Abstract
Gastrin, a gastrointestinal hormone responsible for gastric acid secretion, has been confirmed as a growth factor for gastrointestinal tract malignancies. High expression of gastrin mRNA was observed in pancreatic and colorectal cancer; however, the mechanism is unclear. Epidermal growth factor (EGF) was found to increase gastrin mRNA stability, indicating mRNA turnover regulation mechanism is involved in the control of gastrin mRNA expression. Using biotin-labeled RNA probe pull-down assay combined with mass spectrometry analysis, we identified the heterogeneous nuclear ribonucleoprotein K (hnRNP K) and poly(C) binding protein 1 (PCBP1) bound with the C-rich region in gastrin mRNA 3′ untranslated region. Nucleolin bound with the AGCCCU motif and interacted with hnRNP K were also demonstrated. Under EGF treatment, we observed the amount of nucleolin interacting with hnRNP K and gastrin mRNA increased. Using small interfering RNA technology to define their functional roles, we found hnRNP K, PCBP1, and nucleolin were all responsible for stabilizing gastrin mRNA. Moreover, nucleolin plays a crucial role in mediating the increased gastrin mRNA stability induced by EGF signaling. Besides, we also observed hnRNP K/PCBP1 complex bound with the C-rich region in the gastrin mRNA increased nucleolin binding with gastrin mRNA. Finally, a novel binding model was proposed.
INTRODUCTION
Regulation of gene expression is essential for the homeostasis of an organism. Transcriptional regulation is well documented in controlling gene expression; however, it is becoming increasingly clear that regulation of mRNA decay rates is also an important control point in determining the abundance of cellular transcripts (Wilusz et al., 2001; Wilusz and Wilusz, 2004). The aberrant control of mRNA turnover has been implicated in disease states, including cancer, chronic inflammatory responses, and coronary disease (Hollams et al., 2002; Audic and Hartley, 2004). For example, in myeloma and human T-cell leukemia, the c-myc gene is mutated due to a translocation or loss of the 3′ untranslated regions (UTRs). This renders the c-myc mRNA up to 7 times more stable than the wild type (Hollis et al., 1988; Aghib et al., 1990).
mRNA turnover rate is mainly specified by control elements that are usually located within the 3′UTR of mRNAs and they are recognized by various RNA-binding proteins. The most well studied specific cis-acting element controlling the half-life of mRNA is the adenylate- and uridylate-rich (AU-rich) element (ARE) found in 3′UTR of a variety of short-lived mRNAs, including those encoding cytokines, lymphokines, proto-oncogenes, and growth factors (Chen and Shyu, 1995; Wilusz et al., 2001). A plethora of ARE-binding proteins have been identified, characterized, and cloned, including AUF1, HuR, TIA-1, and tristetraprolin, which exerted either negative or positive effects on mRNA stability (Brennan and Steitz, 2001; Bevilacqua et al., 2003). C-rich element has also been reportedly involved in the regulation of mRNA stability. Three discontinuous C-rich elements within the 3′UTR of α-globin mRNA are linked to its high-level accumulation in erythroid cells (Weiss and Liebhaber, 1995). The poly(C) binding proteins, including PCBP1 (αCP-1), and PCBP2 (αCP-2), were proved to bind with these motifs to prevent mRNA degradation (Wang et al., 1995).
Gastrin is a classical gut peptide hormone, which was identified originally as a stimulant of gastric acid secretion. It is normally produced at high levels by endocrine (G) cells located in the gastric antrum and proximal duodenal mucosa, although lower levels have been detected in the colon and pancreas (Dockray et al., 2005). Besides, gastrin has been proven to play a crucial role in the development of gastric carcinoids. Up-regulation of gastrin gene expression is reported in numerous cancers, including colorectal, pancreatic, and lung cancers (Rehfeld et al., 1989; Aly et al., 2004; Ferrand and Wang, 2006). Overexpression of gastrin in transgenic animals induces increased colonic proliferation and accelerates the development of colon cancer (Wang et al., 1996; Cobb et al., 2004). The higher expression level of gastrin by human lung cancers was associated with a significantly decreased survival in patients (Koh et al., 2004). Recent studies further highlight that the Ras, transforming growth factor-β, and Wnt/β-catenin signaling pathway involved in the gastrin gene expression, indicate that gastrin is in fact a downstream target of multiple oncogenic pathways (Lei et al., 2004; Chakladar et al., 2005).
Although the phenomenon of higher gastrin mRNA expression exists in different kinds of cancers, the detailed mechanism involved in the up-regulation of gastrin is not very clear. It has been reported that epidermal growth factor (EGF) receptor activation is a major regulator of gastrin gene expression and that the transcription factor specificity protein 1 is the most likely target of the EGF receptor signaling cascade that in turn binds to and transactivates the gastrin promoter (Ford et al., 1997). However, no information is known about the posttranscriptional regulation mechanism involved in gastrin gene expression. Here, we found that gastrin mRNA stability increased under EGF treatment. This indicates that mRNA turnover regulation mechanism plays a role in regulating the gastrin mRNA expression. Moreover, we identified heterogeneous nuclear ribonucleoprotein K (hnRNP K), poly(C) binding protein 1 (PCBP1), and nucleolin were involved in the regulation of gastrin mRNA turnover and contributed to the increased gastrin mRNA expression.
MATERIALS AND METHODS
Materials
Human EGF was purchased from Peprotech (Rocky Hill, NJ). Actinomycin D obtained from Sigma-Aldrich (St. Louis, MO). Antibodies purchased from Santa Cruz Biotechnology (Santa Cruz, CA) included mouse rabbit polyclonal anti-human nucleolin (H-250), goat polyclonal anti-human PCBP1 (T-18), and monoclonal anti-hnRNP K antibodies (D6). Rabbit polyclonal anti-human actin antibodies were purchased from Sigma-Aldrich.
Cell Culture
The experiments were performed using the human gastric adenocarcinoma AGS cell line purchased from American Type Culture Collection (Manassas, VA). AGS cells were cultured in F-12K culture medium (Biowest, Loire Valley, France) supplemented with 10% fetal bovine serum, 100 μg/ml streptomycin, and 100 U/ml penicillin. The cultures were incubated at 37°C in a humidified 5% CO2 incubator.
Subcellular Fractionation of AGS Cells
AGS cells were scraped, rinsed with phosphate-buffered saline, and lysed with cytoplasm lysis buffer (10 mM HEPES, pH 8.0, 40 mM KCl, 3 mM MgCl2, 5% glycerol, 2 mM dithiothreitol [DTT], and 0.5% NP-40) supplemented with protease inhibitors (1 mg/ml leupeptin, 1 mg/ml aprotinin, and 0.5 mM phenylmethylsulfonyl fluoride) on ice for 10 min. The cytosolic fraction was prepared by performing high-speed centrifugation (6000 rpm for 5 min at 4°C), and the supernatant was transferred to new Eppendorf and stored at −80°C. The nuclei fraction was prepared by suspended the pellet with nuclei lysis buffer (10 mM HEPES, pH 8.0, 420 mM NaCl, 1.5 mM MgCl2, 25% glycerol, and 0.5 mM DTT) on ice for 10 min. After centrifugation (6000 rpm for 5 min at 4°C), the supernatant was collected as nuclear fraction and stored at −80°C.
mRNA Turnover Analysis
Cells were grown to 70% confluence and treated with or without 10 nM EGF for 4 h, followed by addition of 5 mg/ml actinomycin D. Total RNAs were isolated from the cells at different time points after addition of actinomycin D and analyzed by quantitative real-time reverse transcription-polymerase chain reaction (RT-PCR). The mRNA half-life was determined by linear regression analysis.
Quantitative RT–PCR
The primers used are listed as follows: for gastrin mRNA, forward: 5′ GCAGC GACTATGTGTGTATGTG 3′ and reverse: 5′ TCCATCCATAGGCTTCTTCTTC 3′; and for the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH), forward: 5′ CCCACTCCTCCA CCTTTGAC 3′ and reverse: 5′ TCTCTCTTCCTCTTGTGCTCTTG 3′. Total RNAs were isolated, and the mRNA levels were measured using TITANIUM one-step RT-PCR reagent (Clontech, Mountain View, CA) and analyzed by LightCycler 480 real-time PCR system (LightCycler Instrument; Roche Molecular Biochemicals, Mannheim, Germany).
RNA Electrophoretic Mobility Shift Assay (EMSA)
To perform gel shift analysis, 20–30 μg of proteins were incubated with 50 pM biotin-labeled RNA probes in RNA-EMSA buffer [50 mM KCl, 5% (vol/vol) glycerol, 0.1% (vol/vol) NP-40, 1 mM MgCl2, 1 mM dithiothreitol, 10 mM Tris-HCl, pH 8.0, 0.5 mg/ml heparin, and 0.5 mg/ml yeast tRNA] for 30 min at room temperature. Samples were subjected to electrophoresis on a 6% native acrylamide gel (acrylamide/bisacrylamide ratio of 37.5:1), and transferred to positive-charge nylon membrane. The signals of RNA-EMSA reaction were detected by LightShift chemiluminescent kit (Pierce Chemical, Rockford, IL). For competition assays, RNA homopolymers [20 ng; poly(rU), poly(rC), and poly(rA); GE Healthcare, Chalfont, St. Giles, United Kingdom) were added to the reaction mixture for 10 min at 25°C before incubation with labeled RNA probe.
Biotin Pull-Down Assay and Mass Spectrometry Analysis
AGS cytoplasmic extract (500 μg) was incubated with 50 pM biotin-labeled RNA and rotated for 2 h at 4°C. The sample was placed on ice and irradiated with UV (240-nm UV bulb; Stratagene, La Jolla, CA) for 15 min at 4°C. The biotin-labeled RNA was isolated by streptavidin-conjugated agarose beads (Sigma-Aldrich), and washed with wash buffer (10 mM HEPES, pH 8.0, 40 mM KCl, 3 mM MgCl2, 5% glycerol, 2 mM DTT, and 2% NP-40) supplemented with protease inhibitors (1 mg/ml leupeptin, 1 mg/ml aprotinin, and 0.5 mM phenylmethylsulfonyl fluoride) for three times. The RNA–protein complex was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE), and the gel was fixed for silver staining. The protein bands of interest on the gel were excised, ground manually, and transferred into a siliconized 0.5-ml microcentrifuge tube for further identification by mass spectrometry analysis. The in-gel digestion, mass spectrometry analysis, and database search were performed as described previously (Tyan et al., 2005).
Protein Immunoprecipitation Assay
In total, 100–300 μg of AGS cytosolic extract was incubated with a specific antibody and rotated at 4°C overnight, and then 50 μl of protein A/G-agarose beads was added for 2 h at 4°C. The complexes were washed with wash buffer (10 mM HEPES, pH 8.0, 40 mM KCl, 3 mM MgCl2, 5% glycerol, 2 mM DTT, and 0.5% NP-40) twice, and then they were resuspended in 50 μl of 1× SDS sample buffer for analysis by SDS-PAGE and Western blotting.
RNA Immunoprecipitation Assay
AGS cells were grown to 70% confluence and lysed with lysis buffer (10 mM HEPES, pH 8.0, 40 mM KCl, 3 mM MgCl2, 5% glycerol, 0.5% NP-40, and 1 U/μl RNaseOUT) for 20 min on ice. The cell extract was incubated with nucleolin antibodies (H-250) or hnRNP K (D-6) antibodies, and protein A/G agarose beads at 4°C overnight. Immunoprecipitated complexes were washed three times with lysis buffer, and bound RNAs were extracted by TRIzol reagent (Invitrogen, Carlsbad, CA). Gastrin mRNA was detected by RT-PCR or quantitative real-time PCR.
Short Interfering RNA
The short interfering RNA against hnRNP K and PCBP1 were purchased from Ambion (Austin, TX). The short interfering RNA sequence of hnRNP K was sense: 5′-GCGCAUAUUGAGUAUCAGU-3′ and antisense: 5′ACUGAUACUCAAUAUGCGC-3′. The short interfering RNA sequence of PCBP1 was sense: 5′-CCUCUAGAUGCCUACUCGA-3′ and antisense: 5′-UCGAGUAGGCAUCUAGAGC-3′. The short interfering RNA against nucleolin was purchased from Invitrogen. The sequence was sense: 5′-GGAAGGUCAGCAGUCUUCCAUGAGA-3′ and antisense: 5′-UCUCAUGGAAGACUGCUGACCUUCC-3′. To deliver the siRNA, the AGS cells were plated at ∼40% confluence in a six-well plate, and 50 nM siRNA was transfected to cells using Lipofectamine 2000 (Invitrogen) at 20 h after plating the cells. The cells were changed to serum starvation medium 20 h later, and they continued to incubate for 24 h. After plating the cells for 64 h, the cells were treated with 10 nM EGF for 4 or 8 h, and then cells were lysed and RNAs or proteins were collected for analysis.
Tet-Off mRNA Turnover Assay
The 1.7 kb of luciferase gene from pGL-3 vector (Promega, Madison, WI) was subcloned into pTRE2-hyg vector (Clontech) and fused with gastrin 3′UTR to produce pTRE-luc-gastrin 3′UTR (pTRE-luc-3′UTR) construct. To establish pTet-Off stable AGS cell line, the pTet-Off plasmid was transiently transfected into AGS cells and cultured through 400 μg/ml Geneticin (G-418; Invitrogen) for selection. The stable pTet-Off AGS cells were transfected with 20 μM scramble si-control or si-nucleolin oligonucleotides with 0.2 μg of pTRE-luc-3′UTR by using Lipofectamine 2000. After 36 h of transfection, cells were treated with or without 10 nM EGF for 3 h, and then luciferase mRNA expression was turned off by adding 50 μg/ml doxycycline. To analyze gastrin mRNA turnover rate under specific knockdown condition, the cells were harvested at different time points, and luciferase activity was measured. Results were the mean ± SD of three independent experiments.
RESULTS
EGF Stimulates Gastrin mRNA Expression and Increases the mRNA Stability
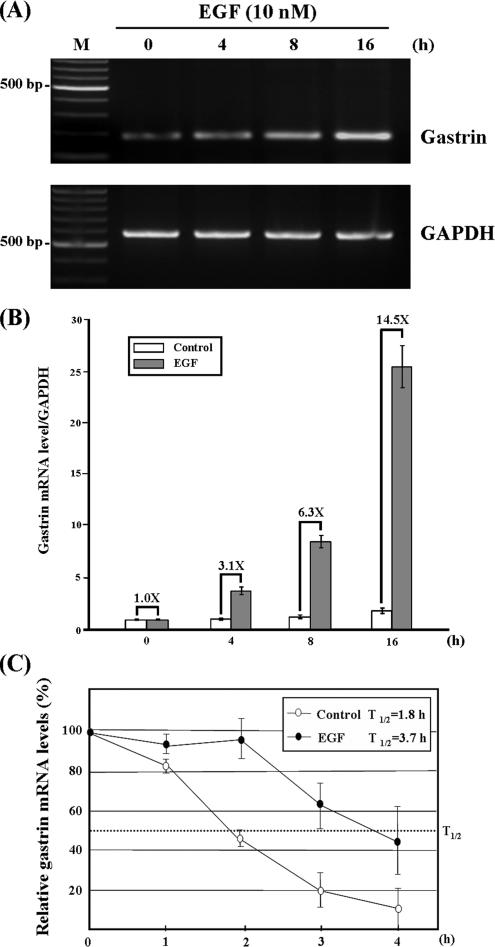
It has been shown that EGF induces human gastrin mRNA expression in a gastric cell line (Ford et al., 1997). To confirm this phenomenon existed in our study system, AGS cells were treated with EGF, and gastrin mRNA levels were detected at different time points using RT-PCR. Consistent with previous results, we observed EGF increased gastrin mRNA expression in a time-dependent manner (Figure 1A). The mRNA expression levels were also quantified by quantitative RT-PCR. As shown in Figure 1B, gastrin mRNA expression levels are elevated to 3.1- and 14.5-fold after EGF treatment for 4 and 16 h, respectively. To check whether the mRNA turnover regulation mechanism was involved in the up-regulation of gastrin mRNA under EGF treatment, AGS cells were incubated with or without EGF for 4 h, and then actinomycin D was added to block the transcriptional activities. The cells were harvested at different time intervals, and the levels of gastrin mRNA were analyzed by quantitative RT-PCR. The half-life of gastrin mRNA was ∼1.8 h in normal conditions. In contrast, when AGS cells were pre-treated with EGF for 4 h, the half-life of gastrin mRNA was extended to ∼3.7 h (Figure 1C). This result indicated that EGF increases the gastrin mRNA stability, and it implies an mRNA turnover regulation mechanism is involved in the gastrin mRNA expression.
Figure 1.
EGF induces the expression and increased the gastrin mRNA stability in AGS cells. (A) EGF up-regulates the gastrin mRNA expression in a time-dependent manner. The AGS cells were plated at ∼40% confluence, and then the incubation medium was changed to serum starvation medium after 20 h. The cells were starved for 24 h and treated with EGF for 4, 8, and 16 h. At different time points, RNA was isolated and analyzed by RT-PCR. (B) Quantitative result of gastrin mRNA expression levels under EGF treatment. The -fold represents the mRNA expression level of EGF-treated cell divided by control cell. (C) EGF increases gastrin mRNA stability. AGS cells treated with or without 10 nM EGF for 4 h before the addition of 5 μg/ml actinomycin D. Total RNA was isolated, and gastrin mRNA level was determined using quantitative RT-PCR and normalized with the mRNA level of GAPDH. Quantitative results were plotted as a percentage of total gastrin mRNA at different time points compared with time point 0 h. Values are the means ± SE from three separate experiments.
Gastrin 3′UTR Associates with Protein Complex
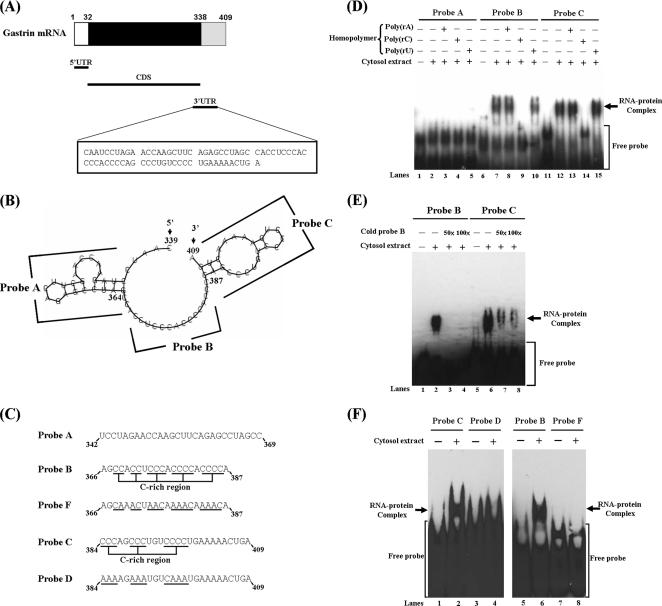
Regarding the mechanism regulated mRNA turnover, protein complexes interacted with 3′UTR play an important role in modulating the mRNA turnover (Parker and Song, 2004). Therefore, we tried to identify the cis-acting elements in gastrin 3′UTR and trans-acting proteins. Figure 2A illustrates the structure of human gastrin mRNA. The length of 3′UTR is only 71 nucleotides, and the structure predicted by Vienna RNA Secondary Structure Prediction program is shown in Figure 2B. It forms two stem-loop regions connected by a spacer region. Three RNA probes (A, B, and C) based on these structures were synthesized (Figure 2C). To identify which probes interacted with proteins, RNA- EMSAs were performed. Only probes B and C interacted with proteins to form observable RNA–protein complexes. (Figure 2D, lanes 7 and 12). The RNA–protein complex disappeared after incubation with poly(rC) homopolymer. This indicates that the binding proteins belong to the family of poly(C) binding proteins (Figure 2D, lanes 9 and 14). To identify whether the same protein complex interacted with probes B and C, we synthesized nonbiotin-labeled probe B to compete in the reaction of protein binding with biotin-labeled probes B and C. We found that it could inhibit the RNA–protein complex formation in a dose-dependent manner (Figure 2E, lanes 2–4 and 6–8). These results suggested that probe B and probe C interact with the same RNA-binding proteins in AGS cells.
Figure 2.
Characterization of the protein complexes interacted with gastrin mRNA 3′UTR. (A) The diagram illustrates the structure of human gastrin mRNA. (B) The structure of human gastrin 3′UTR was predicted by Vienna RNA Secondary Structure Prediction program (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). (C) Sequences of three biotinylated RNA probes (probe A, B, and C) and mutant counterpart probes (probe D and F) used in RNA-EMSA. (D) Biotin-labeled probes A, B, and C were incubated with or without cytosolic extracts of AGS cells, and RNA-EMSA was performed. For competition assay, the reaction mixtures were added with RNA homopolymers [poly(rA), poly(rC), and poly(rU), respectively]. (E) Nonbiotinylated probe B (cold probe B) was added to reaction mixture as a competitor to examine the protein complex existing in probe B and C. (F) No protein complex was present in probe D or F (C-rich regions mutants of wild-type probes C and B, respectively).
To further demonstrate that the RNA-binding proteins belong to the poly(C) binding proteins, probes D and F, the same sequence as probe C and B with disruption of its C-rich region, respectively (Figure 2C), were incubated with the cytosolic extract to check the binding activity. From the Figure 2F, no binding complex formation was observed. These results implied that unknown poly(C) binding proteins bind with the gastrin mRNA 3′UTR and that the C-rich region is important for this interaction.
hnRNP K and PCBP1 Bind with Gastrin mRNA 3′UTR
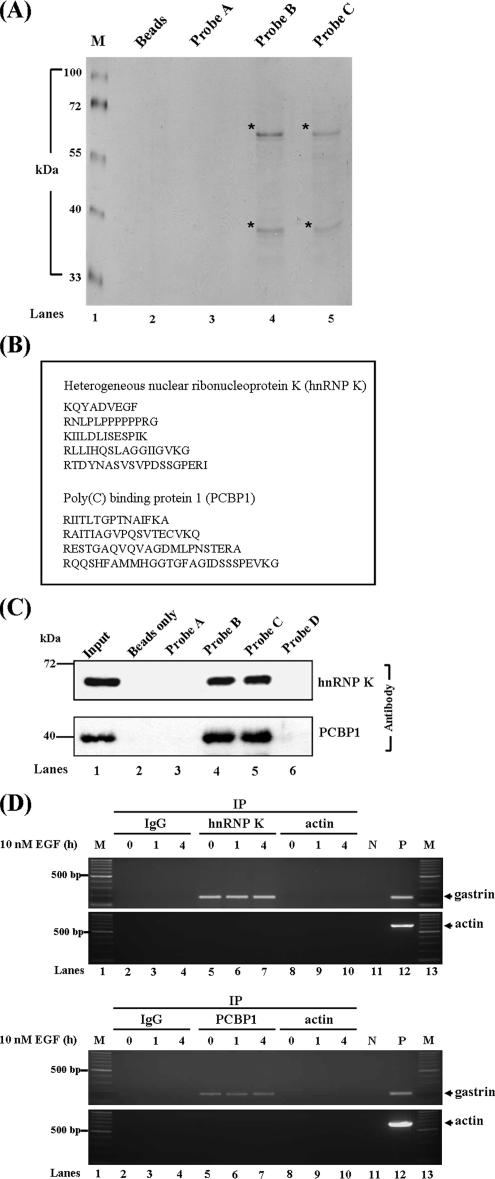
To identify the trans-acting factors that may interact with the gastrin mRNA 3′UTR, the biotin pull-down assay was performed to analyze the potential protein associations with this region. Two major proteins were observed to interact with probe B and probe C (Figure 3A). Using mass spectrometry analysis, hnRNP K and PCBP1, belonging to the poly(C) binding protein family, were identified to be the target proteins interacted with the gastrin mRNA 3′UTR (Figure 3B). To further confirm this finding, UV cross-linking/biotin-labeled probe pull-down combined with Western blot analysis using hnRNP K and PCBP1 antibodies was performed. The data revealed that both hnRNP K and PCBP bound with probe B and C, but not probe A (Figure 3C, lanes 3–5). Also, both hnRNP K and PCBP1 could not bind with probe D, which had a mutated C-rich region (Figure 3C, lane 6).
Figure 3.
Identification of the RNA-binding proteins interacted with the gastrin mRNA. (A) Biotin-labeled probes A, B, and C were incubated with cytosolic extracts and pulled down by streptavidin beads. The mixtures were analyzed by SDS-PAGE combined with silver staining. (B) hnRNP K and PCBP1 were identified as major binding proteins interacted with gastrin mRNA 3′UTR by mass spectrometry analysis. (C) hnRNP K and PCBP1 interacted with probes B and C in vitro. The UV-cross-linking/biotin pull-down analysis and Western blot were performed with hnRNP K and PCBP1 antibodies. Beads represent the streptavidin beads control. (D) hnRNP K and PCBP1 bound to gastrin mRNA in vivo. AGS cells were treated with or without 10 nM EGF for 1 and 4 h, and then the cytoplasm proteins were harvested and incubated with anti-hnRNP K, anti-PCBP1, or anti-actin antibodies. RNA from immunoprecipitated complex was extracted, and the mRNA expression levels of gastrin and actin were measured by RT-PCR analysis (M, marker; P, positive control; N, negative control).
To examine whether the hnRNP K and PCBP1 also bind with gastrin mRNA in vivo, the RNA-immunoprecipitation assay was conducted. Human gastrin mRNA was detected in the hnRNP K and PCBP1 immunoprecipitation complexes under normal and EGF treatment at different time points (Figure 3D, lanes 5–7), but it was not detected in the negative control immunoglobulin (Ig)G immunoprecipitation complex (Figure 3D, lanes 2–4). Overall, these observations support the notion that hnRNP K and PCBP1 bind with gastrin mRNA 3′UTR through the C-rich region.
EGF Signaling Regulates Nucleolin Interacted with Gastrin mRNA and hnRNP K
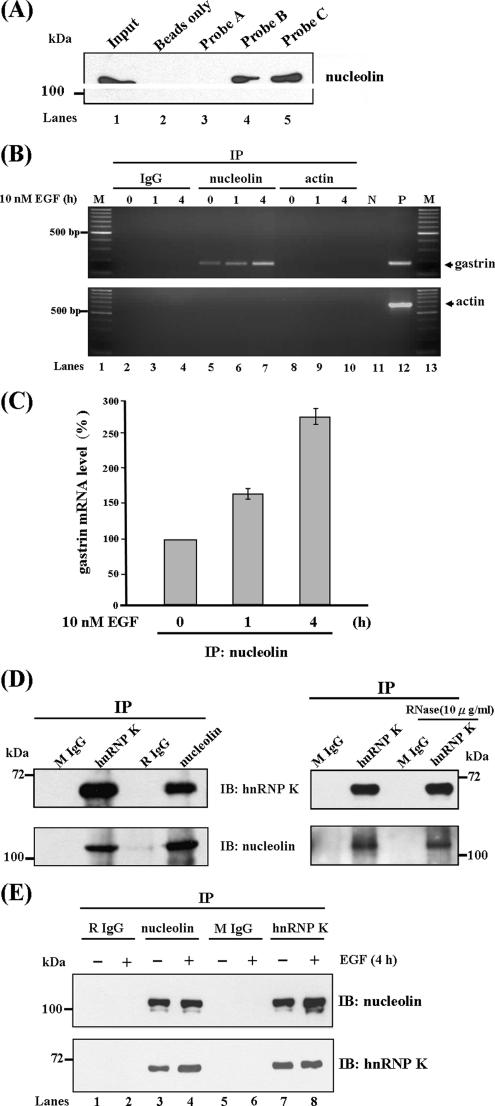
Increased binding between trans-factors with cis-acting elements in mRNA 3′UTR was reported to enhance the mRNA stability under stimulator treatment (Wilusz et al., 2001; Bevilacqua et al., 2003). From Figure 3D, we found HnRNP K and PCBP1 interacted with gastrin mRNA in vivo; however, the level of gastrin mRNA associated with hnRNP K and PCBP1 was unchanged under EGF treatment (compare lanes 5–7). This raises the possibility that other factors are involved in the increase of gastrin mRNA stability induced by EGF. To identify this regulation factor, we examined the possible RNA-binding motifs existing in the gastrin mRNA 3′UTR. Interestingly, we found a motif sequence AGCCCU located between the C-rich region similar to the reverse orientation of nucleolin binding sequence (U/G)CCCG(A/G). Then, the biotin-labeled probe pull-down combined with Western blot analysis was conducted to confirm the interaction between nucleolin and gastrin mRNA. We found nucleolin, a RNA-binding protein involved in many cellular processes, interacted with probes B and C, but not with probe A (Figure 4A, lanes 3–5). This result indicated that the nucleolin associates with gastrin mRNA 3′UTR in vitro.
Figure 4.
Nucleolin binds with gastrin mRNA 3′UTR and hnRNP K. (A) Biotin-labeled RNA probes A, B, and C were incubated with cytosolic extract. The mixtures were analyzed by UV-cross link/biotin pull-down assay, followed by Western blot analysis by using nucleolin antibodies. (B) EGF increases the binding level of nucleolin with gastrin mRNA in vivo. AGS cells were treated with or without 10 nM EGF for 1 and 4 h, and the RNA immunoprecipitation assays were performed with anti-nucleolin or anti-actin antibodies. RNA was extracted and the mRNA expression levels of gastrin and actin were resolved by RT-PCR. (C) Quantitative results of RNA immunoprecipitation assay by using nucleolin antibodies under 10 nM EGF treatment for 0, 1, and 4 h. (D) hnRNP K forms a complex with nucleolin. AGS cell lysates were subjected to immunoprecipitation with anti-hnRNP K and anti-nucleolin antibodies. The immunoprecipitated complexes were treated with or without RNase and analyzed by SDS-PAGE and immunoblotting by using anti-nucleolin and anti-hnRNP K antibodies. The rabbit IgG (RIgG) and mouse IgG (MIgG) were used as negative controls. (E) EGF enhanced the interaction between nucleolin and hnRNP K. AGS cells were treated with or without 10 nM EGF for 4 h, and the protein immunoprecipitation assay was then conducted as described above.
Furthermore, the in vivo association of nucleolin with gastrin mRNA was demonstrated by RNA immunoprecipitation assay. We found that nucleolin bound to gastrin mRNA and slightly increased the binding amounts under EGF treatment for 4 h (Figure 4B, compare lanes 5–7). The quantitative result is shown in Figure 4C. Overall, these results indicated nucleolin really interacts with gastrin mRNA and that it may play a functional role in regulating EGF-induced gastrin mRNA expression.
It is also interesting to know whether nucleolin binds with gastrin mRNA and also whether it interacts with hnRNP K and PCBP1. To address this question, a protein immunoprecipitation assay was performed. As shown in Figure 4D, we found the hnRNP K-immunoprecipitated complex contained the nucleolin protein, and vice versa. To further demonstrate the binding between nucleolin and hnRNP K was through protein–protein interaction, and not due to binding with the same RNA, RNase was added to the reaction mixture to destroy the binding ability through protein–RNA–protein interaction. We found the binding pattern did not change under RNase treatment (Figure 4D, right), which indicated nucleolin and hnRNP K form a protein complex to interact with gastrin mRNA. Furthermore, we observed the interaction between hnRNP K and nucleolin increased under EGF treatment (Figure 4E, compare lanes 3 and 4, immunoblot [IB]: hnRNP K, and lanes 7 and 8, IB: nucleolin). Combining these findings, the increase in binding between nucleolin and hnRNP K seems to contribute to the increased association of nucleolin and gastrin mRNA under EGF treatment.
Nucleolin Binds with AGCCCU Motif in the Gastrin mRNA and This Binding Ability Is Enhanced by hnRNP K/PCBP1 Complex
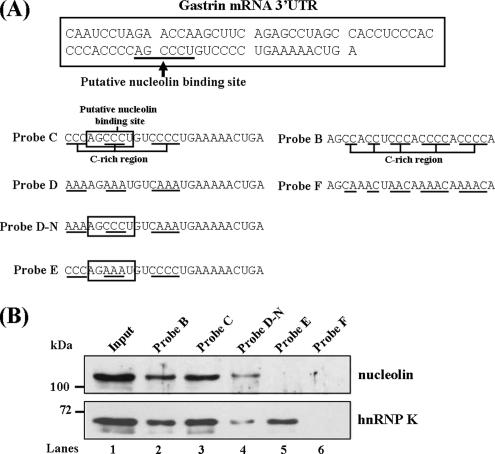
To identify the exact binding site of nucleolin, the putative nucleolin binding site AGCCCU was mutated to AGAAAU to test the binding activity of nucleolin (Figure 5A, probe E). As shown in Figure 5B, we found that nucleolin could not bind with probe E, which lacked the putative nucleolin binding site. These results indicated the AGCCCU motif was the target site for nucleolin to bind with gastrin mRNA. Interestingly, the binding activity decreased with probe D-N. The binding ability of hnRNP K/PCBP1 complex with this probe was abolished due to lack of C-rich region. This result indicated that nucleolin binds with gastrin mRNA not only dependent on the binding sequence but also regulated by hnRNP K/PCBP1 complex interacting with the C-rich region. Besides, we also found the probe B bound with lower level nucleolin protein than probe C. Mutation of the C-rich region of probe B abolished the binding activity of nucleolin (Figure 5B, probe F). It indicated the C-rich region of probe B may contain a minor binding site for nucleolin. But, the putative nucleolin binding site in probe C may be the major interaction element with nucleolin.
Figure 5.
Identified the nucleolin binding sequence in the gastrin mRNA. (A) The diagram shows the putative nucleolin binding site (baseline). Biotinylated RNA probes were synthesized, including two wild-type probes, B and C, and four mutant counterparts: probe D (mutation of three C-rich sites of probe C), probe D-N (mutation of two C-rich sites of probe C and preservation of putative nucleolin binding site), probe E (the putative nucleolin binding site was mutated), and probe F (mutation of the C-rich region of probe B). (B) RNA probes B, C, D-N, E, and F were incubated with cytosolic extract, respectively. The mixtures were analyzed by UV-cross link/biotin pull-down assay combined with Western blot analysis by using nucleolin and hnRNP K antibodies.
Reduction of hnRNP K, PCBP1, and Nucleolin Protein Expression Decreases Gastrin mRNA Expression Level
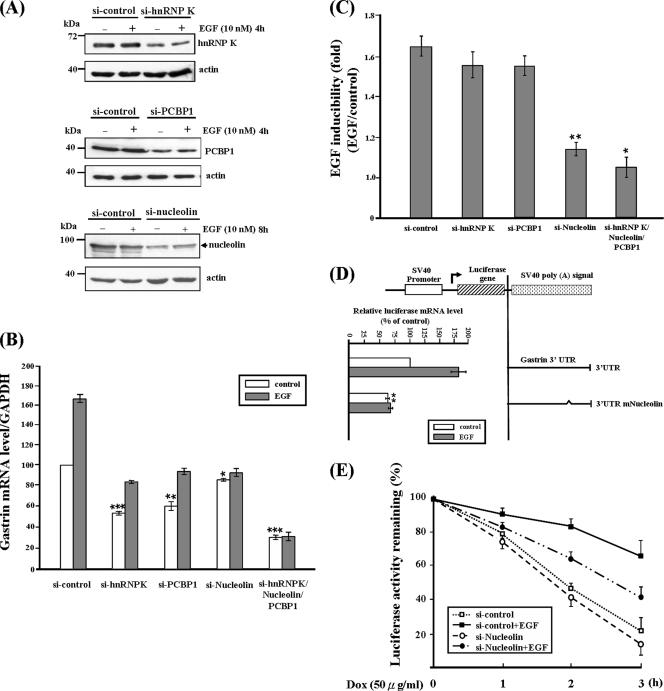
To characterize the functional role of these binding proteins on gastrin mRNA expression, the protein expression of hnRNP K, PCBP1, and nucleolin was knocked down by siRNA, and the gastrin mRNA level was analyzed by quantitative real-time RT-PCR. As shown in Figure 6A, the protein expression of hnRNP K, PCBP1, and nucleolin were significant reduced under normal or EGF treatment by siRNA treatment. The gastrin mRNA expression levels were reduced to ∼60% under the reduced expression of hnRNP K and PCBP1 (Figure 6B). Knocked down nucleolin protein expression only reduced the gastrin mRNA expression level ∼15%. Simultaneous silencing of these three genes had the serious inhibitory effect on gastrin mRNA expression. These results indicated that these factors contribute to stabilization of gastrin mRNA. Meanwhile, hnRNP K and PCBP1 play a more important role than nucleolin in prevention of gastrin mRNA degradation under normal conditions.
Figure 6.
Silencing of hnRNP K, PCBP1, and nucleolin reduces gastrin mRNA expression level. (A) Three different kinds of siRNAs targeted each of hnRNP K, PCBP1, and nucleolin, respectively, were transiently transfected into AGS cells. The whole cell lysate was harvested, and Western blot analysis was performed using anti-hnRNP K, anti-PCBP1, and anti-nucleolin antibodies. (B) The gastrin mRNA expression levels were measured by quantitative real-time RT-PCR under different siRNA conditions. GAPDH mRNA expression level was used as an internal control for calibration. (C) The EGF inducibility of gastrin mRNA expression was calculated by dividing the quantity of gastrin mRNA expression level with EGF treatment to the quantity without EGF treatment under different siRNA conditions. (D) The AGS cells were transiently transfected with luciferase constructs bearing wild-type gastrin 3′UTR and mutated nucleolin binding site (3′UTRmNucleolin). After 24-h serum starvation, transfected cells were treated with or without 10 nM EGF for 6 h. Luciferase mRNA expression levels were measured by quantity real-time RT-PCR. (E) Tet-Off mRNA turnover assay to study the turnover rate with or without EGF treatment under reducing nucleolin expression. *p < 0.05, **p < 0.01, and ***p < 0.001 vs. si-control.
To dissect their functional roles under EGF treatment, the gastrin mRNA expression levels were compared between normal and EGF treatment conditions, and the ratio (EGF:normal) was shown as EGF inducibility (-fold) as indicated in Figure 6C. A minor inhibition effect was observed under hnRNP K and PCBP1 knockdown condition. But reduction of the nucleolin protein expression almost abolished the EGF inducibility. Therefore, this indicated that nucleolin is the key mediator in the increase of the gastrin mRNA expression under EGF treatment.
To further confirm the functional role of nucleolin under EGF treatment, luciferase reporter gene assay was conducted. The wild-type 3′UTR and putative nucleolin binding site mutated 3′UTR, which abolished the nucleolin binding ability (3′UTRmNucleolin), were ligated to a luciferase reporter vector. The AGS cells were transiently transfected with these reporter constructs in the presence or absence of EGF, respectively, and then the luciferase mRNA expression levels were measured by quantitative real-time RT-PCR. From Figure 6D, we found the luciferase mRNA levels of 3′UTRmNucleolin decreased under normal conditions, and, more importantly, the EGF inducibility was also inhibited under EGF treatment (compare the control and EGF treatment of the 3′UTR mNucleolin clone). Besides, the Tet-Off mRNA turnover assay was performed to study the gastrin mRNA degradation rate under normal and reduced nucleolin protein expression conditions. As shown in Figure 6E, reduction of nucleolin expression accelerates the degradation rate of reporter gene fused with gastrin 3′UTR under EGF treatment, but not the normal conditions (compare si-control + EGF and si-Nucleolin + EGF). These results suggested nucleolin plays an important role in mediating the increased mRNA stability under EGF signaling.
DISCUSSION
Gastrin, an important polypeptide hormone responsible for gastric acid secretion, has been found to possess multiple carcinogenic activities in gastrointestinal adenocarcinomas (Ferrand and Wang, 2006). Gastrin can impart antiapoptotic activities through activation of genes associated with cell division, invasion, and angiogenesis, including EGF receptor, regenerating protein, cyclooxygenase-2, and matrix metalloproteinases (Chiba et al., 2000; Wroblewski et al., 2002; Konturek et al., 2003; Clarke et al., 2006). Therefore, to dissect the mechanism that maintains higher expression levels of gastrin in cancer cells is important for understanding the progress of gastrointestinal cancer development. Here, we report a novel mechanism that increases the gastrin expression level through regulating mRNA turnover.
First, we found EGF signal increased the gastrin mRNA stability in AGS cells (Figure 1), indicating that mRNA turnover regulation is involved in the mechanism which maintains the gastrin mRNA expression level. To dissect the molecular mechanism governing gastrin mRNA turnover, we identified the trans-acting factors, hnRNP K and PCBP1, interacting with the C-rich region located at the gastrin mRNA 3′UTR (Figure 3). hnRNP K and PCBP1 belong to the family of poly(C) binding proteins, which are involved in a remarkable array of gene expression from transcription, mRNA processing, mRNA transport, mRNA stability, and translation (Makeyev and Liebhaber, 2002). PCBP1/PCBP2 complex was reported to stabilize α-globin and erythropoietin mRNA by binding with the C-rich region existing in their 3′UTR (Weiss and Liebhaber, 1995; Czyzyk-Krzeska and Bendixen, 1999). The major functional role of hnRNP K, which interacted with 3′UTR was focused on translational regulation. hnRNP K formed a complex with PCBP1/2, binding to the differentiation control element located at the 15-lipoxygenase mRNA 3′UTR and mediating translational silencing effect (Ostareck et al., 1997). Some literature indicates that hnRNP K is involved in regulating renin and neurofilament mRNA stability (Skalweit et al., 2003; Thyagarajan and Szaro, 2004). But, these studies only show the interaction between hnRNP K and mRNAs, and no direct evidence is provided to prove hnRNP K really regulates mRNA turnover in vivo. In this study, the functional role of PCBP1 and hnRNP K was proved by reducing these two protein expressions using siRNA. We found the expression level of gastrin mRNA decreased (Figure 6B). Therefore, we propose that hnRNP K and PCBP1 are responsible for stabilizing the gastrin mRNA.
Nucleolin, a multifunctional RNA binding protein, is involved in regulation of chromatin condensation, rRNA processing, and nuclear-cytoplasmic transport (Ginisty et al., 1999; Mongelard and Bouvet, 2007). Recent studies indicate that nucleolin is also involved in regulation of mRNA turnover, for example, GADD45α mRNA, bcl-2 mRNA, and CD154 mRNA (Sengupta et al., 2004; Singh et al., 2004; Zhang et al., 2006). From our results, we identified that nucleolin, but not hnRNP K and PCBP1, increased association with gastrin mRNA under EGF treatment (Figure 4B). The functional role of nucleolin was demonstrated by reducing nucleolin protein expression by using siRNA technology. We observed that decreased nucleolin protein expression significantly exerted an inhibitory effect on the EGF inducibility (Figure 6C). Using a reporter assay system, we found mutation of the putative nucleolin binding site that abolished the nucleolin binding displayed the same inhibitory effect (Figure 6D). Moreover, reduction of nucleolin expression reduced the stabilization effect exerting by EGF in Tet-Off mRNA turnover assay (Figure 6E). Therefore, the functional role of nucleolin was proved to mediate the effect of increasing the gastrin mRNA stability under EGF signaling.
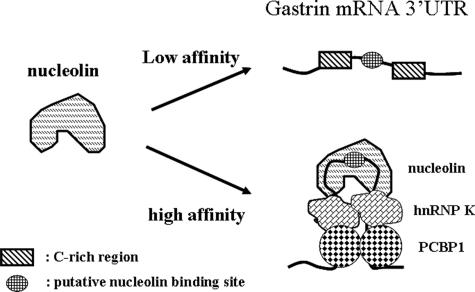
Nucleolin has been shown to bind the consensus sequence (U/G)CCCG(A/G) in the loop region of hairpin structure with a 7- to 14-bp stem region (Bouvet et al., 2001). Other sequence motifs were also reported to bind with nucleolin, such as the 5′CUCUCUUUA3′ motif in interleukin 2 mRNA and the A/U-rich motif in the Bcl-2 mRNA (Chen et al., 2000; Sengupta et al., 2004). In our study, we demonstrate that the AGCCCU motif is the nucleolin binding site, and the C-rich regions are also important for the nucleolin to associate with this motif (Figure 5B). It is interesting to know how the C-rich regions enhance the nucleolin to bind with the putative nucleolin target sequence. From our studies, we observed hnRNP K and PCBP1 bind with the C-rich regions. Other studies report that hnRNP K and PCBP1 can form both homomeric and heteromeric dimers (Kim et al., 2000), and the binding affinity of nucleolin with the hairpin-like structure is higher than with the linear structure (Ghisolfi-Nieto et al., 1996), Therefore, we propose when hnRNP K and PCBP1 bind with the C-rich regions, the linear structure of mRNA may reform to a hairpin-like structure through protein–protein interaction in the stem region and expose the AGCCCU sequence, which is located between the C-rich regions, in the loop region. Nucleolin has higher binding affinity with hairpin structure gastrin mRNA than the linear structure. The novel nucleolin binding model is shown in Figure 7. This model explains our observations, that both the C-rich region and putative nucleolin binding site are important for nucleolin association with gastrin mRNA.
Figure 7.
Binding model of nucleolin interacted with gastrin mRNA. hnRNP K and PCBP1 bind with the C-rich regions of gastrin mRNA to form a hairpin-like structure. The putative nucleolin binding site is exposed in the loop region, and this structure has higher binding affinity than the linear structure with nucleolin.
The importance of these factors contributions to gastrin mRNA expression in cancer development is highlighted by recent studies. It was reported that hnRNP K was overexpressed and associated with poor prognosis in colorectal cancer (Carpenter et al., 2006). The higher expression level of nucleolin in tumors or other rapidly dividing cells was also documented (Srivastava and Pollard, 1999). Examination of the expression profile of nucleolin on different cancers by analysis of expressed sequence tag (EST) counts, using the National Center for Biotechnology Information EST database, we found colorectal cancer and gastrointestinal cancer expressed higher amounts of nucleolin (801 and 411 transcripts per million) than other cancers and normal tissue (190 transcripts per million). According to our novel findings that both hnRNP K and nucleolin protect gastrin mRNA from degradation, these results may explain the higher expression level of gastrin mRNA in colorectal cancer.
hnRNP K and nucleolin are both multifunctional proteins and associate with different kinds of proteins to execute their functional roles. Here, we first describe that hnRNP K associates with nucleolin to play a functional role in maintaining mRNA stability. EGF signaling increases the association between these two proteins and implies posttranslational modification, such as phosphorylation, may be involved in this regulation. CKII and cdc2 kinase were demonstrated to phosphorylate nucleolin in growing cells (Srivastava and Pollard, 1999). hnRNP K was also reported to phosphorylate by Lck and mitogen-activated protein kinase (Bomsztyk et al., 2004). Now, we are studying what kinds of signal pathways regulate this interaction under EGF treatment. This will give us more information about hnRNP K/nucleolin complex in mediating the stabilization of mRNA under growth conditions.
In conclusion, hnRNP K and PCBP1 bound with the C-rich regions of gastrin mRNA 3′UTR facilitates nucleolin to associate with gastrin mRNA. Under EGF treatment, the interaction of nucleolin with hnRNP K/PCBP1/gastrin mRNA complex increased and contributed to the increase of gastrin mRNA stability. This novel mechanism in regulation of gastrin mRNA may provide us another direction to study the mechanism that maintains high expression levels of gastrin mRNA in gastrointestinal cancer.
ACKNOWLEDGMENTS
This work is supported by grant NSC 95-2320-B-006-066-MY3 from the National Science Council of Taiwan, Republic of China.
Abbreviations used:
- 3′UTR
3′ untranslated region
- EGF
epidermal growth factor
- hnRNP K
heterogeneous nuclear ribonucleoprotein K
- PCBP1
poly(C) binding protein 1.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-04-0384) on October 10, 2007.
REFERENCES
- Aghib D. F., Bishop J. M., Ottolenghi S., Guerrasio A., Serra A., Saglio G. A 3′ truncation of MYC caused by chromosomal translocation in a human T-cell leukemia increases mRNA stability. Oncogene. 1990;5:707–711. [PubMed] [Google Scholar]
- Aly A., Shulkes A., Baldwin G. S. Gastrins, cholecystokinins and gastrointestinal cancer. Biochim. Biophys. Acta. 2004;1704:1–10. doi: 10.1016/j.bbcan.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Audic Y., Hartley R. S. Post-transcriptional regulation in cancer. Biol. Cell. 2004;96:479–498. doi: 10.1016/j.biolcel.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Bevilacqua A., Ceriani M. C., Capaccioli S., Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J. Cell. Physiol. 2003;195:356–372. doi: 10.1002/jcp.10272. [DOI] [PubMed] [Google Scholar]
- Bomsztyk K., Denisenko O., Ostrowski J. hnRNP K: One protein multiple processes. Bioessays. 2004;26:629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- Bouvet P., Allain F. H., Finger L. D., Dieckmann T., Feigon J. Recognition of pre-formed and flexible elements of an RNA stem-loop by nucleolin. J. Mol. Biol. 2001;309:763–775. doi: 10.1006/jmbi.2001.4691. [DOI] [PubMed] [Google Scholar]
- Brennan C. M., Steitz J. A. HuR and mRNA stability. Cell Mol. Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter B., McKay M., Dundas S. R., Lawrie L. C., Telfer C., Murray G. I. Heterogeneous nuclear ribonucleoprotein K is overexpressed, aberrantly localised and is associated with poor prognosis in colorectal cancer. Br. J. Cancer. 2006;95:921–927. doi: 10.1038/sj.bjc.6603349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakladar A., Dubeykovskiy A., Wojtukiewicz L. J., Pratap J., Lei S., Wang T. C. Synergistic activation of the murine gastrin promoter by oncogenic Ras and beta-catenin involves SMAD recruitment. Biochem. Biophys. Res. Commun. 2005;336:190–196. doi: 10.1016/j.bbrc.2005.08.061. [DOI] [PubMed] [Google Scholar]
- Chen C. Y., Gherzi R., Andersen J. S., Gaietta G., Jurchott K., Royer H. D., Mann M., Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14:1236–1248. [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y., Shyu A. B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem. Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- Chiba T., Fukui H., Kinoshita Y. Reg protein: a possible mediator of gastrin-induced mucosal cell growth. J. Gastroenterol. 2000;12:52–56. [PubMed] [Google Scholar]
- Clarke P. A., Dickson J. H., Harris J. C., Grabowska A., Watson S. A. Gastrin enhances the angiogenic potential of endothelial cells via modulation of heparin-binding epidermal-like growth factor. Cancer Res. 2006;66:3504–3512. doi: 10.1158/0008-5472.CAN-05-0280. [DOI] [PubMed] [Google Scholar]
- Cobb S., Wood T., Ceci J., Varro A., Velasco M., Singh P. Intestinal expression of mutant and wild-type progastrin significantly increases colon carcinogenesis in response to azoxymethane in transgenic mice. Cancer. 2004;100:1311–1323. doi: 10.1002/cncr.20094. [DOI] [PubMed] [Google Scholar]
- Czyzyk-Krzeska M. F., Bendixen A. C. Identification of the poly(C) binding protein in the complex associated with the 3′ untranslated region of erythropoietin messenger RNA. Blood. 1999;93:2111–2120. [PubMed] [Google Scholar]
- Dockray G., Dimaline R., Varro A. Gastrin: old hormone, new functions. Pflugers Arch. 2005;449:344–355. doi: 10.1007/s00424-004-1347-5. [DOI] [PubMed] [Google Scholar]
- Ferrand A., Wang T. C. Gastrin and cancer: a review. Cancer Lett. 2006;238:15–29. doi: 10.1016/j.canlet.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Ford M. G., Valle J. D., Soroka C. J., Merchant J. L. EGF receptor activation stimulates endogenous gastrin gene expression in canine G cells and human gastric cell cultures. J. Clin. Invest. 1997;99:2762–2771. doi: 10.1172/JCI119466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisolfi-Nieto L., Joseph G., Puvion-Dutilleul F., Amalric F., Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J. Mol. Biol. 1996;260:34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- Ginisty H., Sicard H., Roger B., Bouvet P. Structure and functions of nucleolin. J. Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- Hollams E. M., Giles K. M., Thomson A. M., Leedman P. J. MRNA stability and the control of gene expression: implications for human disease. Neurochem. Res. 2002;27:957–980. doi: 10.1023/a:1020992418511. [DOI] [PubMed] [Google Scholar]
- Hollis G. F., Gazdar A. F., Bertness V., Kirsch I. R. Complex translocation disrupts c-myc regulation in a human plasma cell myeloma. Mol. Cell. Biol. 1988;8:124–129. doi: 10.1128/mcb.8.1.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Hahm B., Kim Y. K., Choi M., Jang S. K. Protein-protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 2000;298:395–405. doi: 10.1006/jmbi.2000.3687. [DOI] [PubMed] [Google Scholar]
- Koh T. J., Field J. K., Varro A., Liloglou T., Fielding P., Cui G., Houghton J., Dockray G. J., Wang T. C. Glycine-extended gastrin promotes the growth of lung cancer. Cancer Res. 2004;64:196–201. doi: 10.1158/0008-5472.can-03-2112. [DOI] [PubMed] [Google Scholar]
- Konturek P. C., Kania J., Kukharsky V., Ocker S., Hahn E. G., Konturek S. J. Influence of gastrin on the expression of cyclooxygenase-2, hepatocyte growth factor and apoptosis-related proteins in gastric epithelial cells. J. Physiol. Pharmacol. 2003;54:17–32. [PubMed] [Google Scholar]
- Lei S., Dubeykovskiy A., Chakladar A., Wojtukiewicz L., Wang T. C. The murine gastrin promoter is synergistically activated by transforming growth factor-beta/Smad and Wnt signaling pathways. J. Biol. Chem. 2004;279:42492–42502. doi: 10.1074/jbc.M404025200. [DOI] [PubMed] [Google Scholar]
- Makeyev A. V., Liebhaber S. A. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongelard F., Bouvet P. Nucleolin: a multiFACeTed protein. Trends Cell Biol. 2007;17:80–86. doi: 10.1016/j.tcb.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Ostareck D. H., Ostareck-Lederer A., Wilm M., Thiele B. J., Mann M., Hentze M. W. mRNA silencing in erythroid differentiation: hnRNP K and hnRNP E1 regulate 15-lipoxygenase translation from the 3′ end. Cell. 1997;89:597–606. doi: 10.1016/s0092-8674(00)80241-x. [DOI] [PubMed] [Google Scholar]
- Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- Rehfeld J. F., Bardram L., Hilsted L. Gastrin in human bronchogenic carcinomas: constant expression but variable processing of progastrin. Cancer Res. 1989;49:2840–2843. [PubMed] [Google Scholar]
- Sengupta T. K., Bandyopadhyay S., Fernandes D. J., Spicer E. K. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J. Biol. Chem. 2004;279:10855–10863. doi: 10.1074/jbc.M309111200. [DOI] [PubMed] [Google Scholar]
- Singh K., Laughlin J., Kosinski P. A., Covey L. R. Nucleolin is a second component of the CD154 mRNA stability complex that regulates mRNA turnover in activated T cells. J. Immunol. 2004;173:976–985. doi: 10.4049/jimmunol.173.2.976. [DOI] [PubMed] [Google Scholar]
- Skalweit A., Doller A., Huth A., Kahne T., Persson P. B., Thiele B. J. Posttranscriptional control of renin synthesis: identification of proteins interacting with renin mRNA 3′-untranslated region. Circ. Res. 2003;92:419–427. doi: 10.1161/01.RES.0000059300.67152.4E. [DOI] [PubMed] [Google Scholar]
- Srivastava M., Pollard H. B. Molecular dissection of nucleolin's role in growth and cell proliferation: new insights. FASEB J. 1999;13:1911–1922. [PubMed] [Google Scholar]
- Thyagarajan A., Szaro B. G. Phylogenetically conserved binding of specific K homology domain proteins to the 3′-untranslated region of the vertebrate middle neurofilament mRNA. J. Biol. Chem. 2004;279:49680–49688. doi: 10.1074/jbc.M408915200. [DOI] [PubMed] [Google Scholar]
- Tyan Y. C., Wu H. Y., Su W. C., Chen P. W., Liao P. C. Proteomic analysis of human pleural effusion. Proteomics. 2005;5:1062–1074. doi: 10.1002/pmic.200401041. [DOI] [PubMed] [Google Scholar]
- Wang T. C., Koh T. J., Varro A., Cahill R. J., Dangler C. A., Fox J. G., Dockray G. J. Processing and proliferative effects of human progastrin in transgenic mice. J. Clin. Invest. 1996;98:1918–1929. doi: 10.1172/JCI118993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Kiledjian M., Weiss I. M., Liebhaber S. A. Detection and characterization of a 3′ untranslated region ribonucleoprotein complex associated with human alpha-globin mRNA stability. Mol. Cell. Biol. 1995;15:1769–1777. doi: 10.1128/mcb.15.3.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss I. M., Liebhaber S. A. Erythroid cell-specific mRNA stability elements in the alpha 2-globin 3′ nontranslated region. Mol. Cell. Biol. 1995;15:2457–2465. doi: 10.1128/mcb.15.5.2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz C. J., Wilusz J. Bringing the role of mRNA decay in the control of gene expression into focus. Trends Genet. 2004;20:491–497. doi: 10.1016/j.tig.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Wilusz C. J., Wormington M., Peltz S. W. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- Wroblewski L. E., Pritchard D. M., Carter S., Varro A. Gastrin-stimulated gastric epithelial cell invasion: the role and mechanism of increased matrix metalloproteinase 9 expression. Biochem. J. 2002;365:873–879. doi: 10.1042/BJ20020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Bhatia D., Xia H., Castranova V., Shi X., Chen F. Nucleolin links to arsenic-induced stabilization of GADD45alpha mRNA. Nucleic Acids Res. 2006;34:485–495. doi: 10.1093/nar/gkj459. [DOI] [PMC free article] [PubMed] [Google Scholar]