Abstract
The Rho-type GTPase Cdc42 is a central regulator of eukaryotic cell polarity and signal transduction. In budding yeast, Cdc42 regulates polarity and mitogen-activated protein (MAP) kinase signaling in part through the PAK-family kinase Ste20. Activation of Ste20 requires a Cdc42/Rac interactive binding (CRIB) domain, which mediates its recruitment to membrane-associated Cdc42. Here, we identify a separate domain in Ste20 that interacts directly with membrane phospholipids and is critical for its function. This short region, termed the basic-rich (BR) domain, can target green fluorescent protein to the plasma membrane in vivo and binds PIP2-containing liposomes in vitro. Mutation of basic or hydrophobic residues in the BR domain abolishes polarized localization of Ste20 and its function in both MAP kinase–dependent and independent pathways. Thus, Cdc42 binding is required but is insufficient; instead, direct membrane binding by Ste20 is also required. Nevertheless, phospholipid specificity is not essential in vivo, because the BR domain can be replaced with several heterologous lipid-binding domains of varying lipid preferences. We also identify functionally important BR domains in two other yeast Cdc42 effectors, Gic1 and Gic2, suggesting that cooperation between protein–protein and protein–membrane interactions is a prevalent mechanism during Cdc42-regulated signaling and perhaps for other dynamic localization events at the cell cortex.
INTRODUCTION
Control of cellular architecture and interaction with the extracellular environment rely on dynamic localization of proteins to the cell cortex. For example, signal transduction is often initiated by the assembly of protein complexes at the plasma membrane, where external stimuli are detected by membrane receptors (Kholodenko et al., 2000; Cho, 2006). Dynamic assembly can be ensured by reversible recruitment of cytoplasmic factors to the membrane-localized complex (Johnson and Cornell, 1999; Teruel and Meyer, 2000). Hence, signaling proteins commonly consist of multiple, modular interaction domains that dictate their localization and assembly behavior (Pawson and Nash, 2003). Membrane recruitment is governed not only by protein–protein interactions but also by protein–membrane interactions, which can be mediated by lipid-binding motifs such as pleckstrin homology (PH) domains as well as PX, C1, C2, FYVE, ENTH, and FERM domains (Lemmon, 2003; Hurley, 2006). These domains have distinct structural folds and often recognize specific phospholipids. Other proteins can interact with membranes via short polybasic motifs, which contain multiple positively charged residues that bind acidic phospholipids through electrostatic interactions and that often act in concert with other interaction domains to promote efficient membrane targeting (Fivaz and Meyer, 2003; Heo et al., 2006). Although there are several physiologically relevant examples, the short length and indefinite sequence of these polybasic motifs hinders their identification and obscures estimates of their prevalence (Fivaz and Meyer, 2003). This study reports the identification of short, basic-rich (BR) membrane-interaction domains in multiple Cdc42 effectors in the budding yeast Saccharomyces cerevisiae.
Cdc42, a member of the Rho family of small GTPases, plays a central role in the regulation of cellular polarity in all eukaryotic cells (Johnson, 1999; Etienne-Manneville, 2004). In budding yeast, Cdc42 controls polarized cell growth by regulating cytoskeleton assembly and membrane trafficking, and it also participates in several signal transduction pathways (Johnson, 1999; Park and Bi, 2007). To regulate these processes, Cdc42 interacts with a number of downstream effectors. One class includes three members of the p21-activated kinase (PAK) family of serine/threonine protein kinases—Ste20, Cla4, and Skm1—which function in signal transduction, polarized growth, and cell cycle control (Leberer et al., 1992; Cvrckova et al., 1995; Holly and Blumer, 1999; Hofken and Schiebel, 2002). Another class includes two redundant cell polarity factors, Gic1 and Gic2, which function in actin polarization and septin assembly (Brown et al., 1997; Chen et al., 1997). Each of these effectors has a conserved Cdc42/Rac-interactive binding (CRIB) domain, which mediates Cdc42 binding and regulates their function in vivo.
Ste20 is perhaps the best understood Cdc42 effector in budding yeast. It regulates multiple mitogen-activated protein kinase (MAPK) pathways that control mating, filamentous growth, and osmotic stress response and also is involved in cell polarity and cell cycle control (Leberer et al., 1992; Liu et al., 1993; Cvrckova et al., 1995; O'Rourke and Herskowitz, 1998; Holly and Blumer, 1999; Raitt et al., 2000; Hofken and Schiebel, 2002). In the mating pathway, Ste20 activates MAPK cascade signaling when mating pheromones bind to membrane receptors (Dohlman and Thorner, 2001). The receptor-activated Gβγ dimer binds Ste20 (Leeuw et al., 1998) and recruits the scaffold protein Ste5 to the plasma membrane (Pryciak and Huntress, 1998), allowing Ste20 to phosphorylate the first in a chain of Ste5-associated kinases that eventually trigger cell cycle arrest, transcription of mating genes, and polarized morphogenesis. Ste20 is recruited to sites of polarized cell growth, such as the tips of buds and mating projections, via binding of its CRIB domain to Cdc42 (Peter et al., 1996; Leberer et al., 1997; Lamson et al., 2002; Ash et al., 2003). Cdc42 binding also regulates Ste20 kinase activity by disrupting an autoinhibitory conformation formed by interactions between the CRIB and kinase domains (Lamson et al., 2002). Point mutations in the CRIB domain can differentially affect Cdc42 binding and autoinhibition. Those that disrupt Cdc42 binding without affecting autoinhibition (S338A H345G) produce a nonfunctional kinase, whereas those that disrupt autoinhibition (L369G) produce a constitutively active, Cdc42-independent kinase (Lamson et al., 2002). Thus, localization and activation of Ste20 are normally coupled by their dependence on Cdc42 binding. Proper localization and function of Ste20 is also promoted by binding of a proline-rich motif in Ste20 to an SH3 domain in the cortical protein Bem1 (Moskow et al., 2000; Winters and Pryciak, 2005).
Here, we show that the localization and function of Ste20 is critically dependent on a previously unrecognized element. We identify a short membrane-binding region in the Ste20 N-terminus that promotes the proper polarized localization of Ste20. This membrane-binding domain is required for Cdc42-dependent regulation of Ste20 and for the in vivo function of the kinase in both MAPK-dependent and -independent pathways. Furthermore, we identify similar membrane-binding motifs in two other Cdc42 targets, Gic1 and Gic2, and show that they are critical for the function of these proteins. Our observations suggest a common theme for Cdc42 effectors in which a membrane-binding domain is required to help target the protein to its activator.
MATERIALS AND METHODS
Strains and Plasmids
Standard yeast media and genetic methods were used (Rose and Fink, 1990). Yeast strains and plasmids are listed in Supplementary Tables 1 and 2, respectively.
Mating, Filamentation, Two-Hybrid, and β-Galactosidase Assays
Mating assays, halo assays of growth arrest, and FUS1-lacZ transcriptional induction assays in response to α factor or galactose-inducible constructs, were performed as described previously (Lamson et al., 2002); all transcription and shmoo formation assays used 5 μM α factor for 2 h. Agar invasion assays used fresh transformants patched onto YPD plates, as described previously (Winters and Pryciak, 2005). Quantitative two-hybrid and β-galactosidase assays used methods described previously (Lamson et al., 2002).
Microscopy
To visualize green fluorescent protein (GFP)-Ste20, Gic1-GFPx3, and Gic2-GFPx3, plasmid-transformed cells were grown at 30°C in -Ura media and were observed without fixation using a Nikon E600 epifluorescence microscope (Melville, NY) with a 50× Plan oil immersion objective. Images were captured using a Hamamatsu Orca-ER digital camera (Bridgewater, NJ) and IPLab Spectrum version 3.5.5 software (Scanalytics, Rockville, MD). To analyze galactose-inducible GFP and glutathione S-transferase (GST)-GFP fusions to isolated protein domains, cells were grown at 30°C in selective raffinose medium and then induced with 2% galactose for 2 h.
Liposome-binding Assays
GST fusion proteins were expressed from pGEX-6P-1 and purified as described previously (Nakanishi et al., 2004) from Escherichia coli strain BL21-RIPL (Stratagene, La Jolla, CA). Phospholipids in chloroform were purchased from Avanti Polar Lipids (Alabaster, AL). To prepare sucrose-laden liposomes (Sciorra et al., 1999), phospholipid mixtures were dried in glass test tubes under a stream of nitrogen, then resuspended in 20 mM HEPES (pH 7.5) with 20 mM NaCl and 0.2 M sucrose, vortexed (∼1 min), and sonicated in a water bath sonicator (four times for 1 min). This suspension (200 μl) was mixed with 700 μl of buffer A (20 mM NaPO4, pH 7.5, and 200 mM NaCl) and centrifuged at 200,000 × g for 15 min at 4°C. Pelleted liposomes (80 or 200 nmol total lipid) were resuspended in buffer A containing 10 μg of bovine serum albumin and then were incubated with 2 μg of purified protein on ice for 30 min in a final volume of 200 μl. Centrifugation was repeated, and protein in the supernatant (precipitated with 4.5% trichloroacetic acid) and pellet fractions were analyzed by SDS-PAGE and Coomassie Blue staining.
Yeast Cell Lysates and Protein Analysis
PPY913 cells carrying Myc-Ste20 plasmids were cultured in -Ura growth medium, and 2 × 107 cells in the log phase (OD660 ∼0.7) were collected by centrifugation. Whole cell lysates were prepared by a post-alkaline extraction method (Kushnirov, 2000) and analyzed by SDS-PAGE and immunoblotting using rabbit anti-Myc antibody (1:200, Santa Cruz Biotechnology, Santa Cruz, CA) and alkaline phosphatase–conjugated secondary antibody (goat anti-rabbit, 1:3000, Bio-Rad, Richmond, CA).
RESULTS
The Ste20 Regulatory Region Contains a Membrane Interaction Domain
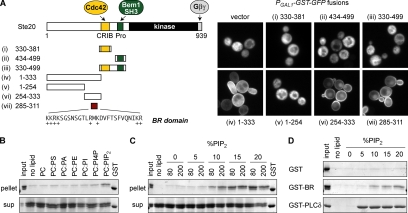
Ste20 is a large protein (939 residues) with a C-terminal kinase domain and an N-terminal “regulatory” region, which comprises over half of the protein and contains the binding sites for both Cdc42 and Bem1 (Figure 1A). To test whether these binding sites could suffice as independent cortical-localization domains, we made a series of GFP fusions to fragments from the Ste20 N-terminus (Figure 1A); these GFP fusions also contained a homodimerizing GST moiety, which can help reveal weak localization determinants by increasing binding avidity (Winters et al., 2005). Despite their role in localization of full-length Ste20, the isolated Cdc42-binding and Bem1-binding domains, or fragments containing both domains, were unable to localize to the cell cortex (Figure 1A, i–iii). Surprisingly, however, a fragment that lacked any previously characterized domains, residues 1-333, localized to the cortex (Figure 1A, iv). (This fragment also affected cell morphology, as discussed in a later section.) Further dissection revealed that a short (27 a.a.) domain, residues 285-311, was sufficient for the cortical localization (Figure 1A, v–vii). This short region contains many basic residues (Figure 1A) and is conserved among fungal Ste20 orthologues (Supplementary Figure S1), and hence we named it the BR domain.
Figure 1.
The Ste20 regulatory region contains a membrane-binding domain. (A) Left, fragments used to map a membrane-targeting domain in the Ste20 N-terminus. Fragment 330-381 spans the minimal Cdc42-binding motif predicted from prior studies (Kim et al., 2000; Morreale et al., 2000; Gladfelter et al., 2001), although it is part of a larger conserved region that also includes the autoinhibitory domain (see Supplementary Figure S1). Right, Ste20 fragments were expressed as GST-GFP fusions (pPP1843, pPP1878, pPP1880, pPP1961, pPP1877, pPP1939, pPP1940, or pPP2428) from a galactose-inducible promoter in wild-type cells (PPY1368). A minimum region for membrane localization maps to residues 285-311, denoted the basic-rich (BR) domain. (B) The Ste20 BR domain binds liposomes containing PIP2. A purified GST-BR fusion (Ste20 residues 285-311) was mixed with 80 nmol of sucrose laden liposomes containing phosphatidylcholine (PC) alone or 80% PC plus 20% (mol/mol) of phosphatidylserine (PS), phosphatidic acid (PA), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylinositol-4-phosphate (PI4P), or PIP2. Liposomes were pelleted, and protein in bound (pellet) and unbound (sup) fractions is shown. “Input” shows 25% of input protein (=0.5 μg). GST was loaded on each gel as a size marker. (C) Dependence of GST-BR binding on concentration and density of PIP2. Binding assays (in 200 μl volume) used 80 or 200 nmol of PC liposomes with varying molar percentage of PIP2 (0–20%). “Input” shows 10% of input protein (=0.2 μg). (D) Comparison of liposome binding by GST alone, GST-BR, and GST-PLCδPH, using 80 nmol of PC-based liposomes with 0–20% PIP2. Only pellet fractions are shown. “Input” shows 12.5% of input protein (=0.25 μg).
The sequence features and localization pattern of the BR domain suggested that it might bind directly to the plasma membrane, with its positively charged residues interacting with negatively charged phospholipids, as we found recently with a domain from another mating pathway protein, Ste5 (Winters et al., 2005). To test this notion, we performed in vitro liposome-binding assays using a bacterially produced GST fusion to the Ste20 BR domain (residues 285-311). This GST-Ste20BR fusion protein bound detectably to liposomes containing phosphatidylinositol-4,5-bisphosphate (PIP2) but not to those containing other phospholipid species (Figure 1B). This binding was sensitive to PIP2 levels (Figure 1C), though it appeared more dependent on the density of PIP2 than on its total concentration (e.g., greater binding for 80 nmol lipid with 10% PIP2 than for 200 nmol lipid with 5% PIP2), which is consistent with a multivalent mode of electrostatic interaction seen previously for polybasic domains and anionic membranes (Wang et al., 2002; Papayannopoulos et al., 2005). When compared with the PH domain from mammalian phospholipase C (PLC)δ (Figure 1D), which is specific for PIP2 (Rebecchi et al., 1992), the binding observed with the GST-Ste20BR fusion was relatively weak, because it required a higher percentage of PIP2, and a smaller fraction of input protein was bound. However, this was not entirely surprising given similar recent observations with analogous domains from other yeast proteins (Nakanishi et al., 2004; Winters et al., 2005). Because several different GST fusions containing the Ste20 BR domain were subject to unusually rapid proteolysis in E. coli cell extracts (data not shown), we did not perform further analysis using the in vitro system. Nevertheless, these results demonstrated that the Ste20 BR domain could bind directly to acidic phospholipid membranes and thereby led us to probe the role of this domain in vivo.
The BR Domain Is Important for Ste20 Localization and Function
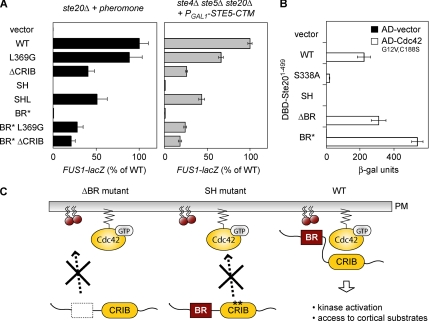
To test the role of the BR domain in full-length Ste20, we initially made two mutations (Figure 2A): one in which all the basic residues were replaced with noncharged residues (BR*), and one in which the entire BR domain was deleted (ΔBR). These mutations were introduced into constructs expressing GFP-tagged Ste20 from its own promoter. Ste20 normally localizes to small and medium buds and to shmoo tips in cells exposed to mating pheromone. We found that neither the BR* mutant nor the ΔBR mutant localized to the bud tip (Figure 2B). When exposed to mating pheromone, ste20Δ cells expressing the BR* or ΔBR mutant could not form mating projections, and GFP-Ste20 localization was diffuse (data not shown). To check localization in cells that could form projections, we repeated the experiment in STE20 cells and found that the BR* and ΔBR mutants failed to localize to the shmoo tip (Figure 2B). Therefore, membrane targeting by the BR domain is not a spurious feature of the isolated domain but is important for normal localization of full-length Ste20.
Figure 2.
The BR domain is important for Ste20 localization and function. (A) Basic residues (+) and mutations in the BR domain (BR* and ΔBR) are shown. (B) Localization of the indicated GFP-Ste20 alleles (pPP538, pPP2204, and pPP2318) was examined in PPY1249 (ste20Δ, −αF) and in PPY1114 (STE20, +αF). The ΔBR and BR* mutations abolish bud tip and shmoo tip localization. (C) Mating pathway phenotypes of ΔBR and BR* mutants. PPY913 (ste20Δ) cells carrying a vector (pRS316) or GFP-Ste20 alleles (pPP538, pPP2318, pPP2204, and pPP1009) or Myc-Ste20 alleles (pPP2927, pPP2929, pPP2930, and pPP2928) were compared for induction of the transcriptional reporter FUS1-lacZ by α factor. Bars, mean ± SD (n = 4). Below, GFP-Ste20 alleles were used for growth arrest and patch mating assays, and Myc-Ste20 alleles were used to check protein expression by anti-myc immunoblot. WT, wild type; SH, S338A H345G. (D) Agar invasion. PPY1209 (Σ1278b ste20Δ) cells carrying the indicated GFP-Ste20 alleles were assayed for invasive growth. Plates are shown before and after nonadherent cells were rinsed off with water. (E) BR domain mutations disrupt the Cla4-redundant essential function of Ste20. Fivefold serial dilutions of KBY211 (ste20Δ cla4Δ YCp-TRP1-cla4-75ts) cells carrying the indicated GFP-Ste20 alleles were spotted onto -Ura plates and incubated for 2 d at 25 or 37°C. (F) ΔBR and BR* mutants cannot support growth of ste20Δ lte1Δ cells. Fivefold serial dilutions of PPY1978 (ste20Δ lte1Δ PGAL1-STE20) cells carrying GFP-Ste20 alleles (as in E) were spotted onto -Ura -His plates containing either glucose or both raffinose and galactose and incubated at 30°C for 5 d (galactose) or 2 d (glucose).
To test if the BR domain is required for the signaling role of Ste20, we first measured pheromone response. Both ΔBR and BR* mutant forms of Ste20 failed to induce the FUS1-lacZ reporter gene in response to α factor (Figure 2C), yielding signaling defects comparable to that caused by S338A H345G mutations (“SH”) in the CRIB domain, which severely disrupt Cdc42 binding (Lamson et al., 2002). Similar defects were observed in cell cycle arrest and mating assays (Figure 2C). Because the GFP-Ste20 fusions were difficult to detect by Western blotting, we introduced the same mutations into a Myc-Ste20 construct; these showed that the signaling defects were not due to reduced protein levels (Figure 2C). It is unlikely that the BR domain mutations disrupt signaling by perturbing a larger structural domain involving the entire N-terminus, because deletion of other regions more N-terminal to the BR domain (e.g., Δ124-270, Δ2-119) did not cause any detectable defect (see Figure 6B). Because Ste20 participates in multiple MAPK pathways, we also tested if these BR mutants could function in the filamentous growth pathway by an agar invasion assay and found that they were defective (Figure 2D). Ste20 also plays a role in pathways that are independent of MAPK signaling but are essential for cell viability; namely, deletion of STE20 is lethal in cells that lack either the related PAK-family kinase Cla4 or the mitotic exit network factor Lte1 (Cvrckova et al., 1995; Hofken and Schiebel, 2002). We found that the Ste20 BR domain mutants were unable to support growth under either of these conditions (Figure 2, E and F). Collectively, these results indicate that the BR domain is important for multiple functions of Ste20 in signaling and viability.
Figure 6.
A motif in the Ste20 N-terminus affects cell morphology. (A) The indicated fragments were expressed as GST-GFP fusions from the GAL1 promoter (pPP1843, pPP2040, pPP2036, pPP2037, pPP2041, pPP2038, pPP2069, and pPP2070) in wild-type cells (PPY1368). The bud elongation phenotype requires residues 72-120 as well as membrane localization via either the native BR domain or a heterologous membrane-targeting sequence (Cpr vs. control sequence Cpr-SS; Pryciak and Huntress, 1998). (B) Mutations in the morphology-altering domain do not affect the function of full-length Ste20 in mating pathway signaling. PPY913 (ste20Δ) cells harboring the indicated GFP-Ste20 derivatives (pPP316, pPP538, pPP2202, pPP2203, pPP2205, pPP2575, pPP2318, pPP1010, or pPP2356) were tested for induction of FUS1-lacZ by α factor. Results (mean ± SD, n = 4–8) were normalized to wild-type (WT; 100% = 53 β-galactosidase units). “87-91 Ala5” denotes a mutant with Ala replacements at residues 87-91 (SLDDP), which form part of a conserved sequence block (see Supplementary Figure S1).
The BR Domain Is Required for Cdc42-dependent Regulation of Ste20
Because the BR domain is required for both localization and function of Ste20, which are normally controlled by Cdc42, we reasoned that membrane binding might allow Ste20 to be activated by membrane-localized Cdc42. To test this notion, we combined the BR* mutation with mutations in the Ste20 CRIB domain, L369G and ΔCRIB (Δ334-369), which disrupt autoinhibition and hence activate the Ste20 kinase independent of Cdc42 (Lamson et al., 2002). Indeed, these additional mutations restored pheromone signaling to the Ste20 BR* mutant (Figure 3A, left), suggesting that the BR* mutation prevents Cdc42 from activating Ste20. Notably, signaling was less efficient for the BR* L369G double mutant than for L369G alone, implying that membrane binding plays an additional role once Ste20 is activated, such as enhancing its ability to phosphorylate cortical substrates. In fact, this parallels previous findings that Cdc42 binding plays two separable roles in activation and localization of Ste20; for example, although both L369G and ΔCRIB mutants are constitutively active, the L369G mutant, which still binds Cdc42, signals more efficiently than ΔCRIB, which cannot bind Cdc42 and is delocalized (Lamson et al., 2002; and Figure 3A, left). To rule out the possibility that the BR mutant signaling defects were due to impaired binding between Ste20 and Gβγ (Leeuw et al., 1998), we activated the mating pathway in a Gβγ-independent manner, using ste4Δ ste5Δ ste20Δ cells that express membrane-targeted Ste5 (Ste5-CTM; Pryciak and Huntress, 1998). The BR* mutant phenotypes matched those seen during pheromone response (Figure 3A, right), implying that the BR domain serves primarily to promote Ste20 regulation by Cdc42, not Gβγ. Nevertheless, when using a two-hybrid assay to measure interaction between the Ste20 N-terminus and Cdc42 (Leberer et al., 1997; Lamson et al., 2002), neither of the BR domain mutations blocked Cdc42 binding (Figure 3B). Therefore, we conclude that separate BR-membrane and CRIB-Cdc42 interactions are jointly required for Cdc42 to regulate Ste20 at the plasma membrane (Figure 3C) and that membrane binding allows Cdc42 to promote both activation of Ste20 and its subsequent signaling.
Figure 3.
The BR domain is required for Cdc42-dependent regulation of Ste20. (A) Mutations that disrupt the autoinhibitory conformation of Ste20 suppress the requirement for the BR domain. At left, response to pheromone was tested in ste20Δ cells (PPY913) harboring the indicated GFP-Ste20 alleles (pRS316, pPP538, pPP1117, pPP1010, pPP1009, pPP1109, pPP2204, pPP2239, or pPP2238). Results (mean ± SD; n = 14) were normalized to WT Ste20 (100% = 56 β-galactosidase units). At right, the ability of the same GFP-Ste20 alleles to mediate pheromone- and Gβγ-independent signaling was tested by expression of PGAL1-STE5-CTM (pPP477) in ste4Δ ste5Δ ste20Δ cells (PPY866). Results (mean ± SD; n = 4) were normalized to WT Ste20 (100% = 127 β-galactosidase units). The L369G and ΔCRIB mutations disrupt Ste20 autoinhibition (Lamson et al., 2002); as controls, the SH (S338A H345G) and SHL (S338A H345G L369G) mutations were also tested to compare the ability of L369G to suppress either CRIB or BR domain mutations. (B) BR domain mutations do not affect Cdc42 binding. An activation domain fusion to Cdc42G12V C188S (pPP1027) or vector (pPP244) was coexpressed with DNA-binding domain fusions to the Ste20 N-terminus (DBD-Ste201-499; from top to bottom, pPP167, pPP1053, pPP1059, pPP1061, pPP2950, and pPP2949) in the two-hybrid reporter strain PPY760. Interaction was measured by quantitative β-galactosidase assay (mean ± SD; n = 4). As controls, mutations in the Ste20 CRIB domain that disrupt Cdc42 binding either moderately (S338A) or severely (SH = S338A H345G; Lamson et al., 2002) were assayed in parallel. (C) Schematic model for the Cdc42-dependent regulation of Ste20 at the plasma membrane. In mutants lacking a functional BR domain (ΔBR) or a functional CRIB domain (SH), membrane-localized activation of Ste20 is impaired. Only when both domains are intact (WT) is Ste20 efficiently localized to the plasma membrane, allowing kinase activation and efficient access to cortical substrates.
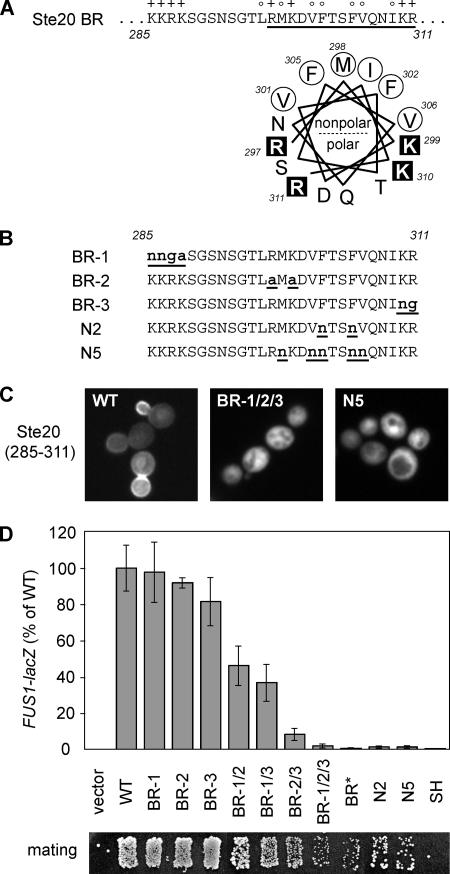
The Function of the BR Domain Requires Both Basic and Hydrophobic Residues
Given the critical functional role of the Ste20 BR domain, we probed its sequence requirements in greater detail. The basic residues in the BR domain are distributed in three small clusters of 2–4 basic residues each (Figure 4A). In addition, the C-terminal half of the BR domain is enriched in hydrophobic residues and has the potential to form an amphipathic α-helix (Figure 4A), suggesting that the BR domain might insert into the inner leaflet of the plasma membrane (Hristova et al., 1999). To compare the role of basic and hydrophobic residues, we made additional mutations (Figure 4B): one set changed each cluster of basic residues, either individually (BR-1, -2, and -3) or in combination; another set used polar Asn residues to replace hydrophobic residues at either two (N2) or five (N5) positions. When introduced into the isolated BR domain (residues 285-311), the BR-1/2/3 and N5 mutations each eliminated membrane targeting (Figure 4C), indicating that both basic and hydrophobic residues are required. In full-length Ste20, mutations in hydrophobic residues (N2, N5) severely disrupted pheromone signaling (Figure 4D). Mutations in basic residues had a milder effect on a per-residue basis, but they showed an additive effect in which signaling became gradually more disrupted as more residues were mutated (Figure 4D). Although they changed fewer basic residues, the BR-2 and -3 mutations had stronger effects on signaling than the BR-1 mutation, either alone or when combined in double mutants (e.g., compare BR-2/3 with BR-1/2 and -1/3). This result correlates with the fact that the BR-2 and -3 clusters lie within the potential amphipathic α-helix and in the more conserved portion of the BR domain (see Supplementary Figure S1). Taken together, these findings show that in addition to basic residues, the hydrophobic residues in the BR domain are critical to both its membrane-targeting ability and its functional role in Ste20 signaling.
Figure 4.
BR domain function requires both basic and hydrophobic residues. (A) At top, basic and hydrophobic residues in the Ste20 BR domain are indicated with + and ○, respectively. At bottom, a helical wheel projection of the underlined sequence shows the potential for forming an amphipathic α-helix. (B) BR-1, -2, and -3 mutants contain changes at individual clusters of 2–4 basic residues each. N2 and N5 mutants contain Asn replacements of 2 or 5 hydrophobic residues, respectively. Additional derivatives that combine mutations at multiple basic clusters are denoted by slashes (BR-1/2, -1/3, -2/3, and -1/2/3) in C and D. (C) Effects of the most extreme mutations (BR-1/2/3 and N5) on localization of the isolated Ste20 BR domain (expressed as a GST-GFP fusion). Plasmids pPP2433, pPP2531, and pPP2529 were analyzed in wild-type (WT) cells (PPY1368). (D) Mating pathway phenotypes. Pheromone response was tested using ste20Δ cells (PPY913) harboring GFP-Ste20 derivatives (pRS316, pPP538, pPP2430, pPP2431, pPP2462, pPP2469, pPP2470, pPP2471, pPP2472, pPP2204, pPP2436, pPP2461, and pPP1009). Results (mean ± SD; n = 4–12) were normalized to wild-type (WT; 100% = 46.4 β-galactosidase units). Below, patch mating tests of the same transformants are shown.
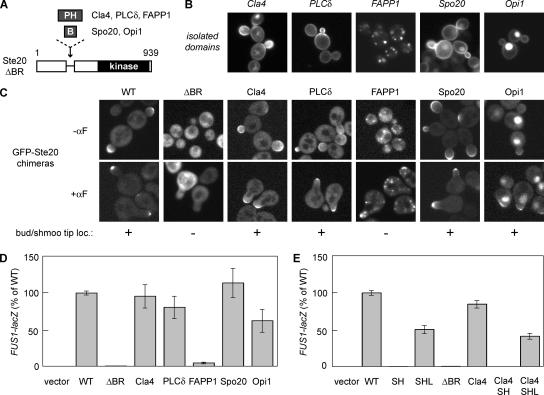
Substitution of the Ste20 BR Domain with Foreign Membrane-binding Domains
To rigorously test whether Ste20 requires the BR domain for membrane binding rather than for binding to an unknown protein partner, we replaced the BR domain with heterologous membrane-binding motifs (Figure 5A). We used PH domains from mammalian PLCδ (which binds PIP2), mammalian FAPP1 (which binds PI4P and PIP2), and yeast Cla4 (which binds multiple phospholipids; Kavran et al., 1998; Levine and Munro, 2002; Wild et al., 2004), as well as BR motifs from yeast Spo20 (which prefers PA) and yeast Opi1 (which binds PA and mediates both membrane and nuclear localization; Loewen et al., 2004; Nakanishi et al., 2004). When expressed as isolated domains tagged with GFP, each of these domains localized in a manner consistent with previous studies (Figure 5B), although in several cases they showed polarized localization that was not reported previously (see Discussion).
Figure 5.
Functional replacement of the Ste20 BR domain with heterologous membrane-binding domains. (A) Schematic diagram of chimeras in which the BR domain of Ste20 was replaced by PH domains from Cla4, PLCδ, and FAPP1, or by basic domains from Spo20 and Opi1. (B) Localization of the isolated membrane-binding domains expressed as galactose-inducible GST-GFP fusions (Cla4, PLCδ, and FAPP1) or GFP fusions (Spo20 and Opi1). PPY1368 cells harbored pPP2407, pPP2480, pPP2943, pPP2426, or pPP2418. (C) Localization of GFP-Ste20 chimeras (pPP538, pPP2318, pPP2317, pPP2455, pPP2456, pPP2459, and pPP2458) in ste20Δ cells (PPY1249) during vegetative growth (−αF), and in wild-type cells (PPY1114) treated with α factor (+αF). (D) Pheromone response of ste20Δ cells (PPY913) carrying GFP-Ste20 chimeras (pRS316, pPP538, pPP2318, pPP2317, pPP2455, pPP2456, pPP2459, and pPP2458). Results (mean ± SD, n = 8) were normalized to wild-type (WT; 100% = 60.8 β-galactosidase units). (E) Signaling by the Ste20Cla4 chimera still requires Cdc42 binding. The effects of mutations in the CRIB domain (SH and SHL) were compared when the native BR domain was either intact or replaced with the Cla4 PH domain. Pheromone response was tested in ste20Δ cells (PPY913) harboring the indicated GFP-Ste20 derivatives (pRS316, pPP538, pPP1009, pPP1109, pPP2318, pPP2317, pPP2325, or pPP2342). Results (mean ± SD; n = 6) were normalized to wild-type (WT; 100% = 64.3 β-galactosidase units).
Chimeras containing these foreign domains in place of the Ste20 BR domain were expressed from the native Ste20 promoter and tested for localization and pheromone response. Except for the FAPP1 domain, which caused strong localization to Golgi membranes, each of the foreign domains restored Ste20 localization to bud tips and to shmoo tips in pheromone-treated cells (Figure 5C). In addition, the Opi1 chimera showed strong nuclear localization, consistent with the dual targeting activity of the isolated Opi1 domain (Loewen et al., 2004). In pheromone response assays, signaling was restored to levels that agreed with the localization of each chimera (Figure 5D). Namely, the FAPP1 chimera signaled very poorly, whereas those that localized to the plasma membrane functioned at or near wild-type levels. Thus, the BR domain of Ste20 is functionally replaceable by other membrane-binding motifs that can mediate plasma membrane localization.
In principle, the foreign domains could rescue the signaling defect of the Ste20ΔBR mutant by disrupting the autoinhibitory conformation, rather than by restoring interaction with Cdc42. To distinguish between these possibilities, we tested whether Cdc42 binding was still required in the Ste20Cla4 chimera, by introducing the CRIB domain mutations S338A H345G. This new mutant (Cla4/SH) was defective at signaling (Figure 5E), whereas this defect was reversed in a further mutant (Cla4/SHL) in which autoinhibition was disrupted by the additional mutation L369G (Lamson et al., 2002). Therefore, as with wild-type Ste20, the chimeras are still autoinhibited and are still activated by Cdc42 binding. A related approach suggests that Golgi mislocalization of the Ste20FAPP1 chimera disrupts both activation by Cdc42 and postactivation signaling efficiency (see Supplementary Figure S2). Taken together, these results show that the role of the Ste20 BR domain is to provide a membrane-binding activity that promotes regulation of Ste20 by membrane-localized Cdc42. They also indicate that the function of the BR domain does not require a specific protein structure or binding to a specific phospholipid, as long as it can localize the kinase to the plasma membrane.
A Motif in the Ste20 N-Terminus Affects Cell Morphology
The original overexpressed Ste20 fragment that led to the identification of the BR domain (1-333) also induced elongated bud morphology (see Figure 1A). On further dissection, we found that this effect required sequences between residues 72 and 120 (Figure 6A), which overlaps a region of strong local sequence conservation among fungal Ste20 orthologues (see Supplementary Figure S1). Interestingly, the morphological phenotype required the Ste20 N-terminus to be targeted to the plasma membrane (Figure 6A), either by the native BR domain or by heterologous membrane-targeting motifs such as a carboxyl-terminal prenylation and palmitoylation motif (Cpr) from yeast Ras2 (Pryciak and Huntress, 1998); mutations in either targeting motif (BR* or Cpr-SS) eliminated the morphological effect (Figure 6A, iv and vii). As with other morphogenesis defects (Lew, 2003), the bud elongation phenotype was dependent on the checkpoint kinase Swe1 (data not shown). A simple explanation for these observations is that a Ste20 domain involving residues 72-120 interacts with an unknown factor at the cell cortex and interferes with its normal function, resulting in defective bud morphogenesis. In mammalian Cdc42 targets of the WASP family, sequences in the analogous position provide binding sites for auxiliary regulatory factors (Zettl and Way, 2002). However, we were unable to identify a candidate binding partner for this Ste20 domain by a two-hybrid library screen (data not shown). Furthermore, a variety of mutations impinging on this N-terminal region had no effect on the activity of full-length Ste20 in pheromone response (Figure 6B) or in other assays such as agar invasion or growth in the absence of Cla4 (data not shown). Therefore, although intriguing, the normal function of this Ste20 domain remains unclear and was not pursued further.
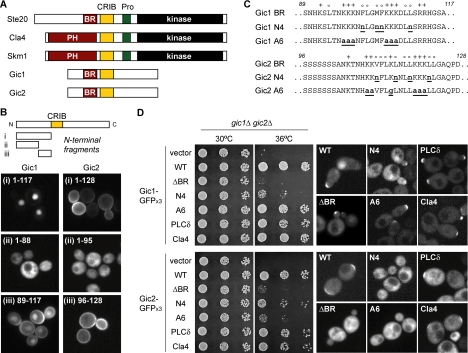
BR Domains in the Yeast Cdc42 Effectors Gic1 and Gic2
The finding that Ste20 function is critically dependent on a small, previously unrecognized membrane-binding domain raised the possibility that similar domains might be required in other yeast Cdc42 binding partners or in other polarized proteins. In S. cerevisiae, there are five Cdc42 effectors with recognizable CRIB domains (Figure 7A). Interestingly, Cla4 and Skm1 have PH domains at a position similar to that of the Ste20 BR domain, and the PH domain of Cla4 is essential for its function (Benton et al., 1997; Wild et al., 2004). Therefore, we wondered if the other two Cdc42 targets, Gic1 and Gic2, might also contain membrane-binding domains in an analogous position. To test this idea, we expressed GST-GFP fusions to N-terminal fragments of Gic1 and Gic2 and examined their membrane-targeting activity in vivo (Figure 7B). Indeed, domains from both proteins conferred membrane localization, and in each case the responsible sequence mapped to a short (29-33 a.a.) motif adjacent to the CRIB domain. As with Ste20, these Gic1 and Gic2 domains were found to be rich in basic and hydrophobic residues (Figure 7C), and hence were named “BR” domains. Notably, the Gic1 BR domain (but not the Gic2 BR domain) also showed nuclear-targeting activity, the strength of which was influenced by adjacent sequences (Figure 7B). Related dual targeting behavior has been found recently for similar domains in Opi1 and Ste5 (Loewen et al., 2004; Winters et al., 2005), and was also weakly apparent for the Ste20 BR domain (see Figure 1A, vi and vii). In addition to their targeting activity as isolated motifs, the BR domains from both Gic1 and Gic2 could potently substitute for the Ste20 BR domain in pheromone response assays (Supplementary Figure S3).
Figure 7.
BR domains in the yeast Cdc42 effectors Gic1 and Gic2. (A) Domain organization of CRIB domain-containing Cdc42 effectors in budding yeast. Data in B–D reveal the presence of functional BR domains in Gic1 and Gic2. (B) Identification of membrane-targeting domains in Gic1 and Gic2. GST-GFP fusions to N-terminal fragments of Gic1 and Gic2 were expressed in wild-type cells (PPY1368). Plasmids used were pPP2528, pPP2533, pPP2481, pPP2419, pPP2535, and pPP2477. (C) Mutations in the BR domains of Gic1 and Gic2. Basic and hydrophobic residues are indicated with + and ○, respectively. (D) The BR domains of Gic1 and Gic2 are essential for their functions. The left panels show fivefold serial dilutions of CCY1033–5D (gic1Δ gic2Δ) carrying various Gic1-GFPx3 and Gic2-GFPx3 alleles, after incubation for 2 d at 30 or 36°C on selective media. The right panels show localization of the same derivatives expressed in wild-type cells (PPY1368). “ΔBR” denotes Gic1Δ89-117 or Gic2Δ96-128. “PLCδ” and “Cla4” denote chimeras with PH domains replacing the Gic1 or Gic2 BR domains. Gic1 plasmids were pPP2640, pPP2642, pPP2913, pPP2914, pPP2671, and pPP2672. Gic2 plasmids were pPP2641, pPP2643, pPP2915, pPP2916, pPP2673, and pPP2674.
To test their functional significance in full-length Gic1 and Gic2, we made deletions of these domains (ΔBR) and point mutations of basic or hydrophobic residues (Figure 7C); these mutants were expressed from their native promoters and tagged with three tandem copies of GFP. When assayed in gic1Δ gic2Δ strains (Figure 7D), the ΔBR mutations in both Gic1 and Gic2 disrupted their ability to support growth at the restrictive temperature (36°C), showing that the BR domains are functionally important. Mutation of basic or hydrophobic residues had distinct effects on the two proteins. In Gic1, mutation of six basic residues (A6) had no evident effect, whereas mutation of four hydrophobic residues (N4) severely reduced function (Figure 7D). Conversely, in Gic2 it was the mutation of basic residues (A6) that had the stronger effect, whereas mutation of hydrophobic residues (N4) caused a weak but detectable defect (Figure 7D). Thus, the BR domains are important for both Gic1 and Gic2, but their dependence on basic versus hydrophobic residues is not identical; this may relate to differences in the competitive effect of nuclear targeting by the two domains (see Discussion). As with Ste20, we found that replacing the BR domains in Gic1 and Gic2 with PH domains from yeast Cla4 or mammalian PLCδ reversed the growth defect of the ΔBR mutants (Figure 7D), suggesting that it is the membrane-targeting activity of the BR domains that is functionally important.
The effects of BR domain mutations on localization were notably different between Gic1 and Gic2. The results with Gic2 clearly support a role for the BR domain in polarized localization, as localization to nascent bud sites was abolished by each of the BR domain mutations (ΔBR, N4, or A6), and was restored in the Gic2PLCδ and Gic2Cla4 chimeras (Figure 7D). Notably, Gic2-GFPx3 levels were actually increased by the BR domain mutations (data not shown), which is consistent with the fact that Cdc42 binding promotes degradation of Gic2 (Jaquenoud et al., 1998). In contrast to these Gic2 phenotypes, none of the Gic1 BR mutations disrupted polarized localization (Figure 7D). However, this result is consistent with previous findings that Gic1 remains localized even when Cdc42 binding is disrupted by mutations in the CRIB domain (Chen et al., 1997); furthermore, as with the previous CRIB domain mutants, the Gic1ΔBR mutant showed enhanced localization to bud necks (data not shown). Thus, while Gic1 clearly must contain other localization information, our results agree with expectations if the Gic1 BR domain serves primarily to facilitate interaction with Cdc42. Interestingly, full-length Gic1 also shows faint nuclear localization (Chen et al., 1997; Iwase et al., 2006), and this was reduced in all mutant and chimeric forms except for the N4 mutant (Figure 7D). This indicates that nuclear localization of full-length Gic1 depends on the basic residues of the BR domain, although the functional significance of this localization is unknown (see Discussion). Despite the complex localization behavior of Gic1, our findings in total demonstrate that multiple yeast Cdc42 effectors contain membrane-binding domains next to their CRIB domains, suggesting a common mechanism for their membrane-localized regulation by Cdc42.
DISCUSSION
Prevalence of Short Membrane-binding Domains
In this study we identify short (∼30 residue) membrane-binding motifs in the budding yeast Cdc42 effectors Ste20, Gic1, and Gic2. These BR domains were previously undetected, and yet they are critical for the in vivo function of each protein. Unlike larger (e.g., 100-120 residue) lipid-binding modules such as PH domains, which have a defined tertiary structure and signature sequence motifs, the short domains found here and in other recent studies (Loewen et al., 2004; Nakanishi et al., 2004; Winters et al., 2005; Heo et al., 2006) are difficult to recognize from sequence alone. Direct comparison of these domains does not yield a clear “consensus” at the primary sequence level, although most are significantly enriched in both basic and bulky hydrophobic groups (see Supplementary Table 3). In some cases the basic residues may be clustered (Heo et al., 2006) and some may form an amphipathic α-helix (this study and Nakanishi et al., 2004; Winters et al., 2005), though the BR domain sequences from Gic1 and Gic2 suggest no propensity for this structure. Algorithms for predicting membrane-seeking peptides on the basis of hydrophobic moment (Phoenix and Harris, 2002) or comparative pattern recognition (Sapay et al., 2006) identify some of the experimentally characterized motifs, but they fail to identify others (S. Takahashi and P. M. Pryciak, unpublished data). In comparison, we have found it relatively straightforward to identify these domains by overexpressing protein fragments as dimerized GFP fusions. Because BR-like domains exist in at least two mating pathway proteins and at least three Cdc42 effectors, we suspect that they may be relatively prevalent among cortical proteins but are underappreciated because of their short, cryptic nature.
Our analysis reveals that all five CRIB-containing Cdc42 effectors in budding yeast possess membrane-binding domains (PH or BR) in the same position. This feature may be widespread among Rho GTPase targets. For example, of the two PAKs in the fission yeast Schizosaccharomyces pombe, Pak2/Shk2 has a PH domain (Sells et al., 1998), and Pak1/Shk1 has an uncharacterized BR sequence adjacent to its CRIB domain. Furthermore, uncharacterized sequences that are rich in basic and hydrophobic residues can be found adjacent to CRIB domains in mammalian PAKs 1-3 (Bokoch, 2003), mammalian Borg proteins (Joberty et al., 1999), and targets of the plant GTPase Rop1 (Wu et al., 2001; see Supplementary Table 3). In addition to these GTPase effectors, a recent study shows that polybasic domains are common in small GTPases themselves (Heo et al., 2006).
Synergistic Protein–Protein and Protein–Membrane Interactions
The overall behavior of the BR domains identified here is similar to a related domain from Ste5 (Winters et al., 2005), the mating pathway scaffold protein. In each case, the membrane-binding domain normally does not function alone but rather in conjunction with binding to a membrane protein such as Cdc42 or Gβγ. Similarly, Cdc42 binding alone may be generally insufficient for localization, as fragments containing only the Cdc42-binding domains from Cla4 (Wild et al., 2004) or Ste20 (this study) cannot localize to the cell cortex. In this regard it is noteworthy that Gic2 “CRIB” fragments used as reporters for active Cdc42 during yeast cell polarization (Ozbudak et al., 2005) include both the CRIB domain and the BR domain identified here.
The mammalian Cdc42 effector N-WASP, a regulator of actin assembly, provides a well-studied paradigm for synergism between protein and phospholipid binding (Prehoda et al., 2000; Rohatgi et al., 2000). N-WASP activation requires Cdc42 to bind in concert with PIP2, which acts through a polybasic domain located similarly to the BR domains of yeast Ste20, Gic1, and Gic2. However, there are notable differences between these domains. First, the N-WASP domain consists almost entirely of basic residues, and its PIP2-binding properties can be mimicked by a run of 10 consecutive lysines (Papayannopoulos et al., 2005), whereas the yeast BR domains require both basic and hydrophobic residues. In fact, the N-WASP domain does not localize to the plasma membrane when expressed in yeast, and it cannot restore signaling when used to replace the Ste20 BR domain (S. Takahashi and P. M. Pryciak, unpublished data). Second, the polybasic domain of N-WASP has been proposed to interact transiently with acidic residues in Cdc42 and thereby accelerate CRIB-Cdc42 binding by “electrostatic steering” (Hemsath et al., 2005). In contrast, it seems unlikely that the yeast BR domains need to directly contact Cdc42, given the ability of PH domains to substitute for their function. Rather, our findings suggest a simpler general model in which the BR domain provides membrane affinity, and this promotes binding between the CRIB domain and membrane-localized Cdc42.
Cooperative binding of proteins to both Cdc42 and a specific phospholipid offers the potential for “coincidence detection,” in which two separate signal inputs are integrated together (Prehoda et al., 2000; Wild et al., 2004). Although an attractive notion, there is little evidence that modulation of phospholipid levels is a significant regulator of Cdc42 targets in vivo, and our chimeric proteins suggest that lipid specificity is not critical for Ste20, Gic1, or Gic2 function. Nevertheless, cooperative binding can still be advantageous even without lipid specificity by allowing the protein–protein interaction affinity to be weak, which may permit more dynamic sampling of the status of the protein partner (Fivaz and Meyer, 2003). Indeed, even during the slow protrusive growth of yeast cells, polarized Cdc42 targets can be extremely dynamic (Ozbudak et al., 2005).
Roles for Charge and Hydrophobicity
We found that multiple basic residues in the Ste20 BR domain contribute additively to its function. A similar additive effect was seen in tests of PIP2 binding by basic domains from mammalian MARCKS and N-WASP (Wang et al., 2002; Papayannopoulos et al., 2005). These behaviors are consistent with multivalent electrostatic interaction between a polycationic protein domain and a polyanionic membrane surface (Papayannopoulos et al., 2005). However, different basic residues can make unequal contributions, as mutation of four basic residues in the first Ste20 cluster (BR-1) had a much weaker effect than mutation of four basic residues in the second and third clusters (BR-2/3), perhaps because the latter region is part of the predicted amphipathic helix. In addition, BR-like domains can be remarkably tolerant of reduced positive charge, because loss of three, four, or even six basic residues in Ste20 or Gic1 (this study) or in Ste5 (Winters et al., 2005) can yield a protein with largely intact function in vivo.
Furthermore, positive charge is often not sufficient. Instead, hydrophobic residues can also play a critical role in membrane binding, most likely by partitioning into the hydrophobic core of the lipid bilayer (Hristova et al., 1999; Wang et al., 2002). They may also help counteract alternative targeting effects of basic regions, such as nuclear targeting. Indeed, mutations that add or remove hydrophobic residues from mixed basic/hydrophobic domains can dramatically alter the partitioning between nucleus and plasma membrane (Winters et al., 2005; Heo et al., 2006). Thus, hydrophobic residues may be especially important when the basic residues confer affinity for other intracellular destinations. This could explain why Ste20 and Gic1 are less tolerant of losing hydrophobic residues compared with Gic2, because both the Ste20 and Gic1 BR domains show nuclear affinity, whereas the Gic2 BR domain does not. It may also help explain why the strong contribution of hydrophobicity to membrane localization in vivo is often poorly reflected during liposome-binding assays in vitro (Loewen et al., 2004; Nakanishi et al., 2004; Winters et al., 2005).
The nuclear targeting activity of several membrane-binding domains reported here and in other recent studies (Loewen et al., 2004; Winters et al., 2005; Heo et al., 2006) is curious, but in most cases the functional significance is unknown. For Ste20, nuclear translocation has been proposed to allow it to phosphorylate histone H2B and promote apoptosis in response to oxidative stress (Ahn et al., 2005), but preliminary tests suggest that neither the BR domain nor the CRIB domain is required for this process (S.-H. Ahn and C. D. Allis, personal communication). Gic1 shows some nuclear affinity, but chimeras that lack this localization still function normally (see Figure 7D). Thus, it remains unclear whether nuclear targeting by BR-like domains is mostly a gratuitous consequence of their basic content or if it provides a mechanism for sampling multiple compartments that is commonly exploited for functional purposes.
Plasma Membrane Targeting
The short membrane-binding domains identified here and elsewhere (Nakanishi et al., 2004; Winters et al., 2005) preferentially target the plasma membrane. As discussed previously (Johnson and Cornell, 1999; Lemmon, 2003), this localization need not imply selectivity for a particular lipid because the plasma membrane is the most acidic due to its enrichment in lipids such as PIP2, PA, and PS (Schneiter et al., 1999; Sprong et al., 2001; Okeley and Gelb, 2004). In vitro, the Ste20 BR domain favors liposomes containing PIP2, which is highly acidic and hence offers a strong electrostatic attraction. Yet when lipid kinase mutants are used to deplete lipid pools in vivo, the Ste20 BR domain is detectably displaced from the plasma membrane only after depletion of both PIP2 and PI(4)P (S. Takahashi and P. M. Pryciak, unpublished data), reminiscent of results with other polybasic domains (Winters et al., 2005; Heo et al., 2006). Although our chimeric proteins suggest that interaction with a specific lipid species is not required for Ste20 function in vivo, the poor signaling by the Golgi-localized Ste20FAPP1 chimera underscores the utility of preferential affinity for the plasma membrane. The degree to which this is important for any given protein may depend on the other interactions that synergistically control its localization. Indeed, when the same FAPP1 PH domain was used to replace a membrane-binding domain in Ste5, the chimera could be diverted to the plasma membrane in response to Gβγ activation (Winters et al., 2005); this difference could be due to a stronger protein–protein interaction or to the presence of a second membrane-binding domain in Ste5 (Garrenton et al., 2006).
Interestingly, many of the isolated membrane-targeting domains localized asymmetrically to the membrane of growing buds. A similar pattern was seen with a domain from Ste5 (Winters et al., 2005), and the new examples show that it is not uncommon; in fact, we often observed this behavior even for domains (e.g., PLCδ, Cla4, and Spo20) that showed uniform localization in previous studies (Stefan et al., 2002; Nakanishi et al., 2004; Wild et al., 2004). The asymmetry may be more apparent in our studies because we assayed newly synthesized proteins shortly after induced expression, which might help prevent saturation of preferred binding sites and/or minimize the effects of diffusion (Valdez-Taubas and Pelham, 2003). It is possible that the asymmetry reflects a polarized distribution of specific lipids such as PIP2, PA, or PS. Alternatively, these domains might tend to bind membranes in the secretory pathway and thereby become transported to sites of polarized growth. In principle, either explanation could contribute to proper cell polarity control, which has been proposed to use a positive-feedback mechanism in which polarity regulators such as Cdc42 are delivered to sites of polarization by directional secretion (Wedlich-Soldner et al., 2004).
In conclusion, this study uncovers several new examples of an emerging group of short membrane-binding motifs that play essential roles in signaling and polarized localization and suggests a common mechanism in which general membrane-targeting motifs work in conjunction with specific protein–protein interactions in order to regulate the function of Cdc42 targets and other cortical factors.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Winters for assistance with early stages of this project, D. McCollum and N. Rhind for comments on the manuscript, and T. Hoefken (University of Kiel, Germany) and C. Chan (University of Texas, Austin, TX) for strains and plasmids, as well as S.-H. Ahn and C. D. Allis for communicating unpublished results. Finally, we thank H. Nakanishi, E. Luna, J. McNally, and J. DiNitto for assistance and advice with liposome assays. This work was supported by a grant from the National Institutes of Health (GM57769) to P.M.P.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0676) on October 3, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Ahn S. H., Cheung W. L., Hsu J. Y., Diaz R. L., Smith M. M., Allis C. D. Sterile 20 kinase phosphorylates histone H2B at serine 10 during hydrogen peroxide-induced apoptosis in S. cerevisiae. Cell. 2005;120:25–36. doi: 10.1016/j.cell.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Ash J., Wu C., Larocque R., Jamal M., Stevens W., Osborne M., Thomas D. Y., Whiteway M. Genetic analysis of the interface between Cdc42p and the CRIB domain of Ste20p in Saccharomyces cerevisiae. Genetics. 2003;163:9–20. doi: 10.1093/genetics/163.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton B. K., Tinkelenberg A., Gonzalez I., Cross F. R. Cla4p, a Saccharomyces cerevisiae Cdc42p-activated kinase involved in cytokinesis, is activated at mitosis. Mol. Cell. Biol. 1997;17:5067–5076. doi: 10.1128/mcb.17.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokoch G. M. Biology of the p21-activated kinases. Annu. Rev. Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- Brown J. L., Jaquenoud M., Gulli M. P., Chant J., Peter M. Novel Cdc42-binding proteins Gic1 and Gic2 control cell polarity in yeast. Genes Dev. 1997;11:2972–2982. doi: 10.1101/gad.11.22.2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. C., Kim Y. J., Chan C. S. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae. Genes Dev. 1997;11:2958–2971. doi: 10.1101/gad.11.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W. Building signaling complexes at the membrane. Sci. STKE. 2006;2006:pe7. doi: 10.1126/stke.3212006pe7. [DOI] [PubMed] [Google Scholar]
- Cvrckova F., De Virgilio C., Manser E., Pringle J. R., Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- Dohlman H. G., Thorner J. W. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 2001;70:703–754. doi: 10.1146/annurev.biochem.70.1.703. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. Cdc42—the centre of polarity. J. Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- Fivaz M., Meyer T. Specific localization and timing in neuronal signal transduction mediated by protein-lipid interactions. Neuron. 2003;40:319–330. doi: 10.1016/s0896-6273(03)00634-2. [DOI] [PubMed] [Google Scholar]
- Garrenton L. S., Young S. L., Thorner J. Function of the MAPK scaffold protein, Ste5, requires a cryptic PH domain. Genes Dev. 2006;20:1946–1958. doi: 10.1101/gad.1413706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A. S., Moskow J. J., Zyla T. R., Lew D. J. Isolation and characterization of effector-loop mutants of cdc42 in yeast. Mol. Biol. Cell. 2001;12:1239–1255. doi: 10.1091/mbc.12.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsath L., Dvorsky R., Fiegen D., Carlier M. F., Ahmadian M. R. An electrostatic steering mechanism of Cdc42 recognition by Wiskott-Aldrich syndrome proteins. Mol. Cell. 2005;20:313–324. doi: 10.1016/j.molcel.2005.08.036. [DOI] [PubMed] [Google Scholar]
- Heo W. D., Inoue T., Park W. S., Kim M. L., Park B. O., Wandless T. J., Meyer T. PI(3,4,5)P3 and PI(4,5)P2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314:1458–1461. doi: 10.1126/science.1134389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofken T., Schiebel E. A role for cell polarity proteins in mitotic exit. EMBO J. 2002;21:4851–4862. doi: 10.1093/emboj/cdf481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holly S. P., Blumer K. J. PAK-family kinases regulate cell and actin polarization throughout the cell cycle of Saccharomyces cerevisiae. J. Cell Biol. 1999;147:845–856. doi: 10.1083/jcb.147.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristova K., Wimley W. C., Mishra V. K., Anantharamiah G. M., Segrest J. P., White S. H. An amphipathic α-helix at a membrane interface: a structural study using a novel X-ray diffraction method. J. Mol. Biol. 1999;290:99–117. doi: 10.1006/jmbi.1999.2840. [DOI] [PubMed] [Google Scholar]
- Hurley J. H. Membrane binding domains. Biochim. Biophys. Acta. 2006;1761:805–811. doi: 10.1016/j.bbalip.2006.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase M., Luo J., Nagaraj S., Longtine M., Kim H. B., Haarer B. K., Caruso C., Tong Z., Pringle J. R., Bi E. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol. Biol. Cell. 2006;17:1110–1125. doi: 10.1091/mbc.E05-08-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquenoud M., Gulli M. P., Peter K., Peter M. The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J. 1998;17:5360–5373. doi: 10.1093/emboj/17.18.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joberty G., Perlungher R. R., Macara I. G. The Borgs, a new family of Cdc42 and TC10 GTPase-interacting proteins. Mol. Cell. Biol. 1999;19:6585–6597. doi: 10.1128/mcb.19.10.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. I. Cdc42, An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol. Mol. Biol. Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. E., Cornell R. B. Amphitropic proteins: regulation by reversible membrane interactions (review) Mol. Membr. Biol. 1999;16:217–235. doi: 10.1080/096876899294544. [DOI] [PubMed] [Google Scholar]
- Kavran J. M., Klein D. E., Lee A., Falasca M., Isakoff S. J., Skolnik E. Y., Lemmon M. A. Specificity and promiscuity in phosphoinositide binding by pleckstrin homology domains. J. Biol. Chem. 1998;273:30497–30508. doi: 10.1074/jbc.273.46.30497. [DOI] [PubMed] [Google Scholar]
- Kholodenko B. N., Hoek J. B., Westerhoff H. V. Why cytoplasmic signalling proteins should be recruited to cell membranes. Trends Cell Biol. 2000;10:173–178. doi: 10.1016/s0962-8924(00)01741-4. [DOI] [PubMed] [Google Scholar]
- Kim A. S., Kakalis L. T., Abdul-Manan N., Liu G. A., Rosen M. K. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- Kushnirov V. V. Rapid and reliable protein extraction from yeast. Yeast. 2000;16:857–860. doi: 10.1002/1097-0061(20000630)16:9<857::AID-YEA561>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Lamson R. E., Winters M. J., Pryciak P. M. Cdc42 regulation of kinase activity and signaling by the yeast p21-activated kinase Ste20. Mol. Cell. Biol. 2002;22:2939–2951. doi: 10.1128/MCB.22.9.2939-2951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Dignard D., Harcus D., Thomas D. Y., Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein βγ subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E., Wu C., Leeuw T., Fourest-Lieuvin A., Segall J. E., Thomas D. Y. Functional characterization of the Cdc42p binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw T., Wu C., Schrag J. D., Whiteway M., Thomas D. Y., Leberer E. Interaction of a G-protein β-subunit with a conserved sequence in Ste20/PAK family protein kinases. Nature. 1998;391:191–195. doi: 10.1038/34448. [DOI] [PubMed] [Google Scholar]
- Lemmon M. A. Phosphoinositide recognition domains. Traffic. 2003;4:201–213. doi: 10.1034/j.1600-0854.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- Levine T. P., Munro S. Targeting of Golgi-specific pleckstrin homology domains involves both PtdIns 4-kinase-dependent and -independent components. Curr. Biol. 2002;12:695–704. doi: 10.1016/s0960-9822(02)00779-0. [DOI] [PubMed] [Google Scholar]
- Lew D. J. The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 2003;15:648–653. doi: 10.1016/j.ceb.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Liu H., Styles C. A., Fink G. R. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- Loewen C. J., Gaspar M. L., Jesch S. A., Delon C., Ktistakis N. T., Henry S. A., Levine T. P. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- Morreale A., Venkatesan M., Mott H. R., Owen D., Nietlispach D., Lowe P. N., Laue E. D. Structure of Cdc42 bound to the GTPase binding domain of PAK. Nat. Struct. Biol. 2000;7:384–388. doi: 10.1038/75158. [DOI] [PubMed] [Google Scholar]
- Moskow J. J., Gladfelter A. S., Lamson R. E., Pryciak P. M., Lew D. J. Role of Cdc42p in pheromone-stimulated signal transduction in Saccharomyces cerevisiae. Mol. Cell. Biol. 2000;20:7559–7571. doi: 10.1128/mcb.20.20.7559-7571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H., de los Santos P., Neiman A. M. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol. Biol. Cell. 2004;15:1802–1815. doi: 10.1091/mbc.E03-11-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke S. M., Herskowitz I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 1998;12:2874–2886. doi: 10.1101/gad.12.18.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okeley N. M., Gelb M. H. A designed probe for acidic phospholipids reveals the unique enriched anionic character of the cytosolic face of the mammalian plasma membrane. J. Biol. Chem. 2004;279:21833–21840. doi: 10.1074/jbc.M313469200. [DOI] [PubMed] [Google Scholar]
- Ozbudak E. M., Becskei A., van Oudenaarden A. A system of counteracting feedback loops regulates Cdc42p activity during spontaneous cell polarization. Dev. Cell. 2005;9:565–571. doi: 10.1016/j.devcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V., Co C., Prehoda K. E., Snapper S., Taunton J., Lim W. A. A polybasic motif allows N-WASP to act as a sensor of PIP2 density. Mol. Cell. 2005;17:181–191. doi: 10.1016/j.molcel.2004.11.054. [DOI] [PubMed] [Google Scholar]
- Park H. O., Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawson T., Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Peter M., Neiman A. M., Park H. O., van Lohuizen M., Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- Phoenix D. A., Harris F. The hydrophobic moment and its use in the classification of amphiphilic structures (review) Mol. Membr. Biol. 2002;19:1–10. doi: 10.1080/09687680110103631. [DOI] [PubMed] [Google Scholar]
- Prehoda K. E., Scott J. A., Mullins R. D., Lim W. A. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science. 2000;290:801–806. doi: 10.1126/science.290.5492.801. [DOI] [PubMed] [Google Scholar]
- Pryciak P. M., Huntress F. A. Membrane recruitment of the kinase cascade scaffold protein Ste5 by the Gβγ complex underlies activation of the yeast pheromone response pathway. Genes Dev. 1998;12:2684–2697. doi: 10.1101/gad.12.17.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raitt D. C., Posas F., Saito H. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 2000;19:4623–4631. doi: 10.1093/emboj/19.17.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecchi M., Peterson A., McLaughlin S. Phosphoinositide-specific phospholipase C-delta 1 binds with high affinity to phospholipid vesicles containing phosphatidylinositol 4,5-bisphosphate. Biochemistry. 1992;31:12742–12747. doi: 10.1021/bi00166a005. [DOI] [PubMed] [Google Scholar]
- Rohatgi R., Ho H. Y., Kirschner M. W. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J. Cell Biol. 2000;150:1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Fink G. R. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Sapay N., Guermeur Y., Deleage G. Prediction of amphipathic in-plane membrane anchors in monotopic proteins using a SVM classifier. BMC Bioinformatics. 2006;7:255. doi: 10.1186/1471-2105-7-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R., et al. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciorra V. A., Rudge S. A., Prestwich G. D., Frohman M. A., Engebrecht J., Morris A. J. Identification of a phosphoinositide binding motif that mediates activation of mammalian and yeast phospholipase D isoenzymes. EMBO J. 1999;18:5911–5921. doi: 10.1093/emboj/18.21.5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells M. A., Barratt J. T., Caviston J., Ottilie S., Leberer E., Chernoff J. Characterization of Pak2p, a pleckstrin homology domain-containing, p21- activated protein kinase from fission yeast. J. Biol. Chem. 1998;273:18490–18498. doi: 10.1074/jbc.273.29.18490. [DOI] [PubMed] [Google Scholar]
- Sprong H., van der Sluijs P., van Meer G. How proteins move lipids and lipids move proteins. Nat. Rev. Mol. Cell Biol. 2001;2:504–513. doi: 10.1038/35080071. [DOI] [PubMed] [Google Scholar]
- Stefan C. J., Audhya A., Emr S. D. The yeast synaptojanin-like proteins control the cellular distribution of phosphatidylinositol (4,5)-bisphosphate. Mol. Biol. Cell. 2002;13:542–557. doi: 10.1091/mbc.01-10-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teruel M. N., Meyer T. Translocation and reversible localization of signaling proteins: a dynamic future for signal transduction. Cell. 2000;103:181–184. doi: 10.1016/s0092-8674(00)00109-4. [DOI] [PubMed] [Google Scholar]
- Valdez-Taubas J., Pelham H. R. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr. Biol. 2003;13:1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Wang J., Gambhir A., Hangyas-Mihalyne G., Murray D., Golebiewska U., McLaughlin S. Lateral sequestration of phosphatidylinositol 4,5-bisphosphate by the basic effector domain of myristoylated alanine-rich C kinase substrate is due to nonspecific electrostatic interactions. J. Biol. Chem. 2002;277:34401–34412. doi: 10.1074/jbc.M203954200. [DOI] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Wai S. C., Schmidt T., Li R. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J. Cell Biol. 2004;166:889–900. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild A. C., Yu J. W., Lemmon M. A., Blumer K. J. The p21-activated protein kinase-related kinase Cla4 is a coincidence detector of signaling by Cdc42 and phosphatidylinositol 4-phosphate. J. Biol. Chem. 2004;279:17101–17110. doi: 10.1074/jbc.M314035200. [DOI] [PubMed] [Google Scholar]
- Winters M. J., Lamson R. E., Nakanishi H., Neiman A. M., Pryciak P. M. A membrane binding domain in the Ste5 scaffold synergizes with Gβγ binding to control localization and signaling in pheromone response. Mol. Cell. 2005;20:21–32. doi: 10.1016/j.molcel.2005.08.020. [DOI] [PubMed] [Google Scholar]
- Winters M. J., Pryciak P. M. Interaction with the SH3 domain protein Bem1 regulates signaling by the Saccharomyces cerevisiae p21-activated kinase Ste20. Mol. Cell. Biol. 2005;25:2177–2190. doi: 10.1128/MCB.25.6.2177-2190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Gu Y., Li S., Yang Z. A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell. 2001;13:2841–2856. doi: 10.1105/tpc.010218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettl M., Way M. The WH1 and EVH1 domains of WASP and Ena/VASP family members bind distinct sequence motifs. Curr. Biol. 2002;12:1617–1622. doi: 10.1016/s0960-9822(02)01112-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.