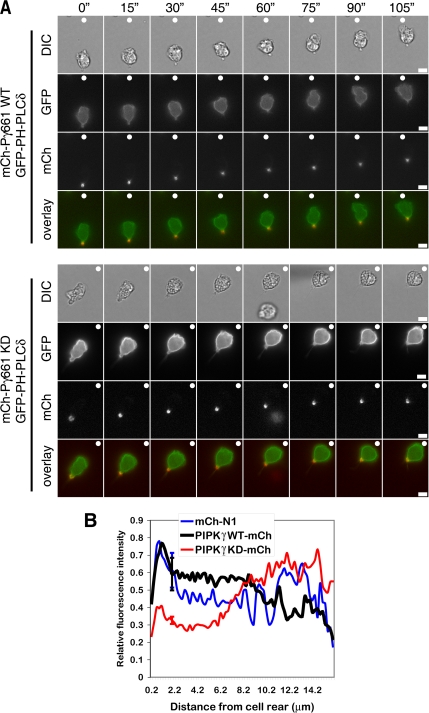
To determine whether PIPK is necessary for the localized generation of PtdIns(4,5)P2 at the uropod during chemotaxis, we examined the effect PIPKIγ661 expression on the distribution of PtdIns(4,5)P2 in primary neutrophils using the GFP-PH-PLCδ probe. mCherry-PIPKIγ661 and kinase-dead mCherry-PIPKIγ661 were coexpressed with GFP-PH-PLCδ in primary murine bone marrow neutrophils. Our findings indicate that expression of kinase-dead PIPKIγ661 impaired the production of PtdIns(4,5)P2 at the uropod compared with expression of either mCherry alone or wild type mCherry-PIPKIγ661 (Figure 8). Taken together, our findings identify PIPKIγ661 as a novel uropod component that regulates rear release via the localized generation of PtdIns(4,5)P2 and suggest that PIPKIγ661-mediated PtdIns(4,5)P2 synthesis and periodic accumulation is important for backness signaling and rear release during neutrophil chemotaxis.
Figure 8.
Kinase-dead PIPKIγ661 expression reduces PtdIns(4,5)P2 localization to the cell rear. (A) Representative DIC and fluorescent images of bone marrow-derived murine neutrophils nucleofected with mCherry-PIPKIγ661 wild type (mCh-PIPKIγ661 WT) and GFP-PH-PLCδ or with mCherry-PIPKIγ661 kinase-dead (mCh-PIPKIγ661 KD) and GFP-PH-PLCδ. Cells were plated on 35-mm glass-bottom dishes that were coated with a mixture of 2.5 μg ml−1 fibrinogen and 10 μg ml−1 fibronectin and exposed to a chemotactic gradient generated by the slow release of C5a from a micropipette. DIC and fluorescent time-lapse images were taken at 15-s (″) intervals as described in Materials and Methods. The direction of chemoattractant source is denoted with a filled white circle (○). Merged image shows enrichment of PIPKIγ661 WT and KD in the cell rear and reduced GFP-PH-PLCδ at the cell rear in cells coexpressing mCh-PIPKIγ661 KD compared with those coexpressing mCh-PIPKIγ661 WT. Scale bars, 5 μm. (B) Fluorescence intensities from the cell rear to the cell front were determined for at least 20 cells in each condition from three different experiments. Shown are the average relative fluorescence intensities for GFP-PH-PLCδ when coexpressed with mCherry control, mCh-PIPKIγ661 WT, or mCh-PIPKIγ661 KD. Note the reduction in rear localization of GFP-PH-PLCδ when coexpressed with GFP-PIPKIγ661 KD compared with coexpression with GFP-PIPKIγ661 WT or mCherry control. These differences in localization at 2 μm from the cell rear were confirmed using repeated-measures ANOVA and subsequent Bonferroni's multiple comparison test and found to have a p < 0.05. Shown are the standard deviations for 2 μm from the cell rear.
DISCUSSION
Although PI3K and its lipid product PtdIns(3,4,5)P3 have emerged as key regulators of directed migration, the role of the type Iγ PIP kinases and its lipid product PtdIns(4,5)P2 during chemotaxis has been relatively unexplored. In this study, we have examined the expression and localization of PIPKIγ during neutrophil chemotaxis and found that PIPKIγ661 is a novel component of the leukocyte uropod. Using time-lapse microscopy, we have examined the localization of PIPKIγ661 during chemotaxis of primary neutrophils and dHL-60 cells. Our results demonstrate that chemoattractant stimulation triggers the rapid recruitment of PIPKIγ661 to the cell rear and that the lipid kinase activity of PIPKIγ661 and localized generation of PtdIns(4,5)P2 is involved in detachment of the trailing edge. Therefore, this study identifies PIPKIγ661 as a novel uropod component and localized PtdIns(4,5)P2 synthesis as a unique factor involved in backness signaling during neutrophil chemotaxis.
During chemotaxis, spatial information about the chemotactic gradient is translated into an internal asymmetry of signaling molecules that elicit either front- or back-specific responses. For example, leading edge components such as Rac, Cdc42, and PI3K are thought to promote the frontness response by triggering accumulation of actin and the phosphoinositide PtdIns(3,4,5)P3 at the leading edge (Meili et al., 1999; Weiner et al., 1999; Servant et al., 2000; Van Keymeulen et al., 2006). In contrast, components of the cell rear such as Rho and ROCK are thought to promote backness by the generation of actomyosin-based contraction and regulation of rear release (Niggli, 1999; Eddy et al., 2000; Xu et al., 2003; Wong et al., 2006). Accordingly, spatial restriction of PIPKIγ661 to the cell rear suggests that PIPKIγ661 plays a role in backness signaling at the trailing edge during neutrophil chemotaxis. Further analysis of PIPKIγ661 function during chemotaxis suggests that PIPKIγ661 promotes backness via localized PtdIns(4,5) P2 production to regulate rear release (Figures 7 and 8), because expression of kinase-dead PIPKIγ661 compromised rear release resulting in the formation of abnormal, elongated, or bicuspid tails, whereas cells that overexpress wild-type PIPKIγ661 generate a more prominent uropod and exhibit efficient rear release.
Previous work has shown that the backness response is mediated by RhoA via downstream signaling to ROCK to initiate actomyosin contraction (Kimura et al., 1996; Yoshinaga-Ohara et al., 2002; Xu et al., 2003). Interestingly, the rear retraction defect noted upon overexpression of kinase-dead GFP-PIPKIγ661 phenocopies previous results observed upon inhibition of RhoA (Alblas et al., 2001; Worthylake et al., 2001), ROCK (Somlyo et al., 2000; Alblas et al., 2001; Worthylake et al., 2001), and myosin IIa (Eddy et al., 2000). Previous work has also shown that PIPKIγ interacts with RhoA (Chong et al., 1994; Ren et al., 1996) and that this interaction increases PtdIns(4,5)P2 production by PIPKI family members (Chong et al., 1994; Weernink et al., 2004). Therefore, PIPKIγ661 may promote rear retraction via integration of PtdIns(4,5)P2 with the backness pathway through the association of PIPKIγ661 with RhoA. Previous work has also shown that PIPKIγ661 associates with AP2 to regulate endocytosis (Bairstow et al., 2006). As endocytosis and recycling of integrins at the cell rear plays an important role in neutrophil rear release (Lawson and Maxfield, 1995), it is intriguing to speculate that PIPKIγ661 may also promote rear detachment via endocytosis of surface receptors at the cell rear. Alternatively, PIPKIγ661 may promote rear detachment via the synthesis of PtdIns(4,5)P2, which regulates the activity of cytoskeleton-associated ERM proteins (Yoshinaga-Ohara et al., 2002; Fievet et al., 2004). A challenge for future investigation will be to define the mechanisms by which PIPKIγ661 and the localized production of PtdIns(4,5)P2 within the uropod regulates rear retraction.
Recent evidence suggests cross talk between front and rear signaling modules in that PtdIns(3,4,5)P3 and Cdc42 signaling at the leading edge feedback to affect RhoA signaling at the back of the cell (Van Keymeulen et al., 2006). However, less is known about how the back of the cell may regulate signaling at the front and modulate cell polarity. There is evidence to support a role for the uropod in regulating cell polarity (Lee et al., 2004), and recent studies suggest that the uropod provides an area where contractile stresses are concentrated (Smith et al., 2007) and may play a more active role in cell motility by regulating integrin clustering through WASP (Zhang et al., 2006). This raises the intriguing possibility that the cell rear may be an active signaling module that regulates cell polarity and provides positive feedback to the front of the cell during directed cell migration. On the basis of our findings, we propose that the uropod component PIPKIγ and its lipid product PtdIns(4,5)P2 play an active role in establishing backness possibly via RhoA/ROCK-mediated signaling during chemotaxis. Future studies should shed light on the role of these pathways in regulating cell polarity and their contribution to positive feedback mechanisms that optimize directed cell migration.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge Jun Zhu for construction of the GFP-PIPKIγ661 fusion constructs; Tamas Balla (NIH, Bethesda, MD) for the GFP-PH-PLCδ and the GFP-PHAKT constructs; Monique Arpin (Institute Curie, France) and Richard Lamb (Institute of Cancer Research, London, United Kingdom) for the GFP-ezrin construct; Rick Horwitz (University of Virginia, Charlottesville, Virginia) for the dsRed-paxillin construct; Richard Anderson and Kun Ling (University of Wisconsin, Madison, WI) for the C-terminal PIPKIγ antibody, PIPKIγ661 cDNA; Garry Nolan (Stanford University, Stanford, CA) for Phoenix cells; Clive Svendsen (University of Wisconsin, Madison, WI) for the pMX retroviral vector; Roger Tsien (University of California at San Diego) for the pRSET-B-mCherry construct; and Kathy Schell, Joel Puchalski, and Dagna Sheerar for their expertise at the Flow Cytometry Facility (University of Wisconsin, Madison, WI). This work was supported by National Institutes of Health Grants R01 GM074827 (A.H.), American Heart Association grant-in-aid (A.H.), and National Institutes of Health Grant RO1 AI68062 (V.O.). P.N. was supported by an American Heart Association predoctoral fellowship, and M.A.S. is supported by a postdoctoral fellowship from the American Heart Association (0625751Z).
Abbreviations used:
- C5a
Complement factor 5a
- DIC
differential interference contrast microscopy
- dHL-60
differentiated HL-60
- fMLP
formyl-Met-Leu-Phe
- PMN
polymorphonuclear cells
- PI3K
phosphoinositide 3-kinase
- PtdIns(3,4,5)P3
phosphatidylinositol (3,4,5)-trisphosphate
- PtdIns(4,5)P2
phosphatidylinositol (4,5)-bisphosphate
- PIPKIγ661
Type Iγ661 phosphatidylinositol phosphate kinase
- KD
kinase-dead
- GFP
green fluorescent protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-05-0428) on October 10, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Alblas J., Ulfman L., Hordijk P., Koenderman L. Activation of Rhoa and ROCK are essential for detachment of migrating leukocytes. Mol. Biol. Cell. 2001;12:2137–2145. doi: 10.1091/mbc.12.7.2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Lebrero J. L., et al. Polarization and interaction of adhesion molecules P-selectin glycoprotein ligand 1 and intercellular adhesion molecule 3 with moesin and ezrin in myeloid cells. Blood. 2000;95:2413–2419. [PubMed] [Google Scholar]
- Bairstow S. F., Ling K., Su X., Firestone A. J., Carbonara C., Anderson R. A. Type Igamma661 phosphatidylinositol phosphate kinase directly interacts with AP2 and regulates endocytosis. J. Biol. Chem. 2006;281:20632–20642. doi: 10.1074/jbc.M601465200. [DOI] [PubMed] [Google Scholar]
- Berdeaux R. L., Diaz B., Kim L., Martin G. S. Active Rho is localized to podosomes induced by oncogenic Src and is required for their assembly and function. J. Cell Biol. 2004;166:317–323. doi: 10.1083/jcb.200312168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong L. D., Traynor-Kaplan A., Bokoch G. M., Schwartz M. A. The small GTP-binding protein Rho regulates a phosphatidylinositol 4-phosphate 5-kinase in mammalian cells. Cell. 1994;79:507–513. doi: 10.1016/0092-8674(94)90259-3. [DOI] [PubMed] [Google Scholar]
- Collins S. J., Gallo R. C., Gallagher R. E. Continuous growth and differentiation of human myeloid leukaemic cells in suspension culture. Nature. 1977;270:347–349. doi: 10.1038/270347a0. [DOI] [PubMed] [Google Scholar]
- Collins S. J., Ruscetti F. W., Gallagher R. E., Gallo R. C. Terminal differentiation of human promyelocytic leukemia cells induced by dimethyl sulfoxide and other polar compounds. Proc. Natl. Acad. Sci. USA. 1978;75:2458–2462. doi: 10.1073/pnas.75.5.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins S. J., Ruscetti F. W., Gallagher R. E., Gallo R. C. Normal functional characteristics of cultured human promyelocytic leukemia cells (HL-60) after induction of differentiation by dimethylsulfoxide. J. Exp. Med. 1979;149:969–974. doi: 10.1084/jem.149.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devreotes P., Janetopoulos C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J. Biol. Chem. 2003;278:20445–20448. doi: 10.1074/jbc.R300010200. [DOI] [PubMed] [Google Scholar]
- Di Paolo G., Pellegrini L., Letinic K., Cestra G., Zoncu R., Voronov S., Chang S., Guo J., Wenk M. R., De Camilli P. Recruitment and regulation of phosphatidylinositol phosphate kinase type 1γ by the FERM domain of talin. Nature. 2002;420:85–89. doi: 10.1038/nature01147. [DOI] [PubMed] [Google Scholar]
- Doughman R. L., Firestone A. J., Anderson R. A. Phosphatidylinositol phosphate kinase puts PI4,5P2 in its place. J. Membrane Biol. 2003;194:77–89. doi: 10.1007/s00232-003-2027-7. [DOI] [PubMed] [Google Scholar]
- Eddy R. J., Pierini L. M., Matsumura F., Maxfield F. R. Ca2+-dependent myosin II activation is required for uropod retraction during neutrophil migration. J. Cell Sci. 2000;113:1287–1298. doi: 10.1242/jcs.113.7.1287. [DOI] [PubMed] [Google Scholar]
- Fievet B. T., Gautreau A., Roy C., Del Maestro L., Mangeat P., Louvard D., Arpin M. Phosphoiniositide binding and phosphorylation act sequentially in the activation mechanism of ezrin. J. Cell Biol. 2004;164:653–659. doi: 10.1083/jcb.200307032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco S. J., Rodgers M. A., Perrin B. J., Han J., Bennin D. A., Critchley D. R., Huttenlocher A. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat. Cell Biol. 2004;6:977–983. doi: 10.1038/ncb1175. [DOI] [PubMed] [Google Scholar]
- Gallagher R., et al. Characterization of the continuous, differentiating myeloid cell line (HL-60) from a patient with acute promyelocytic leukemia. Blood. 1979;54:713–733. [PubMed] [Google Scholar]
- Gomez-Mouton C., Abad J. L., Mira E., Lacalle R. A., Gallardo E., Jimenez-Baranda S., Illa I., Bernad A., Manes S., Martinez A. C. Segregation of leading-edge and uropod components into specific lipid rafts during T cell polarization. Proc. Natl. Acad. Sci. USA. 2001;98:9642–9647. doi: 10.1073/pnas.171160298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Mouton C., Lacalle R. A., Mira E., Jimenez-Baranda S., Barber D. F., Carrera A. C., Martinez A. C., Manes S. Dynamic redistribution of raft domains as an organizing platform for signaling during cell chemotaxis. J. Cell Biol. 2004;164:759–768. doi: 10.1083/jcb.200309101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauert A. B., Martinelli S., Marone C., Niggli V. Differentiated HL-60 cells are a valid model system for the analysis of human neutrophil migration and chemotaxis. Int. J. Biochem. Cell Biol. 2002;34:838–854. doi: 10.1016/s1357-2725(02)00010-9. [DOI] [PubMed] [Google Scholar]
- Kimura K., et al. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- Kunisaki Y., et al. DOCK2 is a Rac activator that regulates motility and polarity during neutrophil chemotaxis. J. Cell Biol. 2006;174:647–652. doi: 10.1083/jcb.200602142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R. F., Ozanne B. W., Roy C., McGarry L., Stipp C., Mangeat P., Jay D. G. Essential functions of ezrin in maintenance of cell shape and lamellipodial extension in normal and transformed fibroblasts. Curr. Biol. 1997;7:682–688. doi: 10.1016/s0960-9822(06)00295-8. [DOI] [PubMed] [Google Scholar]
- Lawson M. A., Maxfield F. R. Ca(2+)- and calcineurin-dependent recycling of an integrin to the front of migrating neutrophils. Nature. 1995;377:75–79. doi: 10.1038/377075a0. [DOI] [PubMed] [Google Scholar]
- Lee J. H., Katakai T., Hara T., Gonda H., Sugai M., Shimizu A. Roles of p-ERM and Rho-ROCK signaling in lymphocyte polarity and uropod formation. J. Cell Biol. 2004;167:327–337. doi: 10.1083/jcb.200403091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling K., Doughman R. L., Firestone A. J., Bunce M. W., Anderson R. A. Type I gamma phosphatidylinositol phosphate kinase targets and regulates focal adhesions. Nature. 2002;420:89–93. doi: 10.1038/nature01082. [DOI] [PubMed] [Google Scholar]
- Ling K., Schill N. J., Wagoner M. P., Sun Y., Anderson R. A. Movin' on up: the role of PtdIns(4,5)P(2) in cell migration. Trends Cell Biol. 2006;16:276–284. doi: 10.1016/j.tcb.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Lokuta M. A., Nuzzi P. A., Huttenlocher A. Calpain regulates neutrophil chemotaxis. Proc. Natl. Acad. Sci. USA. 2003;100:4006–4011. doi: 10.1073/pnas.0636533100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meili R., Ellsworth C., Lee S., Reddy T. B., Ma H., Firtel R. A. Chemoattractant-mediated transient activation and membrane localization of Akt/PKB is required for efficient chemotaxis to cAMP in Dictyostelium. EMBO J. 1999;18:2092–2105. doi: 10.1093/emboj/18.8.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggli V. Rho-kinase in human neutrophils: a role in signalling for myosin light chain phosphorylation and cell migration. FEBS Lett. 1999;445:69–72. doi: 10.1016/s0014-5793(99)00098-8. [DOI] [PubMed] [Google Scholar]
- Niggli V. Signaling to migration in neutrophils: importance of localized pathways. Int. J. Biochem. Cell Biol. 2003;35:1619–1638. doi: 10.1016/s1357-2725(03)00144-4. [DOI] [PubMed] [Google Scholar]
- Nuzzi P. A., Lokuta M. A., Huttenlocher A. Analysis of neutrophil chemotaxis. Methods Mol. Biol. 2007a;370:23–36. doi: 10.1007/978-1-59745-353-0_3. [DOI] [PubMed] [Google Scholar]
- Nuzzi P. A., Senetar M. A., Huttenlocher A. Asymmetric localization of Calpain 2 during neutrophil chemotaxis. Mol. Biol. Cell. 2007b;18:795–805. doi: 10.1091/mbc.E06-09-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent C. A. Making all the right moves: chemotaxis in neutrophils and Dictyostelium. Curr. Opin. Cell Biol. 2004;16:4–13. doi: 10.1016/j.ceb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Ren X. D., Bokoch G. M., Traynor-Kaplan A., Jenkins G. H., Anderson R. A., Schwartz M. A. Physical association of the small GTPase Rho with a 68-kDa phosphatidylinositol 4-phosphate 5-kinase in Swiss 3T3 cells. Mol. Biol. Cell. 1996;7:435–442. doi: 10.1091/mbc.7.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G., Weiner O. D., Herzmark P., Balla T., Sedat J. W., Bourne H. R. Polarization of chemoattractant receptor signaling during neutrophil chemotaxis. Science. 2000;287:1037–1040. doi: 10.1126/science.287.5455.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant G., Weiner O. D., Neptune E. R., Sedat J. W., Bourne H. R. Dynamics of a chemoattractant receptor in living neutrophils during chemotaxis. Mol. Biol. Cell. 1999;10:1163–1178. doi: 10.1091/mbc.10.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. A., Aranda-Espinoza H., Haun J. B., Dembo M., Hammer D. A. Neutrophil traction stresses are concentrated in the uropod during migration. Biophys. J. 2007;92:L58–L60. doi: 10.1529/biophysj.106.102822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somlyo A. V., Bradshaw D., Ramos S., Murphy C., Myers C. E., Somlyo A. P. Rho-kinase inhibitor retards migration and in vivo dissemination of human prostate cancer cells. Biochem. Biophys. Res. Commun. 2000;269:652–659. doi: 10.1006/bbrc.2000.2343. [DOI] [PubMed] [Google Scholar]
- Stauffer J. P., Meyer J. M., Nally J. N. Accuracy of six elastic impression materials used for complete-arch fixed partial dentures. J. Prosthet. Dent. 1976;35:407–415. doi: 10.1016/0022-3913(76)90008-1. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A., Wong K., Knight Z. A., Govaerts C., Hahn K. M., Shokat K. M., Bourne H. R. To stabilize neutrophil polarity, PIP3 and Cdc42 augment RhoA activity at the back as well as signals at the front. J. Cell Biol. 2006;174:437–445. doi: 10.1083/jcb.200604113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varnai P., Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J. Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weernink P. A., Meletiadis K., Hommeltenberg S., Hinz M., Ishihara H., Schmidt M., Jakobs K. H. Activation of type I phosphatidylinositol 4-phosphate 5-kinase isoforms by the Rho GTPases, RhoA, Rac1, and Cdc42. J. Biol. Chem. 2004;279:7840–7849. doi: 10.1074/jbc.M312737200. [DOI] [PubMed] [Google Scholar]
- Weiner O. D., Neilsen P. O., Prestwich G. D., Kirschner M. W., Cantley L. C., Bourne H. R. A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity. Nat. Cell Biol. 2002;4:509–513. doi: 10.1038/ncb811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner O. D., Servant G., Welch M. D., Mitchison T. J., Sedat J. W., Bourne H. R. Spatial control of actin polymerization during neutrophil chemotaxis. Nat. Cell Biol. 1999;1:75–81. doi: 10.1038/10042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K., Pertz O., Hahn K., Bourne H. Neutrophil polarization: spatiotemporal dynamics of RhoA activity support a self-organizing mechanism. Proc. Natl. Acad. Sci. USA. 2006;103:3639–3644. doi: 10.1073/pnas.0600092103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthylake R. A., Lemoine S., Watson J. M., Burridge K. RhoA is required for monocyte tail retraction during transendothelial migration. J. Cell Biol. 2001;154:147–160. doi: 10.1083/jcb.200103048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Wang F., Van Keymeulen A., Herzmark P., Straight A., Kelly K., Takuwa Y., Sugimoto N., Mitchison T., Bourne H. R. Divergent signals and cytoskeletal assemblies regulate self-organizing polarity in neutrophils. Cell. 2003;114:201–214. doi: 10.1016/s0092-8674(03)00555-5. [DOI] [PubMed] [Google Scholar]
- Yoshinaga-Ohara N., Takahashi A., Uchiyama T., Sasada M. Spatiotemporal regulation of moesin phosphorylation and rear release by Rho and serine/threonine phosphatase during neutrophil migration. Exp. Cell Res. 2002;278:112–122. doi: 10.1006/excr.2002.5571. [DOI] [PubMed] [Google Scholar]
- Zhang H., Schaff U. Y., Green C. E., Chen H., Sarantos M. R., Hu Y., Wara D., Simon S. I., Lowell C. A. Impaired integrin-dependent function in Wiskott-Aldrich syndrome protein-deficient murine and human neutrophils. Immunity. 2006;25:285–295. doi: 10.1016/j.immuni.2006.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Loijens J. C., Boronenkov I. V., Parker G. J., Norris F. A., Chen J., Thum O., Prestwich G. D., Majerus P. W., Anderson R. A. Phosphatidylinositol-4-phosphate 5-kinase isozymes catalyze the synthesis of 3-phosphate-containing phosphatidylinositol signaling molecules. J. Biol. Chem. 1997;272:17756–17761. doi: 10.1074/jbc.272.28.17756. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.