Abstract
G-protein coupled receptor kinase-interacting protein (GIT) proteins include an N-terminal Arf GTPase-activating protein domain, and a C terminus that binds proteins regulating adhesion and motility. Given their ability to form large molecular assemblies, the GIT1 protein must be tightly regulated. However, the mechanisms regulating GIT1 functions are poorly characterized. We found that carboxy-terminal–truncated fragments of GIT1 bind their partners with higher efficiency compared with the full-length GIT1. We have explored the hypothesis that GIT1 is regulated by an intramolecular mechanism, and we identified two distinct intramolecular interactions between the N and C terminus of GIT1. The release of these interactions increases binding of GIT1 to paxillin and liprin-α, and it correlates with effects on cell spreading. Analysis of cells plated on fibronectin has shown that different deletion mutants of GIT1 either enhance or inhibit spreading, depending on their subcellular localization. Moreover, although the association between βPIX and GIT1 is insufficient to activate GIT1 binding to paxillin, binding of a PAK1 fragment including the βPIX-binding domain enhances paxillin binding to βPIX/GIT1, indicating that p21-activated kinase can activate the binding of paxillin to GIT1 by a kinase-independent mechanism. The release of the identified intramolecular interaction seems to be an important mechanism for the regulation of GIT1 functions.
INTRODUCTION
The G-protein coupled receptor kinase-interacting protein (GIT) family includes GIT1 and GIT2, two widely expressed proteins with complex domain structure. GIT proteins have binding sites for several proteins, and they are involved in the regulation of cell adhesion, migration, and membrane traffic (Hoefen and Berk, 2006). GIT1 can form homo- and heterodimers (Kim et al., 2003; Paris et al., 2003; Premont et al., 2004), and it has been localized at different sites in the cell, including focal adhesions, endocytic structures, and centrioles (Di Cesare et al., 2000; Zhao et al., 2000, 2005). GIT1 includes an N-terminal Arf GTPase-activating protein (ArfGAP) domain, three ankyrin repeats, a Spa2-homology domain (SHD), a coiled-coil domain including a leucine zipper required for dimerization, and a paxillin-binding site (PBS). In vitro and in vivo data indicate that the N-terminal ArfGAP domain of GIT1 specifically regulates the activity of Arf6 in cells (Vitale et al., 2000; Claing et al., 2001; Albertinazzi et al., 2003; Lahuna et al., 2005; Meyer et al., 2006). One or more binding partners have been identified for some of the other domains of GIT1. The α and βPIX proteins (PAK [p21-activated kinase] interacting exchange factors) are the main binding partners of the SHD domain of GIT1 (Zhao et al., 2000). PIX proteins are homodimeric guanine nucleotide exchange factors (GEFs) for Rac and Cdc42 GTPases (Manser et al., 1998), and endogenous βPIX is found constitutively associated with GIT1 (Botrugno et al., 2006). Because both GIT1 and PIX can form homodimers, they tend to form large oligomers or aggregates when overexpressed together in cells (Paris et al., 2003; Premont et al., 2004). Other binding partners for the SHD domain of GIT1 are the kinase mitogen-activated protein kinase kinase 1 (Yin et al., 2004) and phospholipase Cγ (Haendeler et al., 2003). The focal adhesion protein paxillin interacts with the PBS of GIT proteins via the LD4 motif (Turner et al., 1999; Di Cesare et al., 2000; Zhao et al., 2000). The interaction with paxillin localizes GIT proteins at focal complexes. This localization is prevented by deletion of either the LD4 domain of paxillin, or the PBS domain of GIT (Matafora et al., 2001; West et al., 2001; Brown et al., 2002). Other binding partners of GIT1 include G-protein coupled receptor kinase 2 (Grk2) (Premont et al., 1998), the postsynaptic adaptor protein liprin-α (Ko et al., 2003), and the presynaptic protein Piccolo (Kim et al., 2003). Therefore, GIT1 is able to assemble a variety of molecular complexes devoted to distinct cellular functions.
The mechanisms regulating GIT1 function in the assembly of these complexes remains undefined. Interestingly, experimental evidence suggests that the PBS domain of GIT1 is normally not available for binding to paxillin. Actually, a C-terminal fragment of GIT1 including the PBS region localizes to focal complexes and lamellipodia more robustly compared with the full-length protein (Di Cesare et al., 2000; Manabe et al., 2002). Here, we have used a biochemical approach to identify and characterize an intramolecular interaction between the N- and C-terminal portions of GIT1 that may represent an important mechanism for GIT1 regulation, and we have used morphological analysis to correlate the biochemical data with effects on cell spreading and focal adhesions.
MATERIALS AND METHODS
Plasmid Constructs
Plasmid pFlag-GIT1 (full-length avian GIT1), pFlag-GIT1-N (residues 1–346), pFlag-GIT1-N4 (residues 1–163), pFlag-GIT1-GAP (residues 1–131), pFlag-GIT1-C (residues 346–740), pGEX-GIT1-C2 and pFlag-GIT1-C2 (residues 229-740) (Di Cesare et al., 2000), pFlag-GIT1-K39 (Matafora et al., 2001), pFlag-GIT1-ΔSHD (deletion of residues 258–346), and pFlag-GIT1-LZ and pFlag-GIT1-C2-LZ (Paris et al., 2003) were obtained as described previously. The pFlag-GIT1-ΔAnk plasmid (deletion of residues 132–228) lacking the three ankyrin repeats was obtained by polymerase chain reaction (PCR) on the pBS-GIT1 plasmid with modified primers (5′-CCCAAGCTTCTTGGCGGTGACCCCGTCGTC-3′ and 5′-CCCAAGCTTCGGCTGGCCT TCTACCTGTGC-3′). The PCR product was digested with HindIII to remove the region encoding the ankyrin domains before ligation into the pBS vector. The ApaI and BamHI GIT1-ΔAnk fragment from pBS-GIT1-ΔAnk was then subcloned into pFlag-CMV2 (Kodak). The pFlag-GIT1-N2 plasmid (residues 1–230) was obtained by inserting a fragment of GIT1, obtained by PCR with the oligonucleotides 5′-GCGATATCAATGTCCCGGAAGGCGCA GCGG-3′ and 5′-CATGTCGACTCACAGCCGGTCGGTCAGCTC-3′ and digested with the enzymes EcoRV and SalI, into the pFlag-CMV2 vector digested with the same enzymes. The pFlag-GIT1-N3 plasmid (residues 1–197) was obtained by inserting a fragment of GIT1, obtained by PCR with the oligonucleotides 5′-GCGATATCAATGTCCCGGAAGGCGCAGCGG-3′ and 5′-CATGTCGACTCAGTCGGGCGCACCGGGGTC-3′ and digested with the enzymes EcoRV and SalI, into the pFlag-CMV2 vector digested with the same enzymes. The pFlag-GIT1-(229-431) plasmid was obtained by subcloning the GIT1 fragment digested with Sma1 into the pFlag-CMV2 vector.
The cDNAs for human full-length liprin-α1, and for its F3 fragment (amino acid residues 333–670) including the region binding to GIT1 (Ko et al., 2003), were obtained by reverse transcription-PCR on RNA extracted from human neuroblastoma SKNBE cells. The cDNAs were subcloned into pFlag-CMV2 (Kodak) and pcDNA3.1(−)/Myc-His vectors (Invitrogen, Basel, Switzerland), respectively. The pXJ40-HA-βPIX, pXJ40-HA-βPIX-ΔLZ, pCMV6m-MYC-Pak1, and pCMV6M-MYC-PAK-Pbd (PIX binding domain) plasmids were obtained as described previously (Bokoch et al., 1998; Manser et al., 1998; Za et al., 2006). The pBK-HA-EFA6 construct was obtained by subcloning the cDNA of EFA6 from pSRa-EFA6 (Franco et al., 1999) into a pBK-CMV vector modified to include a sequence coding for a hemagglutinin (HA) tag at the N terminus of the protein. The pEGFP-paxillin plasmid was generously provided by Victor Small (Austrian Academy of Sciences, Vienna, Austria).
Cell Culture and Transfection
COS7 cells were cultured in DMEM with 10% Fetal Clone III (Hyclone PERBIO, Erembodegem, Belgium) and transfected with Lipofectamine 2000 (Invitrogen). We used 100–400 μg of lysates for immunoprecipitation.
Antibodies
The antibodies used in this study were as follows: monoclonal antibodies (mAb) anti-Flag M5 and M2 (Sigma-Aldrich, St. Louis, MO), and anti-vinculin (clone V284; Upstate Biotechnology, Charlottesville, VA), anti-HA 12CA5, anti-Myc (Primm Biotech, Milan, Italy), anti-paxillin (clone 349; BD Biosciences Transduction Laboratories, Lexington, KY), anti-integrin β1 TS2/16 (American Type Culture Collection, Manassas, VA). Polyclonal antibody (pAb) anti-Flag was from Sigma-Aldrich; pAbs anti-βPIX and anti-GIT1 were as described previously (Paris et al., 2003; Za et al., 2006).
Immunoprecipitation, Western Blotting, and Protein Determination
Cells were lysed for 15 min on ice in lysis buffer (150 mM NaCl, 0.5% Triton X-100, 0.5 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 10 mM sodium fluoride, 20 mM Tris-Cl, pH 7.5). For immunoprecipitation, equal amounts of protein were incubated for 2 h at 4°C with the indicated antibodies coupled to protein A-Sepharose beads (GE Healthcare, Chalfont St. Giles, United Kingdom). After washing with lysis buffer containing only 0.1% Triton X-100, samples were boiled in sample buffer, run on SDS-PAGE, blotted onto nitrocellulose membranes (Whatman Schleicher and Schuell, Dassel, Germany), and probed with the indicated antibodies. Proteins were visualized with horseradish peroxidase-labeled secondary antibodies and enhanced chemiluminescence Western blotting detection reagents (GE Healthcare) or with 125I-coupled secondary antibodies or protein A, and then they were exposed to Hyperfilm-MP (GE Healthcare). Protein determination was by Bio-Rad Protein Assay (Bio-Rad, Munich, Germany).
Pull-Down Assays
The fragments GIT1-N2 and GIT1-C2 were subcloned in the two plasmid pET28b (EMD Biosciences, San Diego, CA) and pGEX-4T (GE Healthcare), respectively, to produce the plasmids pET28b-GIT1-N2 and pGEX-4T-GIT1-C2. The His-tagged GIT1-N2 (His-GIT1-N2) and glutathione transferase (gst)-tagged GIT1-C2 (gst-GIT1-C2) fusion proteins were expressed in Escherichia coli BL21(DE3) transformed with each plasmid. After induction overnight at room temperature with 0.1 mM isopropyl β-d-thiogalactoside, bacteria were lysed by sonication. His-GIT1-N2 was purified on Talon beads (Clontech, Mountain View, CA) and eluted at 4°C with 500 mM imidazole, pH 8.0. Gst-GIT1-C2 was purified on glutathione-Agarose beads (Sigma-Aldrich), and eluted at 4°C with 25 mM reduced glutathione in 50 mM NaCl, 100 mM Tris-Cl, pH 8.0.
To test for direct interaction, 3 μg of His-GIT1-N2 and 10 μg of gst-GIT1-C2 (corresponding to 100 pmol of each polypeptide) were diluted to a total volume of 100 μl with binding buffer (300 mM NaCl, 0.1% Triton X-100, and 50 mM Tris-Cl, pH 8.0) and incubated either 3 h or overnight at 4°C with rotation. Controls included each of the two fragments incubated in the absence of the other. Five microliters of anti-GIT1 SI-61 serum against a peptide included in the GIT1-C2 fragment (Paris et al., 2003) preadsorbed to 25 μl of Protein A-Sepharose beads were used to immunoprecipitate gst-GIT1-C2 from each sample. Samples were incubated 3 h at 4°C with rotation for immunoprecipitation. After three washes with binding buffer, immunoprecipitates were run on 12% acrylamide SDS-polyacrylamide gel electrophoresis, and used for immunoblotting to detect the two GIT1 fragments.
Small Interfering RNA (siRNA)
βPIX and control (luciferase) siRNA duplexes were obtained from MWG Biotech (High Point, NC) and Invitrogen. siRNA duplexes correspond to the following target sequence within the coding sequence of rat βPIX mRNA: 5′-CAACAGGAATGACAATCAC-3′. As a control, the following target sequence for luciferase mRNA was used: 5′-CATCACGTACGCGGAATAC-3′. For knockdown of endogenous βPIX, COS7 cells were transfected with 50 nM siRNA oligonucleotides in serum-free Opti-MEM (Invitrogen). siRNA-transfected COS7 cells were incubated in growth medium (DMEM supplemented with serum) for 2 d before lysis, and then they were analyzed by immunoblotting and immunoprecipitation.
Cell Adhesion Assay
We coated 96-well plates not for cell culture (Costar 3590; Corning, New York, New York) with 10 μg/ml fibronectin (BD Biosciences PharMingen, San Diego, CA) in phosphate-buffered saline (PBS) overnight at 4°C. Coated and uncoated wells were incubated for 2 h at 37°C with 1% bovine serum albumin in PBS. COS7 cells were transfected with the indicated plasmids. Eighteen to 24 h after transfection cells were detached from dishes with trypsin, washed in culture medium without serum, and plated at a concentration of 30,000 cells/well in the absence of serum. After culture for 30 min, nonadherent cells were removed, and wells were processed for cell attachment and quantified as described previously (Cattelino et al., 1995). In brief, unattached cells were removed, and adherent cells were fixed with 3% paraformaldehyde, stained with crystal violet (0.5% in 20% methanol), washed with water, and solubilized with 1% SDS. A540 was measured in each well. In all experiments, adhesion to BSA-coated substrates was very low and subtracted from that measured on fibronectin. Four independent experiments were performed for each condition. Values were normalized to those of control cells transfected with pFlag-LacZ, taken as 100% adhesion.
Cell Spreading Assay and Immunofluorescence
COS7 cells transfected with LacZ or with one of the GIT1-derived constructs were trypsinized, and 30,000 cells were plated on each glass coverslip coated overnight at 4°C with 10 μg/ml fibronectin. Cells were cultured at 37°C for 30 and 60 min, gently washed twice with PBS, fixed with 3% paraformaldehyde, and processed for immunofluorescence. After permeabilization with 0.1% Triton X-100, cells were incubated with anti-Flag, anti-GIT1, and/or anti-paxillin antibodies, followed by incubation with secondary antibodies, and/or with fluorescein isothiocyanate-conjugated phalloidin (Sigma-Aldrich). Analysis was performed with an MRC 1024 confocal microscope (Bio-Rad, Hercules, CA). For quantification of the cell areas and of focal adhesions, images were analyzed with the Image-Pro Plus (Media Cybernetics, Silver Spring, MD) and public-domain ImageJ (http://rsb.info.nih.gov/ij/) image processing and analysis software.
RESULTS
Regulated Binding of GIT1 to Paxillin and Liprin-α1
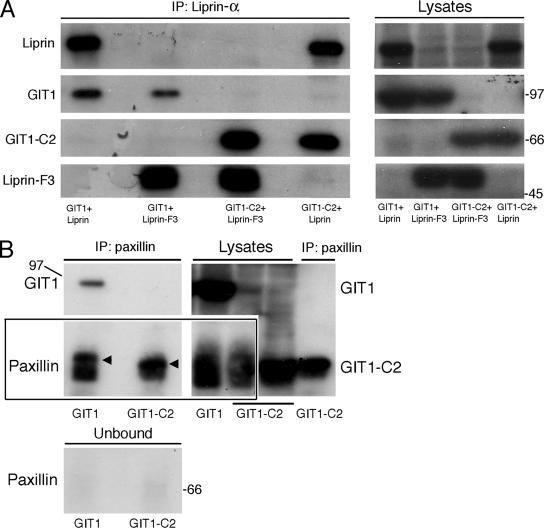
Coimmunoprecipitation experiments from lysates of COS7 cells transfected with differently tagged constructs show that full-length GIT1 interacted weakly with liprin-α1, either full-length or the liprin-α1-F3 fragment including the GIT1-interacting region (Figure 1A), and with endogenous paxillin (Figure 1B). In comparison, we observed a much stronger binding of these proteins to the truncated C-terminal construct GIT1-C2 (Figure 1, A and B) including the binding sites for liprin-α and paxillin (Turner et al., 1999; Di Cesare et al., 2000; Ko et al., 2003). These data prompted us to hypothesize the existence of a conformational switch necessary to activate GIT1 binding to its partners.
Figure 1.
Suboptimal binding of full-length GIT1 to paxillin and liprin-α1. COS7 expressing HA-tagged GIT1 constructs with Flag-liprin-α1 constructs (A) or Flag-tagged GIT1 constructs (B). In each experiment, equal aliquots of lysates were used for immunoprecipitation with anti-Flag (A) or anti-paxillin (B) antibodies. Immunoprecipitates and lysates were blotted, and filters were cut and incubated with specific antibodies to reveal the indicated antigens. Immunoblotting reveals increased binding of both liprin-α1 (A) and endogenous paxillin (B) to GIT1-C2 compared with full-length GIT1. In B, the different mobility of endogenous paxillin between the lanes from cells overexpressing GIT1 and GIT1-C2 (arrowheads) was due to migration of endogenous paxillin on the gel being distorted by the comigrating GIT1-C2 polypeptide. Each immunoprecipitation was from 200 μg (A) and 400 μg (B) of protein from lysates. Lysates in A, 50 μg each; lysates and unbound fractions in B, 100 μg each.
Identification of an Intramolecular Interaction between the N- and C-Terminal Portions of GIT1
One explanation for the increased binding of paxillin and liprin-α to GIT1-C2 is that deletion of the N-terminal part of GIT1 unmasks their binding sites on the C-terminal fragment of GIT1. Therefore, the N-terminal portion of GIT1 may interact intramolecularly with the C-terminal segment, keeping the molecule less accessible for binding to its partners. To test this hypothesis, we have coexpressed differently tagged N-terminal and C-terminal fragments of GIT1 in COS7 cells. Analysis by immunoprecipitation of the C-terminal GIT1-C2 with anti-Flag antibodies showed coprecipitation of N-terminal HA-GIT1-N (Figure 2B). The interaction of GIT1-C2 with GIT1-N was not mediated by indirect binding via endogenous βPIX, because association of endogenous βPIX to the overexpressed proteins in the immunoprecipitates was hardly detectable under our experimental conditions (Figure 2B). GIT1-C2 is known to interact efficiently with βPIX. That we could hardly detect endogenous βPIX in the immunoprecipitates including overexpressed GIT1-C2 is probably due to the fact that most endogenous βPIX is stably associated to endogenous GIT1 (see Figure 9A) and therefore not available for the interaction with the overexpressed GIT1-C2 polypeptide.
Figure 2.
Interaction between N- and C-terminal fragments of GIT1. (A) GIT1-derived constructs used in this study. ANK, ankyrin repeats; LZ, leucine zipper. (B–D) COS7 cells transfected with the indicated constructs were used for immunoprecipitation. Filters were cut and incubated for immunoblotting. Anti-Flag antibodies were used to coimmunoprecipitate either Flag-GIT1-C2 with HA-GIT1-N (B), or Flag-GIT1-C2-LZ with HA-GIT1-N (D). (C) Immunoprecipitation of endogenous PIX did not show any association with transfected HA-GIT1-N. IP, immunoprecipitates with the indicated antibodies, or with protein A-Sepharose beads without antibody (−); Lys, lysate; Ub, unbound fraction after immunoprecipitation. (E) Pull down on control (−) or GST-GIT1-C2–coated beads (C2) from lysates of COS7 cells (400 μg protein/pull down) untransfected (−), or transfected with GIT1-N. (F) Direct interaction between the bacterially purified fusion proteins GIT1-N2 and GIT1-C2 (see Materials and Methods for details). Immunoprecipitates (IP) with anti-GIT1 antibody recognizing GIT1-C2 were blotted, and filters were incubated to detect His-GIT1-N2 and gst-GIT1-C2. Ig-L, light chains of immunoglobulins.
Figure 9.
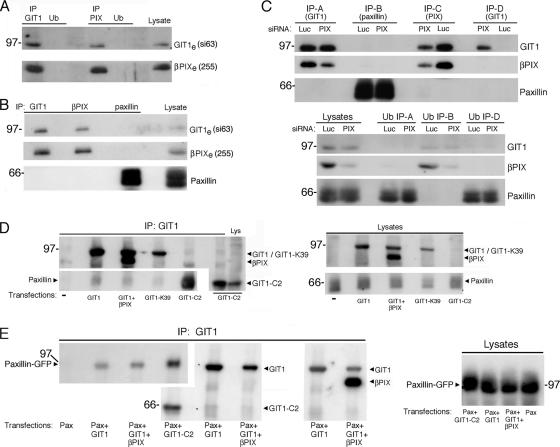
Association of paxillin with the βPIX/GIT1 complex. (A) Immunoprecipitation (IP) with either anti-GIT1 or anti-PIX antibodies depletes both proteins from COS7 lysates. Ub, aliquot of the unbound material after immunoprecipitation. Lysate, 200 μg; 630 μg, lysate/immunoprecipitation. (B) Endogenous paxillin is not associated with the endogenous βPIX/GIT1 complex. Aliquots of 1.4 mg of COS7 lysate were immunoprecipitated with antibodies for GIT1, PIX, or paxillin; immunoprecipitates were analyzed for the presence of the three proteins. Last lane, 200 μg of lysate. (C) Knockdown of βPIX in COS7 cells. Cells were transfected with 50 nM of either control (Luc) or βPIX-specific oligonucleotides. Two days after transfection, cells lysates were used for IP with the indicated antibodies. For IP-D, the unbound fraction after immunoprecipitation with anti-PIX (IP-C) was used for immunoprecipitation with the anti-GIT1 antibody. Filters with immunoprecipitations, lysates (Ly), and unbound (Ub) fractions were cut and incubated with the indicated antibodies. (D) COS7 cells were transfected with the indicated constructs, and same amounts of protein lysate were immunoprecipitated with anti-Flag antibodies recognizing the tagged GIT1, GIT1-K39, and GIT1-C2 polypeptides. Binding of endogenous paxillin was much stronger to GIT1-C2 than to GIT1, GIT1-K39, or βPIX/GIT1 complex. The top blot was incubated to reveal both transfected GIT1 and βPIX polypeptides. (E) COS7 cells were transfected or cotransfected as indicated, and equal amounts of protein (200 μg) were immunoprecipitated for full-length GIT1 or GIT1-C2. Immunoblotting reveals stronger binding of paxillin-GFP to GIT1-C2 than to GIT1 or to the βPIX/GIT1 complex (Pax, paxillin-GFP). Right, immunoprecipitates include part of the filter blotted for GIT1 (last two lanes) that has been reblotted to reveal the cotransfected βPIX protein. Lysates, 50 μg each.
GIT1-N includes a truncated SHD domain that very poorly interacts with βPIX (Di Cesare et al., 2000). To exclude that binding of endogenous βPIX to GIT1-N could bridge the interaction between GIT1-C2 and GIT1-N, we immunodepleted endogenous βPIX with anti-PIX antibodies, and we found no association of the immunoprecipitated endogenous βPIX to transfected GIT1-N (Figure 2C).
GIT1 is a dimeric protein. Dimerization occurs via the leucine zipper located in the C-terminal half of the protein (Figure 2A). We have previously shown that GIT1-C2 is dimeric as the full-length protein and that mutation of the leucine zipper in the GIT1-C2-LZ construct prevented dimerization (Paris et al., 2003). To exclude that endogenous GIT1 or GIT2 may help bridging the transfected GIT1-N and GIT1-C2 proteins, we tested the interaction between GIT1-N and the monomeric form of GIT1-C2. We found that monomeric GIT1-C2-LZ could still interact with GIT1-N, thus excluding bridging via dimerization with the endogenous GIT proteins (Figure 2D).
The interaction between the C- and N-terminal fragments of GIT1 was confirmed by binding in vitro of transfected GIT1-N to purified, bacterially expressed GST-GIT1-C2 (Figure 2E). Finally, the direct interaction between the bacterially purified fragments GIT1-N2 and GIT1-C2 could be demonstrated in vitro by immunoprecipitation of a complex between the two fragments by using antibodies specific for GIT1-C2 (Figure 2F).
To characterize the identified intramolecular interaction, we made use of several different GIT1-derived constructs. GIT1-N and GIT1-C2 overlap for part of the SHD domain. We have used a series of shorter N-terminal constructs to look at the requirements for the interaction with the C-terminal GIT1-C2 polypeptide (Supplemental Figure 1). The nonoverlapping fragments GIT1-N2 and GIT1-C2 could still interact efficiently. Further deletion of two of the three ankyrin repeats present after the ArfGAP domain (GIT1-N4) did not evidently affect the interaction with GIT1-C2, whereas the interaction was clearly decreased for the GIT1-GAP polypeptide including just the ArfGAP domain. Together, these data indicate that GIT1 may exist in a “closed” conformation as the result of the interaction between the N- and C-terminal halves of GIT1 (Figure 3A). Our data also suggest that the intramolecular interaction engages an extended region of the N-terminal portion including both the ArfGAP domain and at least the first ankyrin repeat (Supplemental Figure 1).
Figure 3.
Identification of the domains required for the closed conformation of GIT1. (A) Schematic representation of the closed (top) and open (bottom) conformations of GIT1. Ank, ankyrin repeats; GAP, ArfGAP domain; LZ, leucine zipper. (B–D) Flag-tagged GIT1-derived constructs were coexpressed with HA-GIT1-N in COS7 cells. Lysates were immunoprecipitated with anti-Flag mAb, and filters were probed with anti-Flag and anti-HA antibodies. (E–H) Requirement of the SHD domain of GIT1 for the interaction with the N-terminal portion of GIT1. A monomeric C-terminal polypeptide of GIT1 interacts with GIT1-ΔSHD. Lysates from cells cotransfected with the indicated GIT1-derived constructs were immunoprecipitated with anti-Flag antibodies. In each panel, immunoprecipitates (IP) were blotted with anti-Flag (top filters) and anti-HA (bottom filters) antibodies. Full-length GIT1 can associate with GIT1-C2 to form dimers via the LZ domain (E), but it cannot associate with monomeric GIT1-C2-LZ (F). Monomeric GIT1-C2-LZ can interact with GIT1-ΔSHD (G). (H) The GIT1-(229-431) construct including the SHD domain interacts with GIT1-N. (I) COS7 cells coexpressing Flag-GIT1-(229-431) with either Flag-GIT1 or Flag-GIT1-ΔSHD were lysed, immunoprecipitated with pAb SI-61 and blotted to reveal the transfected polypeptides. Ly, lysates; C, controls (beads without antibody, incubated with lysate).
According to the proposed model, the C-terminal part of the closed full-length protein would be unavailable for binding to a coexpressed GIT1-N fragment, because the C terminus of the full-length protein would be occupied by the binding to the intramolecular N-terminal segment (Figure 3A). We tested by coimmunoprecipitation experiments the availability of the full-length GIT1 for binding to exogenous GIT1-N. As expected, we found that full-length GIT1 was unable to bind GIT1-N (Figure 3B). The same was true for the monomeric full-length GIT1 (GIT1-LZ), and for the GIT1-K39, a dimeric full-length GIT1 protein mutated in arginine 39, a conserved arginine essential for normal GAP activity in several ArfGAPs (Mandiyan et al., 1999; Jackson et al., 2000; Randazzo et al., 2000; Szafer et al., 2000).
Thus, we tested different deletion mutants of GIT1 for their capacity to interact with N-terminal GIT1-N. The aim of this approach was to identify regions of GIT1 necessary to keep the protein in the closed conformation. First, we set to identify N-terminal domains implicated in the intramolecular interaction with the C-terminal part of GIT1, and required for the closed conformation of GIT1. We reasoned that if one of the intramolecular N-terminal domains was essential to keep the protein in the closed conformation, its deletion would release the intramolecular interaction, thus allowing the C-terminal half of the deleted “open” molecule to bind a coexpressed exogenous GIT1-N fragment. We found that deletion of either the ankyrin repeats (GIT1-ΔAnk), or the ArfGAP domain (GIT1-C3) induced binding of the resulting polypeptide to GIT1-N (Figure 3C). Therefore, we can conclude that each of the two N-terminal regions (ArfGAP domain and ankyrin repeats) is necessary for keeping the protein in the closed conformation, and their deletion results in exposure of a C-terminal half competent for binding to GIT1-N.
We next identified the SHD domain as one region of the C-terminal half of GIT1 required for binding to GIT1-N. We postulated that the SHD domain could be essential for the intramolecular interaction responsible for the closed conformation of GIT1. If so, the absence of the SHD domain should cause the release of the intramolecular interaction, and should prevent binding of the resulting open polypeptide to an exogenous GIT1-N fragment. In support of this hypothesis, we found that the deletion of the SHD domain produced a GIT1-ΔSHD polypeptide unable to interact with GIT1-N (Figure 3D). Moreover, in contrast to GIT1-C2, the shorter C-terminal fragment GIT1-C lacking most of the SHD domain could not interact with GIT1-N (Figure 3D). We attribute the lack of binding of GIT1-C to GIT1-N to the fact that GIT1-C, in contrast to GIT1-C2, does not have the SHD domain required for binding to the ankyrin repeats found in the amino-terminal GIT1-N. Therefore, the presence of the SHD domain is essential in mediating the intramolecular interactions between the C-terminal half of GIT1 and the N-terminal part including GAP domain and ankyrin repeats.
To prove that the deletion of the SHD domain was sufficient to induce an open conformation in the GIT1-ΔSHD polypeptide, we compared the ability of full-length GIT1 and GIT1-ΔSHD to interact with the monomeric C-terminal fragment GIT1-C2-LZ. GIT1-C2-LZ differs from GIT1-C2 for the absence of 2 leucines in the LZ domain that are necessary for dimerization (Figure 2A). We have previously shown that this mutant does not form dimers (Paris et al., 2003). As expected, the full-length GIT1 was able to interact with GIT1-C2 to form mixed dimers via the LZ domains (Figure 3E), but not with GIT1-C2-LZ that cannot form mixed dimers. Moreover, the lack of interaction between full-length GIT1 and GIT1-C2-LZ confirmed that the N-terminal half of the full-length GIT1 polypeptide was not available for binding to the coexpressed GIT1-C2-LZ polypeptide (Figure 3F): according to our model, the SHD domain of GIT1-C2-LZ could not interact with GIT1, because the access to the N-terminal part of the full-length protein was hindered by the intramolecular interaction with the C-terminal portion of the full-length molecule. In contrast, the deletion of the SHD domain resulted in the binding of GIT1-ΔSHD to GIT1-C2-LZ, indicating that the deletion of the SHD domain caused the N-terminal part of the mutant GIT1 to become available for binding to the coexpressed C-terminal GIT1-C2-LZ (Figure 3G). These results support the conclusion that the SHD domain is necessary for the intramolecular interaction with the N terminus of GIT1, thus keeping the full-length protein in a closed conformation (Figure 3A).
The weaker interaction between GIT1-C2 and the GIT1-GAP polypeptide (including only the ArfGAP domain; Supplemental Figure 1), and the results from experiments with the GIT1-ΔAnk mutant and with the mutants lacking the SHD domain (GIT1-C and GIT1-ΔSHD; Figure 3D) suggest that two interactions are responsible for the closed conformation of GIT1: an interaction between the ankyrin repeats and the SHD domain, and a second interaction between the ArfGAP domain and part of the C-terminal region following the SHD domain (Figure 3G). Disruption of the first intramolecular interaction by deletion of the SHD domain prevented the binding of GIT1-N to the remaining C-terminal portion in the GIT1-ΔSHD polypeptide (Figure 3D), suggesting that binding of the SHD domain to the ankyrin repeats is essential to keep the closed conformation. This hypothesis is supported by the finding that GIT1-(229-431), a polypeptide including just about the intact SHD domain of GIT1, was able to interact with GIT1-N (Figure 3H). In support of the hypothesis that the SHD domain is important to keep the closed conformation, we also found a clear interaction of GIT1-(229-431) with GIT1-ΔSHD, in contrast with almost no interaction between GIT1-(229-431) and the full-length protein (Figure 3I).
The Release of the Intramolecular Interaction Allows Increased Binding of GIT1 to Paxillin and Liprin-α1
The binding site of different GIT1 ligand proteins has been localized to the C-terminal half of GIT1. Binding of some of these proteins to full-length GIT1 seemed suboptimal compared with binding of the same proteins to truncated GIT1-C2 (Figure 1). We tested whether the release of the intramolecular interaction of GIT1 by more limited deletions was sufficient to increase the binding of GIT1 partners to the C terminus of the protein. We first compared the recovery of endogenous paxillin in immunoprecipitates from different GIT1-derived constructs. Deletion of either the ankyrin repeats or the SHD domain resulted in enhanced binding to endogenous paxillin that was comparable with that observed with GIT1-C2 (Figure 4A). Quantification showed an average 12-fold increase in paxillin binding to either GIT1-ΔSHD or GIT1-ΔAnk compared with full-length GIT1 (Figure 4B). We found a substantial variation in the levels of endogenous paxillin in lysates and immunoprecipitates from the same amount of the different lysates (i.e., same amount of total protein). This can be explained by the finding that overexpression of some of the constructs resulted in a significant fraction of the transfected protein and of endogenous paxillin ending up in the Triton-insoluble pellet (Figure 4A). This was true for GIT1-C2, and even more evident for GIT1-ΔSHD and GIT1-ΔAnk. Together, these data indicate that binding of paxillin to GIT1 requires either deletion of the amino-terminal portion of the protein, or opening of GIT1 by deletion of specific internal domains, to make the carboxy-terminal part available for binding to paxillin. Moreover, our results indicate that opening of GIT1 exposes sites that are prone to contributing in protein aggregation, and they suggest that the regulation by the proposed conformational change may be essential to timely regulate GIT1 function and to prevent harmful effects in the cell by inappropriate exposure of GIT1 sites involved in protein–protein interactions.
Figure 4.
Increased binding of paxillin and liprin-α1 to GIT1 lacking either the SHD or the ankyrin repeats. (A) Aliquots of lysates from transfected cells were immunoprecipitated with anti-paxillin antibodies. Immunoprecipitates (IP), lysates, and Triton-insoluble pellets were blotted to detect endogenous paxillin and GIT1-derived polypeptides. Two parallel immunoprecipitates from lysates of cells transfected with GIT1-C2 were loaded, each blotted either for paxillin or for GIT1-C2, because these polypeptides run very close on gels. (B) Quantification of the ratio between the intensity of GIT1-derived polypeptides and endogenous paxillin in immunoprecipitates from lysates, as shown in A. The ratios were normalized with respect to the ratio between full-length GIT1 and endogenous paxillin (equal to 1). Bars represent average values from at least three experiments ± SEM (C) Lysates from cells cotransfected with the Myc-tagged F3 fragment of liprin-α1 and one of the indicated GIT1-derived constructs were immunoprecipitated with anti-Myc antibodies. Immunoprecipitates, lysates (Lys) and unbound fractions (Ub) were blotted with anti-Myc to detect F3 (bottom parts of blots) and with anti-Flag to detect the GIT1-derived polypeptides (top parts of blots). A faint band for full-length GIT1 is found associated with F3 (asterisk).
To further support our model, we tested the binding of different GIT1 constructs to the F3 fragment of liprin-α1, which includes the GIT1-binding region (Ko et al., 2003). As for paxillin, we found a strong increase in the efficiency of binding of F3 to GIT1-ΔSHD and GIT1-ΔAnk compared with full-length GIT1 (Figure 4C). From these results, we conclude that the disruption of the intramolecular interaction involving the SHD domain and the ankyrin repeats is sufficient to expose the C-terminal regions required for binding to distinct GIT1 partners.
The Release of the Intramolecular Interaction Affects Cell Spreading, but Not Adhesion to Fibronectin
GIT proteins are regulators of cytoskeletal dynamics during cell spreading and migration, and they are considered part of multimolecular complexes localizing signaling components to specific cellular locations, where they regulate cell motility (Frank et al., 2006; Hoefen and Berk, 2006; Yin et al., 2005). We have used cell adhesion and cell spreading as functional assays, with the aim of looking for differential effects between the full-length GIT1, and the GIT1 mutants that bind paxillin efficiently upon release of the intramolecular interaction.
We first evaluated the effects of the expression of the constructs on the adhesion of COS7 cells to fibronectin. We did not find any significant effect on cell adhesion by expressing either the full-length GIT1 or its deletion mutants (Figure 5A). In contrast, significant effects were observed on cell spreading, measured as the average area of cells plated on fibronectin for 30 or 60 min. Although cells overexpressing the full-length GIT1 did not show significant effects on spreading compared with control β-galactosidase–expressing cells, all the deletion mutants tested showed significant effects on cell spreading (Figure 5B). The effect observed on spreading was either inhibitory (GIT1-C2, GIT1-N, GIT1-ΔAnk, and GIT1-ΔSHD) or stimulatory (GIT1-C). In contrast to the effects observed upon expression of the deletion mutants, the lack of evident effects on spreading in cells overexpressing the full-length protein supports the idea that GIT1 needs to be activated to affect cell shape. In contrast, the inhibition of cell spreading by the N-terminal fragment GIT1-N may be a consequence of the activation of the Arf6-GAP, which could cause inhibition of Arf6 activity that is required for cell spreading (Song et al., 1998; Dunphy et al., 2006). In this direction, we found that coexpression of the Arf6 activator EFA6 prevented GIT1-N-induced inhibition of cell spreading (Figure 5C), whereas EFA6 coexpression did not affect significantly the inhibition of cell spreading by the other mutants of GIT1 (Figure 5D). Moreover, overexpression of GIT1-N caused the reduction of Arf6-GTP levels in transfected cells when compared with cells transfected with full-length GIT1, and this reduction could be prevented by coexpression of the Arf6 GEF EFA6 (A. Totaro and I. de Curtis, unpublished data).
Figure 5.
Effects of the expression of GIT1 mutants on cell adhesion and spreading on fibronectin. (A) Adhesion assays were performed by letting cells attach to fibronectin for 30 min. Quantification was performed as described in Materials and Methods. 100%, attachment of control cells expressing β-galactosidase (LacZ). (B) Cell spreading was measured as described in Materials and Methods, by calculating the areas of cells 30 min (black bars) and 60 min (gray bars) after plating on fibronectin. Each bar represents the average value ± SEM (n = 100 cells/bar, from two independent experiments; n = 150 for LacZ). Asterisks indicate the significance of the difference with the corresponding values from cells expressing β-galactosidase, as determined by the Student's t test (p < 0.05). (C) Expression of EFA6 prevents inhibition of cell spreading by GIT1-N. Cell spreading was measured in cells transfected with the indicated constructs and plated for 60 min on fibronectin. Each bar represents the average area ± SEM (n = 100 cells/bar, from 2 independent experiments). The asterisks indicate the significance of the difference with the values from cells expressing β-galactosidase, as determined by the Student's t test (p < 0.05). (D) Expression of EFA6 specifically prevents inhibition of cell spreading by GIT1-N. COS7 cells transfected with the indicated constructs in absence (clear gray) or presence of EFA6 (dark gray) were plated for 60 min on fibronectin. Each bar represents the average area ± SEM (n = 50). The asterisk indicates a significant difference in spreading between cells cotransfected with GIT1-N and EFA6 versus cells transfected with GIT1-N only, as determined by the Student's t test (p < 0.01).
Because all the GIT1 mutants tested, with the exception of GIT1-N, can efficiently bind endogenous paxillin (Figure 4, A and B), and given the proposed role of paxillin in recruiting GIT1 at focal adhesions, we looked for possible reasons to explain the negative effects on cell spreading observed by expressing the three paxillin binding-competent GIT1 constructs GIT1-C2, GIT1-ΔSHD, and GIT1-ΔAnk. We therefore analyzed the morphology of the transfected cells under the same conditions used in the cell spreading assay (Figure 6). Overexpression of either control β-galactosidase, or full-length GIT1 did not induce evident effects on cell shape, nor on the distribution of endogenous paxillin (Figure 6, A and B). In contrast, with the exception of GIT1-C (Figure 6G), all tested mutants clearly showed reduced spreading compared with cells transfected with β-galactosidase or full-length GIT1 (Figure 6, C–F). In particular, cells expressing the PBS-containing constructs GIT1-C2 (Figure 6C), GIT1-ΔAnk (Figure 6E), and GIT1-ΔSHD (Figure 6F) showed a strong decrease in paxillin-positive peripheral focal adhesions compared with control cells or cells transfected with full-length GIT1 (Figure 6, A and B). The decreased presence of paxillin at the cell periphery correlated with reduced spreading on fibronectin (Figure 5B). As already described for GIT1-C2 (Di Cesare et al., 2000), all truncated constructs including the PBS domain, with the exception GIT1-C (Figure 6G), were often found in large cytoplasmic structures, where the transfected proteins colocalized with endogenous paxillin (Figure 6, C, E, and F). Therefore, displacement of paxillin from focal adhesions by these mutants may explain their negative effects on cell spreading. Staining for vinculin (Figure 6) and β1 integrins (Supplemental Figure 2) showed the presence of focal adhesions in cells expressing the GIT1 mutants negatively affecting spreading. Therefore, these mutants specifically affected paxillin localization. In the case of GIT1-N that lacks the PBS domain and therefore cannot interact with paxillin (Di Cesare et al., 2000), paxillin-positive focal adhesions could still be observed throughout the cell, whereas localization of endogenous paxillin at the GIT1-N–positive structures dispersed in the cytoplasm was not obvious (Figure 6D, insets), indicating that this mutant must affect cell spreading by a different mechanism. The enhanced spreading observed in cells expressing the GIT1-C mutant (Figure 5B) correlated with the colocalization of GIT1-C with endogenous paxillin in focal adhesions at the periphery of the transfected cells (Figure 6G). GIT1-C colocalized more clearly with paxillin at peripheral focal adhesions compared with full-length GIT1-expressing cells (Figure 7).
Figure 6.
Effects of the expression of full-length and mutant GIT1 constructs on cell morphology, and paxillin and vinculin distribution. COS7 cells transfected with the indicated constructs were trypsinized and replated on fibronectin-coated coverslips. After 60 min of culture to allow attachment and spreading, cells were fixed and double stained for the transfected protein (red in merges) and for either endogenous vinculin or paxillin (green in merges). In each panel, same fields are shown in the first and second column, and in the third and fourth column, respectively. Asterisks indicate the transfected cells. Bar, 20 μm; 10 μm in the insets in D.
Figure 7.
Colocalization of GIT1-C with endogenous paxillin at the cell edge and at focal adhesions. COS7 cells transfected with the GIT1-C (A) or full-length GIT1 (B) were trypsinized and replated on fibronectin-coated coverslips. After 60 min of colture, cells were fixed and double stained for the transfected protein (red) and for endogenous paxillin (green). Bar, 10 μm.
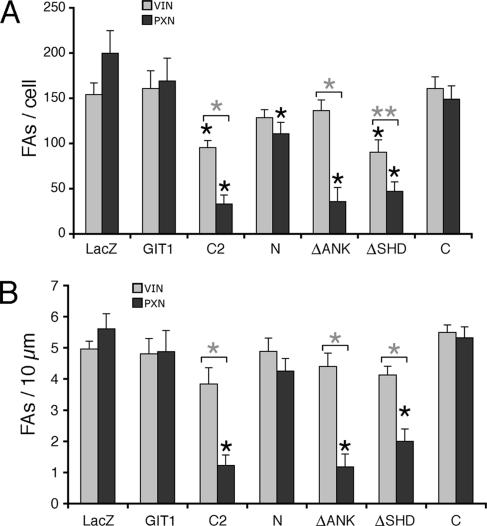
Quantification of the effects of the expression of the different GIT1 constructs on focal adhesions confirmed a statistically significant striking decrease in paxillin-positive focal adhesions in cells expressing the paxillin-binding constructs GIT1-C2, GIT1-ΔSHD, or GIT1-ΔAnk, compared with total focal adhesions stained by vinculin (Figure 8A). This finding correlates with the observed decrease in spreading induced by these mutants. Moreover, the number of total vinculin-positive focal adhesions per cell showed a less marked but significant decrease in cells transfected with GIT1-C2 or GIT1-ΔSHD (Figure 8A). In contrast, no significant difference was observed between paxillin-positive and vinculin-positive focal adhesions in cells expressing GIT1-N, which does not bind paxillin.
Figure 8.
COS7 cells transfected with the indicated constructs were stained for vinculin (light bars, VIN) or paxillin (dark bars, PXN), and then they were analyzed by confocal microscopy for the quantification of the number of focal adhesions (FAs) by using the ImageJ software analysis program. (A) Number of focal adhesions per cell. (B) Number of peripheral focal adhesions per 10 μm, calculated dividing the total number of focal adhesions found all around the cell edge by the perimeter of the cell. Each bar represents the average value ± SEM (8–10 cells for each sample). Asterisks indicate statistical significance by the Student's t test (*p < 0.01; **p < 0.03). The black asterisks indicate a statistically significant difference with respect to the corresponding value for LacZ; the gray asterisks indicate a statistically significant difference between the vinculin-positive and paxillin-positive focal adhesions in cells transfected with the same construct.
During spreading, continuous reorganization of focal adhesions at the cell edge is necessary for protrusion. For this reason, we have considered the density of focal adhesions at the cell edge. Only minor differences were observed in the density of vinculin-positive focal adhesions between GIT1-C2–, GIT1-ΔAnk–, and GIT1-ΔSHD–expressing cells and control cells. On the contrary, the density of paxillin-positive focal adhesions was strongly reduced in cells expressing any of the three paxillin-binding constructs inhibiting cell spreading (Figure 8B). Together, our data indicate that the release of the identified intramolecular interaction represents an important mechanism to regulate GIT1 functions, which include the regulation of the subcellular distribution of paxillin.
PAK-Pbd and βPIX Are Both Required to Induce Paxillin Binding Competent GIT1
The GIT-binding site in the C-terminal portion of PIX proteins interacts with the SHD domain in the central part of the GIT polypeptides. It has been suggested that binding of βPIX to the SHD domain of GIT1 induces a conformational change leading to increased binding of GIT1 to paxillin (Zhao et al., 2000). These studies showed that coexpression of βPIX with GIT1 in COS7 cells increased binding to overexpressed GFP-paxillin. We set to further investigate this issue. Immunoprecipitation with either an anti-GIT1 or an anti-βPIX antibody resulted in the complete recovery of both endogenous proteins in the immunoprecipitates, with consequent immunodepletion from the lysates (Figure 9A). This result demonstrated that endogenous GIT1 and βPIX were associated in stable constitutive complexes in COS7 cells. The analysis of anti-GIT1 or anti-PIX immunoprecipitates by immunoblotting with anti-paxillin antibodies did not show any association of endogenous paxillin with the endogenous βPIX/GIT1 complex (Figure 9B). Accordingly, immunoprecipitation with anti-paxillin antibodies under immunodepleting conditions for endogenous paxillin did not show any association of this protein with the endogenous βPIX/GIT1 complexes. These results demonstrate the lack of detectable association of paxillin with endogenous βPIX/GIT1 complexes in the experimental conditions examined.
To test whether binding of βPIX to GIT1 could prevent binding of GIT1 to paxillin, we down-regulated endogenous βPIX by using βPIX-specific target sequences for siRNA, and then we tested for association between GIT1 and paxillin in the absence of βPIX (Figure 9C). siRNA for βPIX specifically down-regulated the expression of this protein by 85% compared with control siRNA. Immunoprecipitation with either anti-GIT1 or anti-paxillin antibodies from βPIX-depleted lysates to immunodepletion of the respective antigens showed no association between the endogenous proteins (Figure 9C, IP-A and IP-B). To eliminate the residual endogenous βPIX/GIT1 complexes in siRNA-treated cells, we first immunoprecipitated siRNA-treated lysates with anti-PIX antibodies (Figure 9C, IP-C), and then we used the unbound fractions for immunoprecipitation with anti-GIT1, to pull down the residual GIT1 protein free of βPIX; again, no association of paxillin to the GIT1 polypeptide could be detected in the absence of βPIX (Figure 9C, IP-D). Therefore, βPIX association to endogenous GIT1 did not seem to prevent association to paxillin.
Immunoprecipitation from lysates of transfected COS7 cells revealed low levels of endogenous paxillin associated with overexpressed GIT1 (Figure 9D). The association of endogenous paxillin with GIT1 was evidently not affected by coexpression of βPIX, nor by the introduction of a point mutation in the ArfGAP domain of GIT1 (GIT1-K39) that inhibits the ArfGAP activity. In contrast, paxillin was more efficiently recovered in immunoprecipitates of truncated GIT1-C2 (Figure 9D). We repeated this experiment cotransfecting green fluorescent protein (GFP)-tagged paxillin with either GIT1, GIT1 + βPIX, or GIT1-C2 (Figure 9E). Again, the recovery of paxillin-GFP in the immunoprecipitates with anti-GIT1 antibodies was similar in lysates from cells overexpressing GIT1 alone or in combination with βPIX. In contrast, we could detect an increase in the binding of paxillin-GFP to GIT1-C2 (Figure 9E). Similarly, coexpression of βPIX with GIT1 did not alter the efficiency of binding of GIT1 to liprin-α1 (data not shown). Possible experimental differences that may explain the discrepancy between our results and those from Zhao et al. (2000) could not be determined, due to the limited technical information for this experiment in the Zhao et al. (2000) study. In contrast, the various approaches included in our study clearly indicate that the association of βPIX with GIT1 is not sufficient to enhance binding of two ligands, paxillin and liprin-α, to the C-terminal part of full-length GIT1. We therefore postulate that βPIX binding is not sufficient to induce a change in the conformation of GIT1 that is required to increase binding to its partners under all experimental conditions described in this study.
Previous studies indicated that PAK is required for the recruitment of GIT and PIX proteins at sites of adhesion to the extracellular matrix by a kinase-independent mechanism: expression of the PAK regulatory domain (amino acid 1-329) or the autoinhibitory domain (amino acid 83-149) induces GIT2/PKL, PIX, and PAK localization to focal adhesions, indicating a kinase-independent scaffolding role for PAK (Brown et al., 2002). The N-terminal portion of PAK1 contains a proline-rich region (amino acid 184-204) that binds the SH3 domain of PIX (Manser et al., 1998). One possibility is that PAK induces paxillin binding by interacting with βPIX, thereby inducing the release of the intramolecular interaction in GIT1.
To test for a kinase-independent effect of PAK on GIT1 binding to paxillin, we expressed in cells the PAK–Pbd fragment (amino acid 150–250 of PAK1), including the PIX-binding proline-rich region (Za et al., 2006). This construct was used for coimmunoprecipitation experiments, to assess the effects on the binding of GIT1 to endogenous paxillin (Figure 10). Although the formation of the βPIX/GIT1 complex had no effects on paxillin binding (Figures 9D and 10A), coexpression of PAK-Pbd with GIT1 and βPIX or with GIT1 and monomeric βPIX-ΔLZ clearly enhanced binding to endogenous paxillin (Figure 10A). Quantification showed an average 2.5-fold increase in paxillin binding to PAK-Pbd/βPIX/GIT1 complexes compared with βPIX/GIT1 complexes (Figure 10B). Moreover, PAK-Pbd did not induce a significant increase in binding of paxillin to GIT1 in the absence of βPIX (Figure 10, A and B).
Figure 10.
PAK-Pbd enhances binding of the βPIX/GIT1 complex to paxillin. (A) Lysates from cells cotransfected with the indicated constructs were immunoprecipitated with anti-Flag antibodies. Filters with immunoprecipitates (IP), and equal amounts of lysates and unbound fractions after immunoprecipitations, were blotted as indicated, using anti-Flag (for GIT1), anti-HA (for βPIX), anti-Paxillin (for endogenous paxillin), and anti-Myc (for PAK-Pbd) antibodies. ΔLZ = βPIX-ΔLZ. (B) Quantification of the ratio between the intensity of bands for endogenous paxillin and GIT1 in immunoprecipitates with anti-Flag, as shown in A. The ratios were normalized with respect to the ratio (equal to 1) in the immunoprecipitates from cells cotransfected with GIT1 and βPIX. Bars represent average values from at least three experiments ± SEM. The asterisk indicates significant differences with respect to immunoprecipitations from cells transfected with GIT1 and βPIX, as determined by the Student's t test (p < 0.05). (C) Immunoprecipitation of GIT1 with anti-Flag antibodies from lysates of COS7 cells transfected with the indicated constructs. Filters were cut and blotted with the indicated antibodies. (D) Aliquots of three of the lysates shown in C were immunoprecipitated with anti-Myc antibodies to detect full-length PAK and PAK-Pbd. Filters were cut and incubated with the indicated antibodies.
PAK is known to exist in an inactive dimeric state, with the N-terminal autoregulatory domain of one partner interacting with the kinase domain of the other partner (Bokoch, 2003). Binding of the Cdc42/Rac interactive binding sequence within the autoregulatory region of PAK by GTP-bound Rac or Cdc42 “opens” the molecule by exposing the different domains of PAK. Interestingly, in contrast to PAK-Pbd, overexpression of full-length PAK1 could not induce an evident increase in paxillin binding to the βPIX/GIT1 complex (Figure 10C). Moreover immunoprecipitation of the two PAK constructs with anti-Myc antibodies showed lower levels of βPIX/GIT1 associated with full-length PAK1 compared with PAK-Pbd (Figure 10D). These data suggest that activation of PAK is required to expose the βPIX binding site necessary for binding to the βPIX/GIT1 complex, and for the consequent enhancement of GIT1 binding to paxillin.
DISCUSSION
We have used a biochemical approach to identify and characterize an intramolecular interaction between the N-terminal and C-terminal portions of GIT1. We have considered the change in efficiency of association of GIT1 with two of its partners, paxillin and liprin-α, as a measure of the release of the intramolecular interaction. We propose that the release of the identified intramolecular interaction represents an important mechanism for the timely activation of at least some of the functions of GIT1 in the cell.
Given the complexity of the molecular assemblies that can be formed by GIT1 and its partners, the tight regulation of the different interactions becomes a must for proper function of this protein in the cell. One indication of the importance of such regulation comes from the observation that the artificial expression of certain truncated C-terminal fragments of the protein cause large intracellular membrane-bound aggregates (Matafora et al., 2001). Moreover, aggregates containing C-terminal (but not N-terminal) GIT1 fragments are found in huntingtin aggregates of patients (Goehler et al., 2004). The formation of these structures leads to dysregulation of GIT1 function that may affect normal cell function (Za et al., 2006).
The data presented in this study show that binding of either paxillin or liprin-α to GIT1 requires the release of an intramolecular interaction that keeps the protein in a binding-incompetent state. We have shown that the release of this interaction, and the consequent increase in the ability of GIT1 to bind its partners, may be obtained by N-terminal truncations of the GIT1 polypeptide, or by deletion of specific internal domains. The biochemical and functional analysis presented here indicates that the intramolecular inhibitory mechanism may rely on two distinct interactions between the N- and C-terminal portions of GIT1: one interaction implicating the ankyrin region and the SHD domain, and a second interaction between the GAP domain and the C-terminal part of GIT1 (Figure 3A). That the disruption of the intramolecular interaction between the SHD domain prevented the binding of GIT1-N with the remaining C-terminal portion suggests that the interaction between the SHD domain and the ankyrin repeats is probably the main intramolecular interface to keep the closed conformation of GIT1.
The lack of effects on paxillin distribution after the overexpression of the full-length protein supports the model that GIT1 needs to be activated to influence the morphology of the cells. Moreover, our data indicate that although the use of deletion constructs is informative for studying the mechanisms of GIT1 activation, the analysis of the subcellular distribution of the constructs shows that they may alter the localization of endogenous ligands, thus affecting cell spreading. In contrast to the full-length protein, the release of the intramolecular interaction in deletion mutants results in effects on cell spreading and morphology that may be explained by the exposure of specific domains in the mutants.
The lack of effects on the localization of paxillin at focal adhesions in cells expressing GIT1-N suggest that the inhibitory effects on spreading by this mutant need to be explained by different mechanisms. We have tested the hypothesis that overexpression of GIT1-N may down-regulate Arf6 activation, a GTPase implicated in the regulation of cell motility (Brown et al., 2001). Accordingly, we found that coexpression of the Arf6 GEF EFA6 was able to recover specifically the inhibition of cell spreading by GIT1-N, but not by the paxillin-binding mutants (Figure 5), supporting a role of GIT1 in the regulation of Arf6 activity during the protrusive activity of the cell (de Curtis 2001).
The data support the hypothesis that the different outcomes on cell spreading are a consequence of the properties of the distinct paxillin binding-competent mutants. Paxillin is implicated in the regulation of cell spreading and migration. In this respect, the recruitment of paxillin to focal adhesion is necessary for focal adhesion turnover (Webb et al., 2004), and fibroblasts deficient in paxillin show defects in the cortical cytoskeleton, cell spreading and migration (Hagel et al., 2002). Paxillin participates in the recruitment of molecular complexes, including GIT and PIX proteins at focal adhesions and at the cell edge (Manabe et al., 2002; Lamorte et al., 2003). Truncation mutants of GIT1 including either the SHD domain (e.g., GIT1-C2) or the ankyrin repeats (e.g., GIT1-N) form cytoplasmic structures that include endocytic markers, whereas the carboxy-terminal construct GIT1-C lacking the SHD domain shows a diffuse cytoplasmic distribution (Di Cesare et al., 2000). Here, we also found that the expression of GIT1-ΔSHD and GIT1-ΔAnk results in formation of cytoplasmic structures. The inhibitory effects on cell spreading by these mutants correlate with the exposure of different regions of the overexpressed polypeptides (SHD domain, ankyrin repeats) to the cellular environment. The inhibitory effects of GIT1-C2, GIT1-ΔAnk, and GIT1-ΔSHD on cell spreading may be due to the sequestration of a significant fraction of endogenous paxillin that interferes with normal paxillin function at the cell edge during cell spreading (Nishiya et al., 2001; Tsubouchi et al., 2002; Lamorte et al., 2003). The finding that these structures are formed following the expression of the deletion/truncation mutants, but not of the full-length protein, underlines the importance of regulating the exposure of specific domains of GIT1. In contrast, enhancement of cell spreading by the paxillin binding-competent GIT1-C polypeptide may be explained by GIT1-C binding paxillin (Di Cesare et al., 2000) without causing formation of cytoplasmic aggregates. The lack of aggregation correlates with lack of both the SHD domain and the ankyrin repeats in GIT1-C, which therefore may act as an active form of GIT1 that favors paxillin-mediated reorganization at the cell periphery (Figures 6G and 7), required for Rac-dependent protrusion (Di Cesare et al., 2000).
We also found that an N-terminal fragment of PAK was required to enhance binding of paxillin to the full-length βPIX/GIT1 complex, as detected by the specific increase in binding of paxillin to the tripartite complex including GIT1, βPIX, and PAK-Pbd (Figure 10). Our results indicate that PAK may act as a regulator of GIT1 by inducing the release of the autoinhibitory intramolecular interaction, and they suggest that the formation of a trimeric complex including activated PAK, βPIX, and GIT1 is required for conformational changes needed to expose the C terminus of GIT1. Association of PAK-Pbd with the PIX/GIT complex causes the unmasking of the C terminus including the PBS region, thus promoting paxillin binding, which is required for the recruitment of the GIT complex at sites of protrusion. In support of this model, activated Rac1 and Cdc42, which cause PAK to associate with PIX/GIT complexes, induce the translocation of these proteins to focal complexes (Manser et al., 1998; Matafora et al., 2001; Brown et al., 2002; Shikata et al., 2003; Loo et al., 2004). Moreover, a PAK mutant that is unable to bind PIX blocks the localization of PIX/GIT to focal complexes (Brown et al., 2002). Alternatively, several studies point to a role of phosphorylation in the regulation of GIT proteins (Brown et al., 2005; Hoefen and Berk, 2006). Recently, several phosphorylation sites have been identified in the central and C-terminal portions of GIT1 (Webb et al., 2006a) that may participate in the regulation of GIT1 functions. In particular, it has been suggested that phosphorylation on serine 709 within the PBS domain of GIT1 is required to stimulate binding to paxillin, and to regulate protrusive activity in cells (Webb et al., 2006b). Conversely, phosphorylation of paxillin by PAK at serine 273 regulates paxillin–GIT1 interaction and increases migration and protrusion by promoting the localization of a GIT1/PIX/PAK signaling module near the leading edge (Nayal et al., 2006). More work will be required to further define the model for GIT1 regulation, by combining the effects of phosphorylation with the conformational changes described in this study.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Telethon-Italy grant GGP05051, by the European Union MAIN Network of Excellence FP6–502935, and by the Italian Ministry of University and Research within the framework of the Fondo per gli Investimenti della Ricerca di Base project grant FIRB RBNE01WY7P.
Abbreviations used:
- βPIX
p21-activated kinase interacting exchange factor
- ArfGAP
Arf GTPase-activating protein
- GIT
G-protein coupled receptor kinase-interacting protein
- PBS
paxillin-binding site
- SHD
Spa2-homology domain.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-06-0550) on September 26, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Albertinazzi C., Za L., Paris S., de Curtis I. ADP-ribosylation factor 6 and a functional PIX/p95-APP1 complex are required for Rac1B-mediated neurite outgrowth. Mol. Biol. Cell. 2003;14:1295–1307. doi: 10.1091/mbc.E02-07-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokoch M. G. Biology of the p21-activated kinases. Annu. Rev. Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- Bokoch M. G., Reilly M. A., Daniels H. R., King C. C., Olivera A., Spiegel S., Knaus G. U. A GTPase-independent mechanism of p21-activated kinase activation. Regulation by sphingosine and other biologically active lipids. J. Biol. Chem. 1998;273:8137–8144. doi: 10.1074/jbc.273.14.8137. [DOI] [PubMed] [Google Scholar]
- Botrugno A., O., Paris S., Za L., Gualdoni S., Cattaneo A., Bachi A., de Curtis I. Characterization of the endogenous GIT1-betaPIX complex, and identification of its association to membranes. Eur. J. Cell Biol. 2006;85:35–46. doi: 10.1016/j.ejcb.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Brown D. F., Rozelle L. A., Yin L. H., Balla T., Donaldson G. J. Phosphatidylinositol 4,5-bisphosphate and Arf6-regulated membrane traffic. J. Cell Biol. 2001;154:1007–1017. doi: 10.1083/jcb.200103107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., West A. K., Turner E. C. Paxillin-dependent paxillin kinase linker and p21-activated kinase localization to focal adhesions involves a multistep activation pathway. Mol. Biol. Cell. 2002;13:1550–1565. doi: 10.1091/mbc.02-02-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C. M., Cary A. L., Jamieson S. J., Cooper A. J., Turner E. C. Src and FAK kinases cooperate to phosphorylate paxillin kinase linker, stimulate its focal adhesion localization, and regulate cell spreading and protrusiveness. Mol. Biol. Cell. 2005;16:4316–4328. doi: 10.1091/mbc.E05-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattelino A., Longhi R., de Curtis I. Differential distribution of two cytoplasmic variants of the alpha 6 beta 1 integrin laminin receptor in the ventral plasma membrane of embryonic fibroblasts. J. Cell Sci. 1995;108:3067–3078. doi: 10.1242/jcs.108.9.3067. [DOI] [PubMed] [Google Scholar]
- Claing A., Chen W., Miller E. W., Vitale N., Moss J., Premont T. R., Lefkowitz J. R. beta-Arrestin-mediated ADP-ribosylation factor 6 activation and beta 2-adrenergic receptor endocytosis. J. Biol. Chem. 2001;276:42509–42513. doi: 10.1074/jbc.M108399200. [DOI] [PubMed] [Google Scholar]
- de Curtis I. Cell migration: GAPs between membrane traffic and the cytoskeleton. EMBO Rep. 2001;2:277–281. doi: 10.1093/embo-reports/kve072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cesare A., Paris S., Albertinazzi C., Dariozzi S., Andersen J., Mann M., Longhi R., de Curtis I. p95-APP1 links membrane transport to Rac-mediated reorganization of actin. Nat. Cell Biol. 2000;2:521–530. doi: 10.1038/35019561. [DOI] [PubMed] [Google Scholar]
- Dunphy L., J., Moravec R., Ly K., Lasell K. T., Melancon P., Casanova E. J. The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of beta1 integrins. Curr. Biol. 2006;16:315–320. doi: 10.1016/j.cub.2005.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco M., Peters J. P., Boretto J., van Donselaar E., Neri A., D'Souza-Schorey C., Chavrier P. EFA6, a sec7 domain-containing exchange factor for ARF6, coordinates membrane recycling and actin cytoskeleton organization. EMBO J. 1999;18:1480–1491. doi: 10.1093/emboj/18.6.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank R. S., Adelstein R. M., Hansen H. S. GIT2 represses Crk- and Rac1-regulated cell spreading and Cdc42-mediated focal adhesion turnover. EMBO J. 2006;25:1848–1859. doi: 10.1038/sj.emboj.7601092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehler H., et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- Haendeler J., Yin G., Hojo Y., Saito Y., Melaragno M., Yan C., Sharma K. V., Heller M., Aebersold R., Berk C. B. GIT1 mediates Src-dependent activation of phospholipase Cγ by angiotensin II and epidermal growth factor. J. Biol. Chem. 2003;278:49936–49944. doi: 10.1074/jbc.M307317200. [DOI] [PubMed] [Google Scholar]
- Hagel M., George L. E., Kim A., Tamimi R., Opitz L. S., Turner E. C., Imamoto A., Thomas M. S. The adaptor protein paxillin is essential for normal development in the mouse and is a critical transducer of fibronectin signaling. Mol. Cell. Biol. 2002;22:901–915. doi: 10.1128/MCB.22.3.901-915.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefen J. R., Berk C. B. The multifunctional GIT family of proteins. J. Cell Sci. 2006;119:1469–1475. doi: 10.1242/jcs.02925. [DOI] [PubMed] [Google Scholar]
- Jackson R. T., Brown D. F., Nie Z., Miura K., Foroni L., Sun J., Hsu W. V., Donaldson G. J., Randazzo A. P. ACAPs are arf6 GTPase-activating proteins that function in the cell periphery. J. Cell Biol. 2000;151:627–638. doi: 10.1083/jcb.151.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., et al. The GIT family of proteins forms multimers and associates with the presynaptic cytomatrix protein Piccolo. J. Biol. Chem. 2003;278:6291–6300. doi: 10.1074/jbc.M212287200. [DOI] [PubMed] [Google Scholar]
- Ko J., Kim S., Valtschanoff G. J., Shin H., Lee R. J., Sheng M., Premont T. R., Weinberg J. R., Kim E. Interaction between liprin-alpha and GIT1 is required for AMPA receptor targeting. J. Neurosci. 2003;23:1667–1677. doi: 10.1523/JNEUROSCI.23-05-01667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahuna O., Quellari M., Achard C., Nola S., Meduri G., Navarro C., Vitale N., Borg J. P., Misrahi M. Thyrotropin receptor trafficking relies on the hScrib-betaPIX-GIT1-ARF6 pathway. EMBO J. 2005;24:1364–1374. doi: 10.1038/sj.emboj.7600616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamorte L., Rodrigues S., Sangwan V., Turner E. C., Park M. Crk associates with a multimolecular Paxillin/GIT2/beta-PIX complex and promotes Rac-dependent relocalization of Paxillin to focal contacts. Mol. Biol. Cell. 2003;14:2818–2831. doi: 10.1091/mbc.E02-08-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo H., T., Ng W., Y., Lim L., Manser E. GIT1 activates p21-activated kinase through a mechanism independent of p21 binding. Mol. Cell. Biol. 2004;24:3849–3859. doi: 10.1128/MCB.24.9.3849-3859.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manabe R., Kovalenko M., Webb J. D., Horwitz R. A. GIT1 functions in a motile, multi-molecular signaling complex that regulates protrusive activity and cell migration. J. Cell Sci. 2002;115:1497–1510. doi: 10.1242/jcs.115.7.1497. [DOI] [PubMed] [Google Scholar]
- Mandiyan V., Andreev J., Schlessinger J., Hubbard R. S. Crystal structure of the ARF-GAP domain and ankyrin repeats of PYK2-associated protein beta. EMBO J. 1999;18:6890–6898. doi: 10.1093/emboj/18.24.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser E., Loo H. T., Koh G. C., Zhao S. Z., Chen Q. X., Tan L., Tan I., Leung T., Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol. Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- Matafora V., Paris S., Dariozzi S., de Curtis I. Molecular mechanisms regulating the subcellular localization of p95-APP1 between the endosomal recycling compartment and sites of actin organization at the cell surface. J. Cell Sci. 2001;114:4509–4520. doi: 10.1242/jcs.114.24.4509. [DOI] [PubMed] [Google Scholar]
- Meyer Z. M., Deliot N., Chasserot-Golaz S., Premont T. R., Bader F. M., Vitale N. Regulation of neuroendocrine exocytosis by the ARF6 GTPase-activating protein GIT1. J. Biol. Chem. 2006;281:7919–7926. doi: 10.1074/jbc.M600081200. [DOI] [PubMed] [Google Scholar]
- Nayal A., Webb J. D., Brown M. C., Schaefer M. E., Vicente-Manzanares M., Horwitz R. A. Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J. Cell Biol. 2006;173:587–589. doi: 10.1083/jcb.200509075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiya N., Tachibana K., Shibanuma M., Mashimo I. J., Nose K. Hic-5-reduced cell spreading on fibronectin: competitive effects between paxillin and Hic-5 through interaction with focal adhesion kinase. Mol. Cell. Biol. 2001;21:5332–5345. doi: 10.1128/MCB.21.16.5332-5345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paris S., Longhi R., Santambrogio P., de Curtis I. Leucine-zipper-mediated homo- and hetero-dimerization of GIT family p95-ARF GTPase-activating protein, PIX-, paxillin-interacting proteins 1 and 2. Biochem. J. 2003;372:391–398. doi: 10.1042/BJ20030047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premont T. R., Claing A., Vitale N., Freeman L. J., Pitcher A. J., Patton A. W., Moss J., Vaughan M., Lefkowitz J. R. Beta2-Adrenergic receptor regulation by GIT1, a G protein-coupled receptor kinase-associated ADP ribosylation factor GTPase-activating protein. Proc. Natl. Acad. Sci. USA. 1998;95:14082–14087. doi: 10.1073/pnas.95.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premont T. R., Perry J., S., Schmalzigaug R., Roseman T., J., Xing Y., Claing A. The GIT/PIX complex: an oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors. Cell Signal. 2004;16:1001–1011. doi: 10.1016/j.cellsig.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Randazzo A. P., Andrade J., Miura K., Brown T. M., Long Q. Y., Stauffer S., Roller P., Cooper A. J. The ARF GAP ASAP1 regulates the actin cytoskeleton. Proc. Natl. Acad. Sci. USA. 2000;97:4011–4016. doi: 10.1073/pnas.070552297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikata Y., Birukov G. K., Garcia G. J. S1P induces FA remodeling in human pulmonary endothelial cells: role of Rac, GIT1, FAK, and paxillin. J. Appl. Physiol. 2003;94:1193–1203. doi: 10.1152/japplphysiol.00690.2002. [DOI] [PubMed] [Google Scholar]
- Song J., Khachikian Z., Radhakrishna H., Donaldson G, J. Localization of endogenous ARF6 to sites of cortical actin rearrangement and involvement of ARF6 in cell spreading. J. Cell Sci. 1998;111:2257–2267. doi: 10.1242/jcs.111.15.2257. [DOI] [PubMed] [Google Scholar]
- Szafer E., Pick E., Rotman M., Zuck S., Huber I., Cassel D. Role of coatomer and phospholipids in GTPase-activating protein-dependent hydrolysis of GTP by ADP-ribosylation factor-1. J. Biol. Chem. 2000;275:23615–23619. doi: 10.1074/jbc.M003171200. [DOI] [PubMed] [Google Scholar]
- Tsubouchi A., Sakakura J., Yagi R., Mazaki Y., Schaefer E., Yano H., Sabe H. Localized suppression of RhoA activity by Tyr31/118-phosphorylated paxillin in cell adhesion and migration. J. Cell Biol. 2002;159:673–683. doi: 10.1083/jcb.200202117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner E. C., Brown C. M., Perrotta A. J., Riedy C. M., Nikolopoulos N. S., McDonald R. A., Bagrodia S., Thomas S., Leventhal S. P. Paxillin LD4 motif binds PAK and PIX through a novel 95-kD ankyrin repeat, ARF-GAP protein: a role in cytoskeletal remodeling. J. Cell Biol. 1999;145:851–863. doi: 10.1083/jcb.145.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale N., Patton A. W., Moss J., Vaughan M., Lefkowitz J. R., Premont T. R. GIT proteins, a novel family of phosphatidylinositol 3,4, 5-trisphosphate-stimulated GTPase-activating proteins for ARF6. J. Biol. Chem. 2000;275:13901–13906. doi: 10.1074/jbc.275.18.13901. [DOI] [PubMed] [Google Scholar]
- Webb J. D., Donais K., Whitmore A. L., Thomas M. S., Turner E. C., Parsons T. J., Horwitz F. A. FAK–Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- Webb J. D., Mayhew W. M., Kovalenko M., Schroeder J. M., Jeffery D. E., Whitmore L., Shabanowitz J., Hunt F. D., Horwitz F. A. Identification of phosphorylation sites in GIT1. J. Cell Sci. 2006a;119:2847–2850. doi: 10.1242/jcs.03044. [DOI] [PubMed] [Google Scholar]
- Webb J. D., Kovalenko M., Whitmore L., Horwitz F. A. Phosphorylation of serine 709 in GIT1 regulates protrusive activity in cells. Biochem. Biophys. Res. Commun. 2006b;346:1284–1288. doi: 10.1016/j.bbrc.2006.06.036. [DOI] [PubMed] [Google Scholar]
- West A. K., Zhang H., Brown C. M., Nikolopoulos N. S., Riedy C. M., Horwitz F. A., Turner E. C. The LD4 motif of paxillin regulates cell spreading and motility through an interaction with paxillin kinase linker (PKL) J. Cell Biol. 2001;154:161–176. doi: 10.1083/jcb.200101039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G., Haendeler J., Yan C., Berk C. B. GIT1 functions as a scaffold for MEK1-extracellular signal-regulated kinase 1 and 2 activation by angiotensin II and epidermal growth factor. Mol. Cell. Biol. 2004;24:875–885. doi: 10.1128/MCB.24.2.875-885.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin G., Zheng Q., Yan C., Berk C. B. GIT1 is a scaffold for ERK1/2 activation in focal adhesions. J. Biol. Chem. 2005;280:27705–27712. doi: 10.1074/jbc.M502271200. [DOI] [PubMed] [Google Scholar]
- Za L., Albertinazzi C., Paris S., Gagliani M., Tacchetti C., de Curtis I. βPIX controls cell motility and neurite extension by regulating the distribution of GIT1. J. Cell Sci. 2006;119:2654–2666. doi: 10.1242/jcs.02996. [DOI] [PubMed] [Google Scholar]
- Zhao S. Z., Manser E., Loo H. T., Lim L. Coupling of PAK-interacting exchange factor PIX to GIT1 promotes focal complex disassembly. Mol. Cell. Biol. 2000;20:6354–6363. doi: 10.1128/mcb.20.17.6354-6363.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S. Z., Lim P. J., Ng W. Y., Lim L., Manser E. The GIT-associated kinase PAK targets to the centrosome and regulates Aurora-A. Mol. Cell. 2005;20:237–249. doi: 10.1016/j.molcel.2005.08.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.