Abstract
A novel gene, prom-1, was isolated in a screen for Caenorhabditis elegans mutants with increased apoptosis in the germline. prom-1 encodes an F-box protein with limited homology to the putative human tumor suppressor FBXO47. Mutations in the prom-1 locus cause a strong reduction in bivalent formation, which results in increased embryonic lethality and a Him phenotype. Furthermore, retarded and asynchronous nuclear reorganization as well as reduced homologous synapsis occur during meiotic prophase. Accumulation of recombination protein RAD-51 in meiotic nuclei suggests disturbed repair of double-stranded DNA breaks. Nuclei in prom-1 mutant gonads timely complete mitotic proliferation and premeiotic replication, but they undergo prolonged delay upon meiotic entry. We, therefore, propose that prom-1 regulates the timely progression through meiotic prophase I and that in its absence the recognition of homologous chromosomes is strongly impaired.
INTRODUCTION
Meiosis is the specialized nuclear division in which the diploid chromosomal complement is reduced to a haploid gametic chromosome set to compensate for the duplication of chromosome number that occurs at fertilization. During meiosis, pairs of homologous chromosomes (one from each parent) separate in a first round of nuclear division (meiosis I), and each chromosome splits into its two sister chromatids during a second round of nuclear division (meiosis II). Before homologous chromosomes disjoin at meiosis I, they reciprocally exchange corresponding portions, which results in each chromosome becoming a mosaic consisting of parts of the original parental chromosomes. This process, which is called crossing over, contributes to the genetic diversity of the four meiotic products, and as a consequence, to the variability of offspring. To allow for the orderly segregation of homologues at meiosis I and crossing over to occur, chromosomes have to pair during meiotic prophase.
Meiotic pairing is a multistep process during which homologous chromosomes first recognize each other, then align, and finally engage in an intimate association along their entire lengths. The final, close apposition is mediated by a ribbon-like proteinaceous structure, referred to as the synaptonemal complex (SC). Crossing over, which takes place within the framework of the SC, produces chiasmata. Together with the cohesion of sister chromatids, chiasmata connect homologues after the disassembly of the SC, allowing them to persist as bivalents until their separation in the first anaphase of meiosis (for review, see Petronczki et al., 2003).
The establishment of bivalents needs the accurate coordination of all the events that are taking place during prophase I. Many aspects of the meiotic cell cycle are regulated by the interplay between cyclin-dependent kinase activity and ubiquitin-mediated proteolysis of inhibitory factors similar to the mitotic cell cycle (Harper et al., 2002; Henderson et al., 2006). To date, little is known about the orchestration and cell cycle dependence of meiotic events of prophase I such as chromatin condensation, homologue recognition, axis formation, and SC formation.
In C. elegans, various defects in meiotic pairing and recombination lead to the elimination of a major portion of affected cells by germ cell apoptosis (Gartner et al., 2000; Bhalla and Dernburg, 2005). Here, we describe a novel gene, progression of meiosis (prom-1), which was originally identified in a screen for increased apoptosis in the germline and which plays a role in the orderly progression of meiotic prophase I and in full homologous chromosome pairing and recombination.
MATERIALS AND METHODS
Worm Strains and Culture Conditions
The wild-type N2 Bristol, the Hawaiian strain CB4856, and strain DC1079 [ces-1(n703) qDf8/hT2[bli-4(e937) let-?(q728) qls48] (I;III)] were obtained from the Caenorhabditis Genetics Center (University of Minnesota, St. Paul, MN). The prom-1 deletion allele ok1140 (strain RB1183) was generated by the C. elegans Gene Knockout Consortium. AV106 spo-11(ok79) (Dernburg et al., 1998) and AV393 htp-1(gk174) (Martinez-Perez and Villeneuve, 2005) were kindly provided by Anne M. Villeneuve. Worms were grown on NGM plates with Escherichia coli OP50 (Brenner, 1974). To create prom-1/qDf8 heterozygotes, prom-1(ok1140) males were mated to DC1079 hermaphrodites. F1 progeny, which do not carry the myo-2::GFP, were selected.
Screen for Mutants with Increased Meiotic Cell Death
A synchronized population of wild-type worms at the L4 larval stage was mutagenized with ethyl methanesulfonate (EMS) under standard conditions, and it was subsequently allowed to grow to adulthood. These adults were bleached and the F2 generation was stained with acridine orange (Gartner et al., 2004), and live animals were screened under a standard fluorescence dissection microscope for increased apoptosis.
Candidate mutants were outcrossed four to five times and tested for the viability of offspring and for a high incidence of males (Him) among the progeny. Mapping of the mutant alleles was performed with single-nucleotide polymorphism (SNP) markers as described in Wicks et al. (2001). The mutation was mapped to an interval between an SNP on cosmid F14B4 (position 13524) and an SNP on M04C9 (position 11470), and the remaining candidate open reading frames were sequenced.
Protein Depletion by Double-stranded RNA (dsRNA) Interference (RNAi)
F54D5.9p and CUL-1 proteins were depleted by double-stranded RNAi by feeding hermaphrodites with dsRNA-producing bacteria (Timmons et al., 2001). Target sequences were polymerase chain reaction (PCR)-amplified from C. elegans genomic DNA and cloned into feeding vector pPD129.36. The following primers were used: MJ959 5′-ctccttggaggatttgcgag-3′ and MJ960 5′-catttggaagctggaagcct-3′ for F54D9.5, and MJ901 5′-atgacaaatagcgaaccgag-3′ and MJ902 5′-acaatcttcgtctgcaactc-3′ for cul-1 (D2045.6).
Measurement of Genetic Recombination in the Oocytes
To assay crossing over in nonviable prom-1 mutant progeny, we tested recombination between loci polymorphic for SNP markers as in Hillers and Villeneuve (2003). First, a strain homozygous for prom-1Δ and heterozygous for two SNP markers on chromosome V was created by crossing the Bristol prom-1Δ strain to a prom-1Δ strain carrying Hawaiian chromosome V. The markers polymorphic for DraI restriction sites were at position base pairs 7973 on cosmid F36H9 (primers 5′-CGGAAAATTGCGACTGTC-3′ and 5′-ATTAGGACTGCTTGGCTTCC-3′) and at position base pairs 92865 on cosmid Y51A2A (primers 5′-TTTATCCGCAGGGACTTGAC-3′and 5′-TCTCCTCTCCCTCATGGTTAAC-3′). Hermaphrodites were individually mated to Bristol males carrying a green fluorescent protein (GFP) transgene. The eggs laid by these individuals were collected at ∼4-h intervals to avoid decomposition of dead eggs, and they were individually lysed. PCR was performed on the lysates and restriction digests were performed. Products were run on 2% agarose gels to detect restriction patterns diagnostic of recombination between the SNP markers. Only eggs positive for GFP or PROM-1 (contributed by the males) were evaluated.
In Situ Detection of DNA Replication by Incorporation of dUTP
Gonads of immobilized animals were injected with a solution of 300 pM Cy3-labeled dUTP (GE Healthcare, Chalfont St. Giles, United Kingdom) in phosphate buffer, pH 7.0 (Mello et al., 1991). Worms were allowed to recover in M9 buffer (0.3% KH2PO4, 0.6% Na2HPO4, 0.5% NaCl, and 1 mM MgSO4) for 30 min, and then they were transferred to a fresh plate. Gonads were dissected and 4,6-diamidino-2-phenylindole (DAPI)-stained (see below) after ∼1.5 h of exposure to the nucleotides. The incorporation of labeled nucleotides into nuclear DNA was directly observed by fluorescence microscopy.
Cytological Preparation of Gonads
Hermaphrodites were cut open to release the gonads in 5 μl of M9 buffer on a microscope slide and fixed by the addition of an equal volume of 7.4% formaldehyde. The material was immediately covered with a coverslip, and gentle pressure was applied. The coverslip was removed after freezing the preparations in liquid nitrogen and the slides were then transferred to 1× phosphate-buffered saline (PBS). For the staining of chromatin and chromosomes, slides were mounted in Vectashield anti-fading medium (Vector Laboratories, Burlingame, CA) containing 2 μg/ml DAPI.
Immunostaining
For immunostaining, gonads prepared in M9 buffer on a microscope slide were fixed in a series of methanol, methanol:acetone (1:1), and acetone for 5 min each at −20°C, and then they were immediately transferred to 1× PBS without drying. Preparations were transferred to fresh 1× PBS twice for 5 min each and were blocked with 3% bovine serum albumin (BSA) in 1× PBS for 30 min at 37°C in a humidity chamber. The primary antibody was applied and the specimen was incubated overnight at 4°C in a humidity chamber. Antibodies were diluted in 1× PBS containing 3% BSA as follows: 1:100 anti-REC-8 (Pasierbek et al., 2001), 1:100 anti-RAD-51 (Colaiácovo et al., 2003), 1:50 anti-SYP-1 (MacQueen et al., 2002), 1:100 anti-HIM-3 (Zetka et al., 1999), and 1:500 anti HIM-8 (Phillips et al., 2005). After washing three times in 1× PBS plus 0.1% Tween 20, secondary antibodies were applied at the following dilutions: anti-rabbit Cy3 (1:250), anti-rabbit fluorescein isothiocyanate (FITC) (1:500) or anti-rat FITC (1:500). After 60-min incubation at room temperature, slides were washed and mounted in Vectashield supplemented with DAPI (see above). Fixation and staining for phosphorylated histone H3 was performed as described previously (Jones et al., 1996).
Fluorescence In Situ Hybridization (FISH)
Pooled cosmids C53D5 and R119, and PCR-amplified 5S rDNA, were used as probes for the left end of chromosome I and the right arm of chromosome V, respectively (Pasierbek et al., 2001). Cosmid DNA was directly labeled with Cy3 by using the BioNick Labeling System (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. The 5S rDNA was labeled by PCR with digoxigenin-11-dUTP during PCR amplification.
Slides prepared according to the above-mentioned protocol were dehydrated by incubation in increasing ethanol concentrations, dried on air, and kept in the freezer until use for FISH. Labeled probe DNA and cytological preparations were denatured separately before the probe was added to the slides for hybridization. The protocol is described in detail in Pasierbek et al. (2001). Hybridized digoxigenin-labeled probes were detected with FITC-conjugated anti-digoxigenin, and FITC (green) and Cy3 (red) fluorescence was emitted by excitation with light of appropriate wavelengths. Slides were mounted in Vectashield containing 2 μg/ml DAPI.
Microscopy and Evaluation
Preparations were examined with a Zeiss Axioskop epifluorescence microscope. Images were recorded with a cooled charge-coupled device camera (Photometrics, Tucson, AZ). Evaluation of cytological phenotypes was performed in animals kept at 20°C 16–24 h after larval stage 4 (L4). For multicolor immunostaining and FISH pictures, monochrome images were captured separately for each emission wavelength. Artificial coloring and merging were undertaken using IPLab Spectrum software (Scanalytics, Fairfax, VA). In some cases, three-dimensional stacks of images were taken (MetaVue software; Molecular Devices, Sunnyvale, CA), deconvolved (AutoDeblur software; AutoQuant Imaging, Troy, NY), and projected (Helicon Focus software; http://helicon.com.ua/heliconfocus/).
RESULTS
Isolation and Characterization of prom-1 Mutant Alleles
In animals, defects in meiotic recombination and/or meiotic pairing can lead to the elimination of affected cells by apoptotic cell death (Gartner et al., 2000). We exploited this phenotype for the screening for C. elegans meiotic mutants, taking advantage of the preferential uptake of the dye acridine orange by apoptotic corpses (Gartner et al., 2004;Schumacher et al., 2005). Two alleles, op240 and jf3, of a novel gene (prom-1) were identified. In addition to increased apoptosis, these mutants exhibited high embryonic lethality and a high incidence of males (Him) phenotype (see below). After backcrossing the mutant animals, the corresponding gene (prom-1) was mapped to open reading frame F26H9.1 on chromosome I by using morphological and SNP markers. Sequencing of these alleles revealed a D242 to N missense mutation in op240 and a Q363 to premature stop codon mutation in jf3 (see Supplemental Figures S1 and S2 for cDNA sequence, intron–exon boundaries, and mutation sites). A third allele, ok1140, was provided by the C. elegans Gene Knockout Consortium. Complementation testing confirmed that jf3, op240, and ok1140 are indeed allelic (data not shown). ok1140 is a deletion of 1229 base pairs encompassing the first five and a half exons and 63 base pairs of the promoter region. Because prom-1(ok1140) is the most severely mutated allele, most quantitative mutant analyses were performed using progeny of homozygous backcrossed ok1140 (hereafter prom-1Δ) mothers, unless indicated otherwise. Similar phenotypes, and in particular strong phenotypic variability between individuals (see below) were observed in the other alleles as well (data not shown).
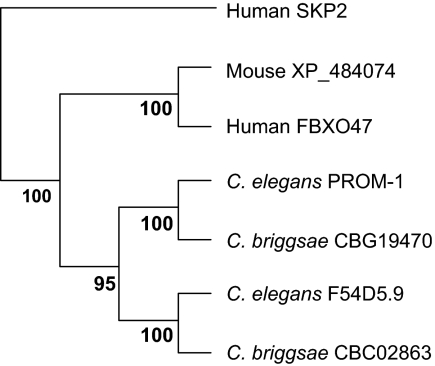
The F26H9.1 transcript shows a three- to fourfold germline enrichment relative to somatic tissues (Reinke et al., 2000). It encodes a 524-amino acid (aa) protein with an F-box motif (Kipreos and Pagano, 2000; Supplemental Figure S1). A C. elegans paralogue, F54D5.9, with an amino acid identity of 22% and a similarity of 39% is listed in WormBase (www.wormbase.org). F54D5.9 and, to a lesser degree, F26H9.1 show homology to the human F-box protein 47 (FBXO47), which is a putative tumor suppressor (Simon-Kayser et al., 2005) and to FBXO47-related proteins in various vertebrates. Although there seems to exist only one FBXO47-like protein in all vertebrates examined, this locus seems to be duplicated in nematodes (Figure 1). Intriguingly, we were unable to find homologues of FBXO47 in Drosophila, plants, or yeast. An RNAi phenotype was not detected for F54D5.9 byKamath et al. (2003) and Rual et al. (2004), whereas Piano et al. (2002) reported 10–29% embryonic lethality. We found RNAi of F54D5.9 to result in embryos with hyperproliferating tissues, whereas meiosis was not notably affected (data not shown).
Figure 1.
Phylogenetic tree of C. elegans PROM-1 and related proteins. The human F-box protein SKP2 is displayed as a member of the outgoup. The phylogenetic tree was derived from a multiple protein sequence alignment by the ClustalX program (Thompson et al., 1997). The resulting tree was tested by resampling (bootstrapping) by using the programs sequboot, protpars, and consense from the PHYLIP package (Felsenstein, 1989), executed on a server of the Department of Molecular Evolution, Uppsala University (http://artedi.ebc.uu.SE/programs/). The numbers at the forks indicate in how many trees (out of 100) the grouping to the right of the fork occurred.
Mutation of prom-1 Causes Reduced Fertility and Increased Production of Males
Brood size was strongly reduced in the mutants, and it was highly variable from animal to animal, with all three backcrossed alleles. prom-1Δ homozygous hermaphrodites laid 143 ± 70 (SD) embryos (ranging from 44 to 256; n = 17 animals) compared with an average brood size of 282 ± 22 (n = 8) of the wild type. prom-1Δ worms also displayed high embryonic lethality. The average viability was 10.2% (70 of 688 laid eggs developed further than to the L1 stage) compared with 99.8% in the wild type (n = 2257 eggs), but it was highly variable between individual hermaphrodites, ranging from 1.4 to 54.2%. Of the surviving mutant offspring, 22% (72 of 328 L3–adult worms in a different experiment) were males. Most of the viable progeny were vigorous and morphologically normal, suggesting that embryonic lethality was due to chromosomal defects in the wake of disturbed meiosis (see below), and that animals receiving a balanced, intact chromosome set were not affected. Deletion of prom-1, therefore, seems to have only a mild, if any, somatic phenotype.
To test whether both female and male meioses are affected and whether the combined defects contribute to the lethality of hermaphrodite progeny, we crossed prom-1Δ hermaphrodites to wild-type (WT) males and fog-2(oz40) female-only worms to prom-1Δ males. Progeny from prom-1Δ hermaphrodites × WT males showed 33% viability (1610 eggs scored). Progeny from fog-2 females × prom-1Δ males showed 28% viability (1309 eggs scored), which is not significantly different (t test). (fog-2 females × WT males produced 99% viable offspring; n = 1622 eggs.) Thus, the involvement of either prom-1 mutant eggs or sperm in zygote formation considerably reduces viability, suggesting that formation of both types of gametes is compromised in prom-1Δ hermaphrodites.
Apoptosis Is Increased in the prom-1Δ Mutant
We scored for apoptotic nuclei by differential interference contrast microscopy in animals 16–24 h past L4. In accordance with the apoptosis phenotype by which prom-1 was initially identified, up to 21 (mean ± SD, 11.6 ± 4.7; n = 30) nuclei in the ovary of the prom-1Δ mutant, at any given time, show morphological features indicative of apoptosis, compared with a maximum of three nuclei (1.4 ± 0.6; n = 23) in the wild type. We tested whether impaired repair of meiotic double-strand breaks (DSBs) contributed to increased germ cell apoptosis by scoring for apoptotic nuclei in a prom-1 spo-11 double mutant. We found that in the absence of SPO-11 (which catalyzes meiotic DSB formation; Dernburg et al., 1998), on average, only 1.9 nuclei ± 1.9 (n = 19) per gonad were apoptotic. Thus, the apoptosis phenotype was bypassed in the absence of DSBs, indicating that incomplete or delayed repair of meiotic DSBs, which is a possible consequence of delayed meiotic progression (see below) may trigger DNA damage checkpoint-dependent apoptosis in a prom-1Δ mutant. Moreover, we found that apoptosis was reduced to almost wild-type levels in a prom-1 cep-1 double mutant (2.7 ± 2.2 apoptotic nuclei per gonad, n = 25). The dependency of apoptosis on CEP-1 (the p53 homologue of the worm) also suggests that it is not due to a recently discovered checkpoint that monitors meiotic chromosome synapsis (Bhalla and Dernburg, 2005). It is conceivable that nonhomologous synapsis (which occurs in the prom-1 mutant; see below) is sufficient to escape this checkpoint (Penkner et al., 2007).
Mutation of prom-1 Causes a Transient Arrest of Nuclei on Meiotic Entry
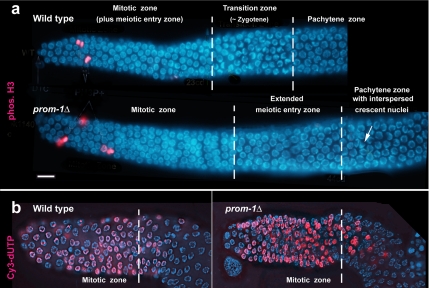
Cytological inspection of mutant gonads revealed that in addition to elevated apoptosis, the normal progression of meiotic stages was perturbed (Figure 2a). In wild-type ovaries, the distal region of the gonad where germ cells are produced by a series of mitoses, is characterized by spherical DAPI-stained nuclei. It is commonly referred to as the mitotic zone, although its most proximal nuclei start to express meiotic genes (MacQueen et al., 2002; Hansen et al., 2004; Crittenden et al., 2006; see below) and hence satisfy the classical cytological definition of leptotene. This “meiotic entry zone” (marking the switch from proliferation to meiotic determination (Hansen et al., 2004) possesses somewhat larger nuclei than the genuine mitotic zone. It is followed by several rows of cells that comprise the transition zone, where leptotene nuclei become replaced by nuclei with chromatin arranged in a crescent, which correspond to early zygotene. The region next to the transition zone is occupied by late zygotene and pachytene nuclei, followed by cells in diplotene and diakinesis (Schedl, 1997).
Figure 2.
Organization of gonads in the wild type and in prom-1Δ (ok1140). (a) In the wild type, the mitotic zone of the germline (with mitotic nuclei immunostainined for phosphorylated histone H3 depicted in red) and a narrow meiotic entry zone (see text) is followed by the transition zone containing crescent-shaped nuclei and then by the pachytene zone. In the prom-1Δ mutant, the mitotic zone is followed by an extended meiotic entry zone with large nuclei that are not crescent shaped. The pachytene zone contains some crescent-shaped nuclei (arrow). The vertical broken lines indicate the boundaries between different regions, defined by DAPI staining (see text). (b) Incorporation of Cy3-labeled dUTP (red) is limited to the mitotic zone in wild-type and mutant gonads. Chromatin is stained blue with DAPI. Bar, 10 μm.
In prom-1 mutants, the zone with small DAPI-bright nuclei, resembling the wild-type mitotic zone, extended for 19.69 ± 4.40 (n = 56) nuclear diameters from the distal tip cell (DTC), which is comparable with the 20.75 ± 1.62 (n = 56) nuclear diameters from the DTC in the wild type. Within this mitotic zone, mitoses were identified by immunostaining for phosphorylated histone H3 (Figure 2a). It was followed by a zone with slightly larger and less bright nuclei, ∼16 nuclear diameters in width (Figure 2a), which was missing in the wild type. This zone did not contain mitoses (Supplemental Figure S3). To determine whether DNA replication was ongoing in these putative postmitotic cells, we monitored DNA synthesis by the incorporation of Cy3-labeled dUTP. Young adult hermaphrodites were injected with high concentrations of Cy3-labeled dUTP, and incorporation was scored in fixed DAPI-stained gonads. After 1.5-h exposure to Cy3-labeled dUTP, it was found that labeled nucleotides had been incorporated for the same number of distal cell rows both in the wild type (27.2 ± 2.1; n = 13) and in the mutant (27.5 ± 3.4; n = 13) (Figure 2b). Further down in the gonad, only stray cells with some Cy3 label were detected. This indicates that DNA synthesis takes place mostly within the first ∼27 cell rows of the wild type and the mutant, and we assume that it corresponds to S phase in proliferative cells (∼20 cell rows) plus a few adjacent cell rows where cells undergo meiotic DNA replication (compare Crittenden et al., 2006). This result suggests that the mitotic zone is normal and that meiotic S phase is essentially complete in the additional cell rows that follow this zone in the mutant.
A distinct transition zone was missing in the mutant. Instead, sporadic nuclei with chromatin arranged in a crescent were scattered over the proximal area (the pachytene zone) of the gonad (Figure 2a). (For simplicity, the area of the prom-1Δ mutant gonad that corresponds to the pachytene zone of the wild type will be also called pachytene zone throughout the article, despite the fact that chromosome pairing may be incomplete; see below.) The occurrence of such transition zone-like nuclei in the pachytene zone of the gonad may reflect delayed and asynchronous initiation of homologous chromosome pairing in the prom-1Δ mutant (see below).
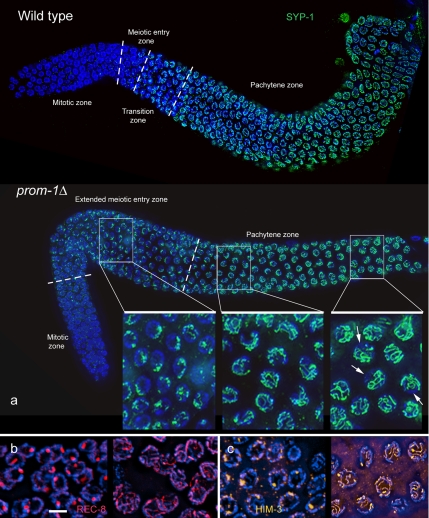
To further determine the nature of the cells following the zone where mitoses and DNA replication take place, we studied the expression of SC proteins. In the wild type, SCs form between pairs of homologous chromosomes along their entire length, and they can be visualized as thread-like structures corresponding to the haploid chromosome number when immunostained for SC components or associated proteins such as SYP-1 (MacQueen et al., 2002), HIM-3 (Zetka et al., 1999), and REC-8 (Pasierbek et al., 2001) (Figure 3; Supplemental Figure S4). In the prom-1Δ mutant, SYP-1, a protein that connects the synapsed partners; HIM-3, an axial element component; and the cohesin subunit REC-8 were present in a few patches inside nuclei proximal to the mitotic zone. This observation confirms our interpretation that these nuclei have entered the meiotic program and hence they represent an extended meiotic entry zone (Figure 3, b and c), and it suggests that PROM-1 functions very early in meiotic prophase.
Figure 3.
(a) Immunostaining of SC component SYP-1 (green) in the wild type and in the prom-1Δ mutant. In the images of whole ovaries, the broken lines mark the borders between the zones with distinguishable nuclear morphologies (see Figure 2a), namely, the mitotic zone, the meiotic entry zone and the pachytene zone. In the wild type, small spots of SYP-1 first occur in a narrow meiotic entry zone just upstream of the transition zone, and then they develop into continuous lines along the pachytene zone. In the prom-1Δ mutant, the meiotic entry zone with SYP-1 spots is greatly expanded; SYP-1 starts to form lines in the distal pachytene zone. Insets are enlarged details of the regions indicated. By the end of the pachytene zone, SC formation in some nuclei is quite extensive, as is evident from the formation of long SYP-1 lines, but incomplete (arrows denote DAPI-positive chromatin regions devoid of SYP-1). (For blow up images of corresponding regions of the wild-type gonad, see Supplemental Figure S4.) (b) REC-8 in the extended meiotic entry zone (left) and the pachytene zone (right) of the prom-1Δ mutant showing increasing formation of linear structures. (c) HIM-3 in the extended meiotic entry zone (left) where it only forms spots, and the pachytene zone (right) where in some nuclei thick HIM-3 lines suggest the presence of synapsed regions in the prom-1Δ mutant. Bar, 5 μm (b, c, and insets in a are shown at the same magnification).
Reduced Levels of Bivalent Formation and Crossing Over in the prom-1Δ Mutant
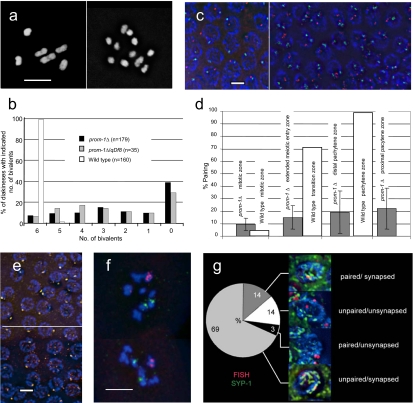
In the pachytene zone of the prom-1Δ mutant, all three SC-associated proteins progressively developed into long lines (Figure 3, b and c), indicating the occurrence of rather extensive although incomplete synapsis. Consistent with partial synapsis, we found a high level of univalents at diakinesis of mutant hermaphrodites. Whereas six bivalents are present in the wild type, in the prom-1Δ animals a range of six to 12 DAPI-positive entities were detected (Figure 4, a and b). Of the 1074 homologue pairs in the 179 diakineses scored, 367 formed bivalents (i.e., the frequency of bivalents was 34%), amounting to an average of approximately two bivalents per meiosis.
Figure 4.
Reduced homologous pairing and bivalent formation in the prom-1Δ mutant. (a) A variable number of univalents is formed in the prom-1Δ mutant. Although six bivalents are formed in diakinesis in the wild type (left), univalents are abundant in the mutant. In this particular prom-1Δ mutant cell (right), 10 univalents and one bivalent are present (DAPI staining). (b) Bivalent frequencies in prom-1Δ mutant, prom-1Δ/qDf8 heterozygous, and wild-type diakineses. Shown is the percentage of diakinesis nuclei with the number of bivalents indicated. n = number of diakineses scored. (c and d) Homologous pairing was tested by FISH of a locus near the left end of chromosome I (Cy3, red) and of the 5S rDNA region on the right arm of chromosome V (FITC, green). (c) In the pachytene zone of the wild type (left), a single signal or a closely associated pair of signals is mostly observed for both loci. In the pachytene zone of the prom-1Δ mutant (right), homologous loci on autosomes remain mostly unpaired. (d) Homologous pairing frequencies of the rDNA locus in different zones of the wild-type and prom-1Δ gonads as indicated by the association of FISH signals. The percentages of nuclei with paired loci (represented by a single dot or two dots touching each other) are shown. The raw data for this figure are given in Supplemental Table S6. (e) Immunostaining of the X chromosome-associated protein HIM-8 (orange) produces mostly a single signal in wild-type (top) and in prom-1Δ mutant (bottom) pachytenes. (f) Examples of prom-1Δ mutant diakinesis nuclei with X chromosomes (red, FISH signal) and chromosomes V (green, FISH signal). The X chromosomes are more often involved in bivalent formation (as indicated by signals sharing a DAPI-positive entity) than the autosomes (see text). (g) Frequencies of homologous pairing and synapsis of the 5S rDNA locus determined by simultaneous FISH and SYP-1 staining. This experiment shows that most of the time synapsis is nonhomologous. Bars, 5 μm.
The number of bivalents was highly variable. Although 39% of diakinesis cells had no bivalents, a complete set of six bivalents was formed in 7% of cells (Figure 4b). To exclude the possibility that the product of the close paralogue F54D5.9 could partially substitute PROM-1, we performed RNAi for F54D5.9 in the prom-1Δ mutant. Depletion of the F54D5.9 protein caused >90% embryonic lethality, but it did not further reduce the frequency of bivalents (data not shown), indicating that the two genes do not function redundantly in meiotic progression. Because reverse transcription-PCR showed a transcript of the remaining open reading frame in the prom-1(ok1140) mutant, there exists the possibility that the mutant expresses low levels of a severely truncated product which could exert some residual function and perhaps cause variable expression of the mutant phenotype. We, therefore, studied the consequence of halving its dosage in a prom-1(ok1140) prom-1(qDf8) heterozygote. qDf8 is a deletion from position 1.14–4.67 cM on chromosome I, which encompasses the prom-1 locus. Bivalent formation was still highly variable and not significantly different (t test, p > 0.05) from the homozygous mutant (Figure 4b).
Because bivalent formation was reduced in the female germline of the prom-1Δ mutant, we wanted to compare it to the crossover frequency in female meioses. Recombination frequencies calculated from rare viable offspring may be biased due to the preferential survival of embryos derived from germ cells in which crossing over was highest and hence faithful segregation of most chromosomes was ensured. Recombination was, therefore, assayed using freshly laid embryos by scoring for the production of recombinant progeny of parents carrying SNPs. To this end, we crossed prom-1Δ/prom-1Δ hermaphrodites heterozygous for two SNP markers on chromosome V to prom-1+/prom-1+ males homozygous for both SNP loci. (Mating was confirmed by PCR testing the presence of a paternal GFP marker or a wild-type copy of prom-1 in the embryos; see Materials and Methods). All recombination scored in this experiment must have taken place in the ovaries of prom-1Δ mothers. The frequency of recombination was calculated by counting the fraction of embryos where a recombination event caused homozygosity of one of the two SNP markers in the progeny (Table 1; Supplemental Figure S5). The control experiment was done with prom-1+/prom-1+ hermaphrodites. The reduction in recombination in prom-1Δ female meiosis to ∼32% of the wild type corresponds reasonably well with the reduction in chromosome V bivalent formation, with bivalents found in 24% of diakinesis nuclei (see below).
Table 1.
Recombination frequencies observed on embryos between SNP markers in cosmids F36H9 and Y51A2A on chromosome V
| No. of recombinants | No. of embryos tested | % | |
|---|---|---|---|
| prom-1Δ | 9 | 65 | 13.8 |
| Wild type | 48 | 112 | 42.8 |
The number of recombinants corresponds to the number of embryos homozygous for the wild-type (Bristol) or the Hawaiian SNP pattern on either side of the interval.
Reduced Homologous Pairing and Nonhomologous Synapsis in the prom-1Δ Mutant
Univalent formation during diakinesis can be caused by defects in 1) homologous chromosome pairing, 2) chromosome synapsis, 3) recombination, and/or in 4) chiasmata maintenance. We performed experiments to assess for potential defects in these processes in prom-1Δ.
To quantify homologous pairing, we highlighted homologous chromosomal loci by FISH (Figure 4, c and d) with probes corresponding to loci near the left end of chromosome I and the right end of chromosome V. In the wild type, there was almost no association of homologous FISH signals in the mitotic zone, but 72 and 99% of FISH signal doublets were homologously associated in the transition zone and pachytene zones, respectively (Figure 4d and Supplemental Table S6). In the mutant, we evaluated the frequencies of associated FISH signals in the mitotic zone, in the extended meiotic entry zone, in the distal pachytene zone just proximal to the extended meiotic entry zone, and in the proximal pachytene zone just proximal to the first diplotene nuclei. Homologous pairing of autosomes was found to vary considerably between gonads, but it was, in general, reduced compared with the wild type (Figure 4d and Supplemental Table S6). C. elegans chromosomes possess a pairing center near one end that has been implicated in the maintenance of pairing and promotion of SC formation (McKim, 2005). We did not observe an obvious difference in the pairing of a chromosome region near the pairing center (5S rDNA locus) and a region at the left end of chromosome I which is distant from it (data not shown).
Next, we studied the pairing of X chromosomes by immunostaining of the HIM-8 protein that is associated with the pairing centers on the X chromosomes (Phillips et al., 2005) (Figure 4e). We scored the association of HIM-8 foci in a total of 350 nuclei (in 6 different gonads) from the proximal pachytene zone. In 69% of prom-1Δ nuclei, we found a single focus or two associated foci (WT control: 99% single or associated; n = 208), suggesting that the pairing of X chromosomes is affected to a lesser degree than that of autosomes. Likewise, when we determined bivalent formation in diakinesis for specific chromosomes by FISH (Figure 4f), it turned out that the X chromosomes were involved in bivalent formation more often than the autosomes. X chromosome bivalents were found in 74% of the nuclei (n = 38), whereas chromosome V bivalents were present in only 24% of the nuclei (n = 21).
Because SYP-1 thread formation indicated extensive synapsis, whereas the association of homologous FISH signals was limited, we tested to which extent synapsis in the prom-1Δ mutant was homologous. To this end, simultaneous FISH of 5S rDNA loci on chromosome V and SYP-1 immunostaining were performed (compare Couteau et al., 2004). In the wild type, paired loci were associated with SYP-1 (i.e., synapsed) tracts in 73 of 74 nuclei scored. In the prom-1Δ mutant, however, one or both unpaired homologous loci were associated with SYP-1 tracts in 69% of the nuclei (n = 213 pachytene nuclei from 7 different gonads), indicating that they were synapsed with a nonhomologous region (Figure 4g). This suggests that nonhomologous synapsis is extensive in the prom-1Δ mutant.
Defective DSB Processing in the prom-1Δ Mutant
The recombination protein, RAD-51, assembles at sites of meiotic DSBs (Shinohara et al., 1992), and immunostaining of RAD-51 produces foci in late zygotene to midpachytene nuclei, just proximal to the transition zone in wild-type C. elegans meiosis (Alpi et al., 2003; Colaiácovo et al., 2003). In the prom-1Δ mutant, RAD-51 foci appeared near the beginning of the pachytene zone and they did not disappear before diplotene (Figure 5). Moreover, their number greatly increased toward the end of the pachytene zone. The mean number of foci in nuclei within the RAD-51–positive region of the prom-1Δ mutant was 22.0 ± 10.0 (n = 234 nuclei from 7 different gonads), whereas in the wild type this number was only 6.0 ± 3.5 (n = 263 nuclei from 5 different gonads). We cannot exclude the possibility that more DSBs than in the wild type are formed in the first place, but the persistence of RAD-51 foci suggests that DSBs may be formed normally but accumulate because of their delayed repair in the absence of PROM-1. Moreover, the ∼4 times increased mean number of RAD-51 foci is comparable with other mutants where the repair of DSBs is delayed or abolished (Alpi et al., 2003; Colaiácovo et al., 2003).
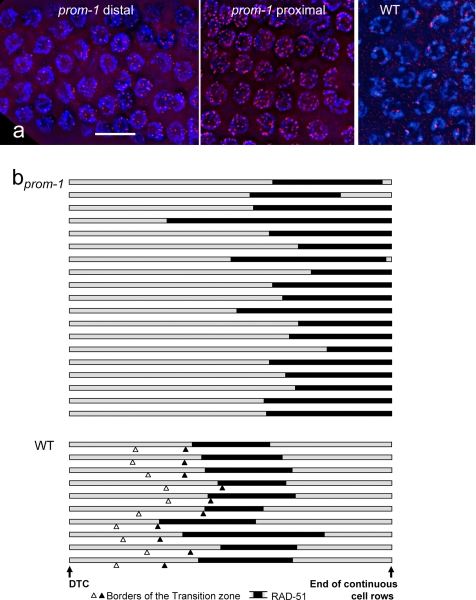
Figure 5.
The abundance and distribution of recombination sites visualized by immunostaining of RAD-51. (a) RAD-51 foci (red) in the prom-1Δ mutant and wild-type (WT) animals. For the prom-1Δ mutant, the distal and the proximal border of a RAD-51–positive region are shown, where RAD-51 foci appear and are most abundant, respectively. In the wild type, there is not very much variation in the number of RAD-51 foci along the RAD-51–positive region of which a representative section is shown. (b) The position and extent of the RAD-51–positive region (black bars) in the gonads (shaded bars) of prom-1Δ mutant and wild-type (WT) animals. Gonads were measured from the DTC to the end of continuous cell rows (roughly coinciding with the end of the pachytene zone) and normalized, and the borders of the RAD-51–positive region were entered. This region is striking due to the abundance of RAD-51 foci. In quantitative terms, it is defined as the zone in which >90% of nuclei have more than two prominent foci by RAD-51 immunostaining. The open and solid arrowheads indicate the distal and proximal borders of the transition zone of the wild type, respectively (for a classification of meiotic stages in the gonad; see MacQueen et al., 2002), whereas the transition zone is absent from the prom-1Δ mutant gonads. Bar, 10 μm.
prom-1 Acts Earlier Than htp-1
prom-1 mutants resemble htp-1 mutants with respect to nonhomologous synapsis, with the difference that htp-1 loads SC components prematurely (Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005). We produced a prom-1Δ htp-1 double mutant to study the genetic interaction between prom-1 and htp-1. REC-8 staining highlighted the aggregations of SC proteins typical for the extended meiotic entry zone (Figure 6). In the prom-1Δ htp-1 double mutant, the extended meiotic entry zone of prom-1Δ was still present. This means that the delayed entry into zygotene cannot be suppressed by inducing premature synapsis in the htp-1 mutant, and it confirms that prom-1 acts early in the leptotene/zygotene commitment upstream of htp-1. The scattered transition zone-like nuclei found in the pachytene zone of prom-1Δ were missing (Figure 6) and only univalents were formed during diakinesis of the double mutant. This supports our assumption that the limited homologous pairing observed in prom-1Δ is initiated in these transition zone-like nuclei.
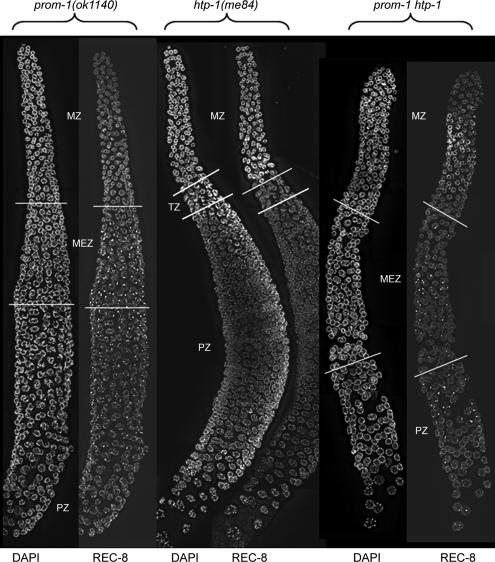
Figure 6.
Analysis of the epistatic relationship between prom-1 and htp-1. Dissected prom-1, htp-1 and prom-1 htp-1 gonads are shown. MZ, mitotic zone; TZ, transition zone; MEZ, meiotic entry zone; PZ, pachytene zone. In the prom-1 mutant, a transition zone is missing, but there is an extended meiotic entry zone. This zone is characterized by the expression of SC proteins, but they do not organize into axial elements but form nuclear spots, such as seen here for REC-8. In the pachytene zone axial elements and SCs do form but some of the spots remain. The pachytene zone is interspersed with nuclei with chromatin arranged in a crescent shape. In the htp-1 mutant, there is onset of axial element formation and precocious synapsis immediately downstream of a narrow and poorly developed transition zone. There are no nuclei with crescent-shaped chromatin in the pachytene zone. In the prom-1 htp-1 double mutant, the extended meiotic entry zone is present and spots of SC proteins remain in pachytene. Nuclei with crescent-shaped chromatin are no longer found.
DISCUSSION
prom-1Δ Mutants Display a Complex Meiotic Phenotype
The prom-1Δ mutation causes aberrant meiotic progression in the ovary. Following the mitotic zone of the gonad, there was an elongated tract of cells (an expanded meiotic entry zone) in which cells were transiently arrested right after entry into the meiotic pathway. These cells had completed mitotic and presumably also premeiotic DNA synthesis, and they expressed the meiosis-specific structural proteins HIM-3 and SYP-1. A transition zone with cells synchronously initiating chromosome pairing was missing, instead, the presence of transition zone-like nuclei in the proximal region of the gonad seems to reflect asynchronous and retarded onset of homology search. Homologous pairing and synapsis were reduced and the number of RAD-51 foci was increased, indicating that the repair of DSBs was abnormal. The increased incidence of apoptotic germline cells and the reduced brood size suggest that in prom-1 mutants a substantial portion of faulty pachytene nuclei is discarded.
By the very end of the pachytene zone the RAD-51 foci disappeared (Figure 5). This, together with the fact that ∼10% of embryos are viable, suggests that although the repair of DSBs was delayed, it must eventually have taken place efficiently. It has been proposed that DSB repair via the sister chromatid becomes possible at the pachytene–diplotene transition once a barrier (possibly created by lateral elements) has disappeared (Colaiácovo et al., 2003). Therefore, it is possible that in the prom-1Δ mutant where chiasma formation and recombination are reduced to about one third of the frequency in the wild type, most of the repair occurs via the sister chromatid.
A remarkable feature of the prom-1 mutants, including the deletion mutant, was the extreme variability of phenotypes between individuals with respect to brood size and the viability of their offspring, between gonads with respect to homologous pairing, and between the cells of a gonad with respect to bivalent formation. All these parameters were evaluated under controlled conditions and in descendants from homozygous prom-1Δ mothers to avoid a possible maternal rescue as a source of variability. (In fact, the viability of embryos laid by homozygous prom-1Δ hermaphrodites, which directly derived from heterozygous mothers was notably higher (21% of laid eggs; n = 1813), which suggests some maternal rescue in the first generation.) The tested parameters continued to vary among later descendants with no obvious tendency to recapitulate high or low brood size, hatch rate, pairing, or bivalent formation of their mothers (data not shown). We do not have at present an explanation for the variable expressivity of the mutant genotype.
PROM-1 Has a Role in Meiotic Chromosome Pairing
In prom-1Δ hermaphrodites, homologous pairing was reduced and only few nuclei with crescent chromatin arrangement indicative of initial chromosome pairing were scattered in the pachytene zone of the gonad. Thus, PROM-1 adds to the list of proteins that have been identified as essential regulators of the establishment of initial pairing in C. elegans. The protein kinase CHK-2, for example, promotes homologous pairing (MacQueen and Villeneuve, 2001). In contrast to prom-1Δ however, absence of CHK-2 also inhibits SC formation, suggesting that the CHK-2 kinase is not directly involved in homology search and recognition but rather plays a central role as a more upstream regulator of meiosis, which allows it to govern both homology recognition and SC formation, which are two independent processes in C. elegans. Also, deletion of him-3, which encodes a meiosis-specific axial element protein causes a pairing defect (Couteau et al., 2004). It is, therefore, conceivable that chromosomal axis components have to be loaded and/or modified in time to establish a chromosomal organization, which supports homology search. HTP-1 is another protein with a role in initial chromosome pairing. It was shown that in addition to its more prominent function in coordinating pairing with SC formation, HTP-1 contributes to pairing because in a htp-1 syp-2 double mutant initial homologous alignment is reduced compared with a syp-2 mutant in which alignment occurs but is not enforced by synapsis (Martinez-Perez and Villeneuve, 2005).
Recently, the chromosome binding ZIM proteins (ZIM-1, -2, and -3 and HIM-8) were identified as factors that facilitate homologous contacts and SC formation. Because individual ZIM proteins are responsible for the pairing of more than one chromosome pair, it is unlikely that the ZIM proteins confer the specificity for the recognition of homologous chromosomes (Phillips and Dernburg, 2006). Together with the ZIM proteins, SUN-1 and ZYG-12 seem to impose a certain chromosomal arrangement within the nucleus as a precondition for pairing (Penkner et al., 2007). It remains to be shown how PROM-1 interacts with the mentioned factors to ensure the encounter of homologues or its manifestation as homologous alignment.
Coordinated Meiotic Entry Is Essential for Downstream Meiotic Events
In the wild type, only few cells of the hermaphrodite gonad pass through the meiotic entry zone at any given time. Hence, the older literature failed to recognize the meiotic nature of the cells immediately upstream of the transition zone and assigned them to the “mitotic zone.” Only more recent accounts (MacQueen et al., 2002; Hansen et al., 2004;Crittenden et al., 2006) associate these cells with early meiotic prophase. The lack of informative mutants aggravated detailed analysis of nuclei at this stage. To date, only the protein kinase CHK-2 was ascribed functions already at very early steps in meiosis that are crucial for downstream meiotic events culminating in faithful chromosome segregation (MacQueen and Villeneuve, 2001).
Germ cells of prom-1Δ mutants readily proceed through mitosis and premeiotic DNA replication. Although they are competent for timely entering the meiotic program as judged by the expression of meiosis-specific proteins, prom-1Δ nuclei display a pronounced delay in the meiotic entry stage resulting in a dramatic extension of this zone. Once prom-1 nuclei have passed the meiotic entry zone, axial and central components of the SC are loaded and chromosome condensation, homologous interactions, and recombination are initiated asynchronously, consistent with the lack of a distinct transition zone. As a consequence, prom-1 mutants undergo extensive nonhomologous synapsis, a feature shared with the weak him-3(vv6) and the htp-1 null mutants (Couteau et al., 2004; Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005). In cases (such as in haploids) where homologous chromosomes are not available but where the SC machinery is intact, it has been observed that the failure to find a homologous partner leads to indiscriminate nonhomologous synapsis (Loidl and Jones, 1986; Loidl, 1994). We think that SC polymerization along chromosomes is a default process that can readily involve nonhomologous chromosomes if homologous alignment has not been achieved by the time SC development starts (Loidl, 1991). Thus, HTP-1 and HIM-3 prevent nonhomologous synapsis, because they may sufficiently delay the onset of synapsis or otherwise coordinate alignment and synapsis to ensure that only prealigned homologues will synapse (Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005). Although untimely synapsis in the htp-1 and him-3 mutants precedes the homologous pairing step, synapsis is delayed in the prom-1Δ mutant. The prom-1Δ htp-1Δ double mutant shows the Prom-1 delayed synapsis phenotype, which indicates that the early onset nonhomologous synapsis in the htp-1Δ mutant requires PROM-1.
A notable difference between the prom-1Δ and the htp-1 mutant is that in the latter RAD-51 foci do not accumulate. Whereas in the prom-1Δ mutant, a possible alternative DSB repair via the sister might be implemented only at the pachytene–diplotene transition after the axial elements become disassembled (see above), in the htp-1 (and in the him-3 null) mutant, the sister chromatid may be efficiently used for DSB repair in pachytene (Martinez-Perez and Villeneuve, 2005).
It is worth mentioning that bivalent formation of the X chromosomes is less affected compared with the autosomes in the prom-1 mutant. This behavior of the X resembles the one in weak him-3 mutants (Couteau et al., 2004; Nabeshima et al., 2004) and in the htp-1 null mutant (Couteau and Zetka, 2005; Martinez-Perez and Villeneuve, 2005), and it suggests redundant or less susceptible pathways ensuring X chromosome pairing. A different regulation of X chromosome pairing is also revealed by the fact that the concentration at the nuclear envelope of the X chromosome specific pairing protein HIM-8 is independent of CHK-2, whereas the behavior of its autosomal counterparts is affected in a chk-2 mutant (Phillips and Dernburg, 2006).
PROM-1 May Play a Role in the Ubiquitination of a Meiotic Regulator
Many F-box proteins have been implicated to act as the substrate-recognizing component of Skp1-Cullin-F-box protein ubiquitin–protein ligase complexes (e.g., Schulman et al., 2000; Hochstrasser, 2002). PROM-1 contains an F-box–like motif, and it is possible that it is involved in ubiquitination. Indeed, components of the ubiquitination pathway have been implicated in meiotic prophase control in various organisms. In budding yeast, meiosis is impaired in mutants of the E2 ubiquitin conjugating enzyme RAD6 (Borts et al., 1986). In mice, loss of ubiquitin-conjugating activity by HR6B, one of the two mammalian RAD6 homologues, results in a damaged SC structure and other meiotic anomalies (Baarends et al., 2003). Likewise, homologous synapsis is impaired by a mutation in the ASK1 gene of Arabidopsis, which encodes a Skp1p homologue (Wang et al., 2004). In C. elegans, depletion of Skp-related proteins 1 and 2 results in univalent formation at diakinesis (Nayak et al., 2002), as does depletion of CUL-1 (cullin) (our preliminary data). A mutation in cul-1 was previously shown to cause hyperplasia of all tissues (Kipreos et al., 1996). Thus, the pairing defect displayed by C. elegans prom-1Δ mutant animals could well be explained by a defect in ubiquitin-dependent degradation of one or several meiotic regulators yet to be identified.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Michael Hengartner (University of Zurich) in whose laboratory the op240 allele was initially isolated. Materials were generously provided by Abby Dernburg (University of California, Berkeley), Andrew Fire (Stanford University, CA), Adriana La Volpe (Istituto di Genetica, Consiglio Nazionale delle Ricerche, Naples), Jill Schumacher (University of Texas, Houston), Anne Villeneuve (Stanford University, CA), and Monique Zetka (McGill University, Montreal). We thank the C. elegans Gene Knockout Consortium for the deletion allele ok1140 and the C. elegans Genetic Center (University of Minnesota) for strains. We are grateful to Michael Jantsch (University of Vienna), Maria Novatchkova (Research Institute of Molecular Pathology, Vienna), and Maria Siomos (Gregor Mendel Institute, Vienna) for valuable suggestions and to Christian Pflügl for technical assistance. This work was supported by grant P-17329 of the Austrian Science Fund (Fonds zur Förderung der wissenschaftlichen Forschung), by a WWTF (Wiener Wissenschafts-, Forschungs- und Technologiefonds) grant (to V.J.), by a Cancer Research UK career development award (to A.G.), and by National Institutes of Health grant GM-063310 (to T.S.).
Abbreviations used:
- DSB
double-strand break
- FISH
fluorescence in situ hybridization
- SC
synaptonemal complex
- SNP
single-nucleotide polymorphism.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-03-0243) on October 3, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Alpi A., Pasierbek P., Gartner A., Loidl J. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma. 2003;112:6–16. doi: 10.1007/s00412-003-0237-5. [DOI] [PubMed] [Google Scholar]
- Baarends W. M., Wassenaar E., Hoogerbrugge J. W., van Cappellen G., Roest H. P., Vreeburg J., Ooms M., Hoeijmakers J.H.J., Grootegoed J. A. Loss of HR6B ubiquitin-conjugating activity results in damaged synaptonemal complex structure and increased crossing-over frequency during the male meiotic prophase. Mol. Cell Biol. 2003;23:1151–1162. doi: 10.1128/MCB.23.4.1151-1162.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla N., Dernburg A. F. A conserved checkpoint monitors meiotic chromosome synapsis in Caenorhabditis elegans. Science. 2005;310:1683–1686. doi: 10.1126/science.1117468. [DOI] [PubMed] [Google Scholar]
- Borts R. H., Lichten M., Haber J. E. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics. 1986;113:551–567. doi: 10.1093/genetics/113.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaiácovo M. P., MacQueen A. J., Martinez-Perez E., McDonald K., Adamo A., La Volpe A., Villeneuve A. M. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 2003;5:463–474. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- Couteau F., Nabeshima K., Villeneuve A., Zetka M. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 2004;14:585–592. doi: 10.1016/j.cub.2004.03.033. [DOI] [PubMed] [Google Scholar]
- Couteau F., Zetka M. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes Dev. 2005;19:2744–2756. doi: 10.1101/gad.1348205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden S. L., Leonhard K. A., Byrd D. T., Kimble J. Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol. Biol. Cell. 2006;17:3051–3061. doi: 10.1091/mbc.E06-03-0170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M., Villeneuve A. M. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94:387–398. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Gartner A., MacQueen A. J., Villeneuve A. M. Methods for analyzing checkpoint responses in Caenorhabditis elegans. Methods Mol. Biol. 2004;280:257–274. doi: 10.1385/1-59259-788-2:257. [DOI] [PubMed] [Google Scholar]
- Gartner A., Milstein S., Ahmed S., Hodgkin J., Hengartner M. O. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell. 2000;5:435–443. doi: 10.1016/s1097-2765(00)80438-4. [DOI] [PubMed] [Google Scholar]
- Hansen D., Hubbard E.J.A., Schedl T. Multi-pathway control of the proliferation versus meiotic development decision in the Caenorhabditis elegans germline. Dev. Biol. 2004;268:342–357. doi: 10.1016/j.ydbio.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Harper J. W., Burton J. L., Solomon M. J. The anaphase-promoting complex: it's not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- Henderson K. A., Kee K., Maleki S., Santini P. A., Keeney S. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell. 2006;125:1321–1332. doi: 10.1016/j.cell.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillers K. J., Villeneuve A. M. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 2003;13:1641–1647. doi: 10.1016/j.cub.2003.08.026. [DOI] [PubMed] [Google Scholar]
- Hochstrasser M. New proteases in a ubiquitin stew. Science. 2002;298:250–252. doi: 10.1126/science.1078097. [DOI] [PubMed] [Google Scholar]
- Jones A. R., Francis R., Schedl T. GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 1996;180:165–183. doi: 10.1006/dbio.1996.0293. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kipreos E. T., Lander L. E., Wing J. P., He W. W., Hedgecock E. M. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Kipreos E. T., Pagano M. The F-box protein family. Genome Biol. 2000;1:3002.1–3002.7. doi: 10.1186/gb-2000-1-5-reviews3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J. Coming to grips with a complex matter. A multidisciplinary approach to the synaptonemal complex. Chromosoma. 1991;100:289–292. doi: 10.1007/BF00360526. [DOI] [PubMed] [Google Scholar]
- Loidl J. Cytological aspects of meiotic recombination. Experientia. 1994;50:285–294. doi: 10.1007/BF01924012. [DOI] [PubMed] [Google Scholar]
- Loidl J., Jones G. H. Synaptonemal complex spreading in Allium. I. Triploid A. sphaerocephalon. Chromosoma. 1986;93:420–428. [Google Scholar]
- MacQueen A. J., Colaiácovo M. P., McDonald K., Villeneuve A. M. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002;16:2428–2442. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A. J., Villeneuve A. M. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes Dev. 2001;15:1674–1687. doi: 10.1101/gad.902601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Perez E., Villeneuve A. M. HTP-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 2005;19:2727–2743. doi: 10.1101/gad.1338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K. S. When size does not matter: pairing sites during meiosis. Cell. 2005;123:989–992. doi: 10.1016/j.cell.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve A. M., Hillers K. J. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics. 2004;168:1275–1292. doi: 10.1534/genetics.104.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak S., Santiago F. E., Jin H., Lin D., Schedl T., Kipreos E. T. The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol. 2002;12:277–287. doi: 10.1016/s0960-9822(02)00682-6. [DOI] [PubMed] [Google Scholar]
- Pasierbek P., Jantsch M., Melcher M., Schleiffer A., Schweizer D., Loidl J. A Caenorhabditis elegans cohesion protein with functions in meiotic chromosome pairing and disjunction. Genes Dev. 2001;15:1349–1360. doi: 10.1101/gad.192701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkner A., Tang L., Novatchkova M., Ladurner M., Fridkin A., Gruenbaum Y., Schweizer D., Loidl J., Jantsch V. The nuclear envelope protein matefin/SUN-1 is required for homologous pairing in C. elegans meiosis. Dev. Cell. 2007;12:873–885. doi: 10.1016/j.devcel.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Petronczki M., Siomos M. F., Nasmyth K. Un ménage à quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Phillips C. M., Dernburg A. F. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell. 2006;11:817–829. doi: 10.1016/j.devcel.2006.09.020. [DOI] [PubMed] [Google Scholar]
- Phillips C. M., Wong C., Bhalla N., Carlton P. M., Weiser P., Meneely P. M., Dernburg A. F. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell. 2005;123:1051–1063. doi: 10.1016/j.cell.2005.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piano F., Schetter A. J., Morton D. G., Gunsalus K. C., Reinke V., Kim S. K., Kemphues K. J. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Science. 2002:1959–1964. doi: 10.1016/s0960-9822(02)01301-5. [DOI] [PubMed] [Google Scholar]
- Reinke V., et al. A global profile of germline gene expression in C. elegans. Mol. Cell. 2000;6:605–616. doi: 10.1016/s1097-2765(00)00059-9. [DOI] [PubMed] [Google Scholar]
- Rual J.-F., et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedl T. Developmental Genetics of the Germ Line. In: Riddle D. L., Blumenthal T., Meyer B. J., Priess J. R., editors. C. elegans II. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 241–269. [PubMed] [Google Scholar]
- Schulman B. A., Carrano A. C., Jeffrey P. D., Bowen Z., Kinnucan E. R., Finnin M. S., Elledge S. J., Harper J. W., Pagano M., Pavletich N. P. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- Schumacher B., Hanazawa M., Lee M.-H., Nayak S., Volkmann K., Hofmann R., Hengartner M., Schedl T., Gartner A. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell. 2005;120:357–368. doi: 10.1016/j.cell.2004.12.009. [DOI] [PubMed] [Google Scholar]
- Shinohara A., Ogawa H., Ogawa T. Rad51 protein involved in repair and recombination in Saccharomyces cerevisiae is a RecA-like protein. Cell. 1992;69:457–470. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- Simon-Kayser B., Scoul C., Renaudin K., Jezequel P., Bouchot O., Rigaud J., Bezieau S. Molecular cloning and characterization of FBXO47, a novel gene containing an F-box domain, located in the 17q12 band deleted in papillary renal cell carcinoma. Genes Chromos. Cancer. 2005;43:83–94. doi: 10.1002/gcc.20170. [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Wang Y. X., Wu H., Liang G. Q., Yang M. Defects in nucleolar migration and synapsis in male prophase I in the ask1-1 mutant of Arabidopsis. Sex. Plant Reprod. 2004;16:273–282. [Google Scholar]
- Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., Plasterk R.H.A. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Zetka M. C., Kawasaki I., Strome S., Muller F. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 1999;13:2258–2270. doi: 10.1101/gad.13.17.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.