Abstract
The major keratins in the pancreas and liver are keratins 8 and 18 (K8/K18), but their function seemingly differs in that liver K8/K18 are essential cytoprotective proteins, whereas pancreatic K8/K18 are dispensable. This functional dichotomy raises the hypothesis that K8-null pancreata may undergo compensatory cytoprotective gene expression. We tested this hypothesis by comparing the gene expression profile in pancreata of wild-type and K8-null mice. Most prominent among the up-regulated genes in K8-null pancreas was mRNA for regenerating islet-derived (Reg)-II, which was confirmed by quantitative reverse transcription-polymerase chain reaction and by an anti-Reg-II peptide antibody we generated. Both K8-null and wild-type mice express Reg-II predominantly in acinar cells as determined by in situ hybridization and immunostaining. Analysis of Reg-II expression in various keratin-related transgenic mouse models showed that its induction also occurs in response to keratin cytoplasmic filament collapse, absence, or ablation of K18 Ser52 but not Ser33 phosphorylation via Ser-to-Ala mutation, which represent situations associated with predisposition to liver but not pancreatic injury. In wild-type mice, Reg-II is markedly up-regulated in two established pancreatitis models in response to injury and during the recovery phase. Thus, Reg-II is a likely mouse exocrine pancreas cytoprotective candidate protein whose expression is regulated by keratin filament organization and phosphorylation.
INTRODUCTION
Intermediate filaments (IFs), microfilaments, and microtubules are the three major cytoskeletal protein groups of mammalian cells (Bershadsky and Vasiliev, 1988; Ku et al., 1999). All IFs share a common prototype structure consisting of a coiled-coil α-helical rod domain that is interrupted by linkers and flanked by non-α-helical N-terminal “head” and C-terminal “tail” domains (Fuchs and Cleveland, 1998; Herrmann et al., 2003; Coulombe and Wong, 2004; Herrmann and Aebi, 2004). Within the IF family, keratins are the largest subfamily, and they make up the IFs of epithelial cells, whereas other IFs are characteristic of unique cell types (e.g., desmin in myocytes, neurofilaments in neuronal cells). This tissue-specific expression reflects the involvement of IFs in a broad range of tissue-selective human diseases (Fuchs and Cleveland, 1998; Omary et al., 2004). Keratins (K) include the type I and type II IFs, and all epithelial cells express at least one type I and one type II keratins as obligate noncovalent heteropolymers (Moll et al., 1982; Schweizer et al., 2006). In simple-type epithelia, as found in the liver, pancreas, and intestine, the major keratins are K18/K19/K20 (type I) and K7/K8 (type II), with the K8/K18 pair being dominant depending on the tissue (Moll et al., 1982; Ku et al., 1999; Herrmann et al., 2003; Zhou et al., 2003).
Keratin networks are versatile structures that undergo dynamic reorganization in response to a variety of intra- and extracellular cues. Posttranslational modifications and keratin-binding partners are essential in mediating such responses (Coulombe and Omary, 2002; Omary et al., 2006). Among the posttranslational modifications, phosphorylation is the best studied, and several K8/K18 in vivo phosphorylation sites have been identified, including K8 S23/S73/S431 and K18 S33/S52 (Omary et al., 2006). An increase in K8/K18 phosphorylation occurs in response to numerous stimuli, including stress, apoptosis, and mitosis, and such phosphorylation regulates keratin filament reorganization and interaction with binding proteins (Omary et al., 1998, 2006). Furthermore, site-specific keratin phosphorylation serves an important cytoprotective function in the liver, as demonstrated using transgenic mouse models that overexpress the phosphomutant keratins K18 S52A (Ku et al., 1998; Ku and Omary, 2006) or K8 S73A (Ku and Omary, 2006), and it is a marker of progression of human liver disease (Toivola et al., 2004).
The use of keratin-related transgenic mouse models has provided a powerful tool in helping clarify the important cytoprotective function of keratins that would otherwise have been difficult to demonstrate using cell culture systems. For example, overexpression of mutant K14 in transgenic mice causes a blistering skin disease (Vassar et al., 1991) that led to the subsequent identification of K14 mutations as a cause of epidermolysis bullosa simplex (Coulombe et al., 1991). For simple-type epithelial keratins, the liver is the major disease-related target of keratin mutation or absence, likely due to the expression of only K8/K18 in hepatocytes (unlike other epithelia, e.g., intestinal that express additional keratins), coupled with the obligate heteropolymeric nature of keratins. For example, transgenic mice that overexpress K18 R89C (Ku et al., 1996), and K8-null or K18-null mice (Loranger et al., 1997; Toivola et al., 1998; Caulin et al., 2000; Zatloukal et al., 2000; Ku and Omary, 2006) are highly susceptible to drug-induced liver injury, and their hepatocytes are exquisitely fragile upon liver perfusion. K18 Arg89 is a highly conserved IF residue whose mutation causes disruption of the hepatocyte cytoplasmic keratin filament network. These findings, coupled with studies of patients with liver disease of multiple etiologies, collectively show that K8/K18 serve an essential protective role in the liver and support the importance of KRT8 and KRT18 genes as susceptibility markers for liver disease progression (Ku et al., 2001, 2005; Strnad et al., 2006).
In contrast to the liver, the functional significance of K8/K18 in the pancreas is very different despite a relatively comparable keratin expression profile in both mouse organs. For example, ductal cells of both organs share similar keratin expression patterns, whereas pancreatic acinar cells consist of two different keratin filament compartments: cytoplasmic filaments that consist predominantly of K8/K18, and apicolateral-proximal filaments that contain K8/K18/K19 (Toivola et al., 2000a) and low levels of K20 (Zhong et al., 2004). Surprisingly, K8-null mice, which lack pancreatic acinar cell keratins (due to degradation of K18/K19/K20 in the absence of their binding partner K8), and K18 R89C mice (which have a disrupted cytoplasmic filament network) had similar susceptibility to injury as wild-type mice in two independent pancreatitis models (Toivola et al., 2000a,b). This apparent cytoprotective dispensability of keratins in the pancreas raised the hypothesis that keratin mutation or absence triggers a unique compensatory response in the pancreas that allows the keratin-mutant mice to cope with insults directed to the exocrine pancreas. We tested this hypothesis by using an unbiased expression profiling approach, and we noted marked up-regulation of several regenerating islet-derived (Reg) family members. The Reg proteins are established stress-inducible proteins that ameliorate pancreatic injury (Graf et al., 2002; Bimmler et al., 2004; Zhang et al., 2004) and may therefore functionally compensate for the lack of keratins in the pancreas. We also examined changes in Reg expression in keratin mouse models that have altered keratin filament organization or phosphorylation.
MATERIALS AND METHODS
Antibodies
The primary antibodies (Abs) used in this study were sheep anti-mouse Reg-I (R&D Systems, Minneapolis, MN), rabbit anti-mouse Reg-II (see below), rabbit anti-mouse/human K8/K18 (Ab 4668) (Ku et al., 2004a), rat anti-mouse K19 (Troma III; Developmental Studies Hybridoma Bank, Iowa City, IA), rabbit anti-human K18 pS52 (Ab 3055; Liao et al., 1995), and Abs that recognize human K18 (DC10), mouse heat-shock protein 60 (Hsp60), and actin (NeoMarkers, Fremont, CA).
Mouse Strains and Acute Pancreatitis Models
We used K8, K18, or K19 null mice and transgenic mouse lines that overexpress wild-type (WT) human (h) K18 or K18 R89C (Ku et al., 1995), K18 S33A (Ku et al., 2002), or K18 S52A (Ku et al., 1998). For the K18 S33A and S52A mice, two independent mouse lines for each mutation were used (S33A1 and S33A2; S52A1 and S52A2). All mice are in an FVB/n genetic background except for the K18-null mice, which are in a mixed 129/Sv and MF-1 background (Magin et al., 1998). Nontransgenic FVB/n and BALB/c mice (Taconic Farms, Germantown, NY) were also used. K8-null (Baribault et al., 1994) and K18-null (Magin et al., 1998) mice (both available through The Jackson Laboratory), and K19-null (Hesse et al., 2000; Tamai et al., 2000) mice (−/−) and their wild type (+/+) littermates were generated by interbreeding of heterozygous (+/−) mice and subsequent polymerase chain reaction (PCR) genotype screening. Transgenic mice of the various strains were obtained by breeding heterozygous males with FVB/n females. PCR screening was done using primers specific for hK18 as described previously (Ku et al., 1995).
Two well-established mouse experimental pancreatitis models were used (Toivola et al., 2000a; Zhong and Omary, 2004; Zhong et al., 2004), and in all comparative cases age and sex-matched animals were used. For caerulein-induced pancreatitis (Jensen et al., 1980), mice were fasted (solid food only; 12–16 h) and then injected intraperitoneally seven times hourly with saline (vehicle control) or with 50 μg/kg caerulein (Research Plus, Bayonne, NJ). For the choline-deficient diet (CDD)-induced pancreatitis (Lombardi et al., 1975), female BALB/c mice weighing 14–19 g (typically 21–25 d old) were fasted (solid food only; 12–16 h) and then fed CDD (Harlan Teklad, Madison, WI) supplemented with 0.5% dl-ethionine (Sigma-Aldrich, St. Louis, MO) or normal Formulab 5008 chow (control group) (Deans Animal Feeds, Redwood City, CA) for 3 d and then switched to a standard diet. At the indicated times, animals were euthanized using CO2 inhalation, and pancreata were immediately removed for subsequent analysis.
Peptide Synthesis and Anti-Regenerating Islet-derived (Reg)-II Antibody Generation
Two rabbit Abs were generated (Anaspec, San Jose, CA) by immunizing with keyhole limpet hemocyanin conjugated to the peptide CHDPKSNRRWHWSSGS. This peptide is relatively conserved among Reg-I and Reg-II, but not among other Reg family members (Figure 3A). A Cys residue (underlined; which is not part of the Reg protein) was introduced to the N terminus to facilitate coupling to the carrier. Two antibodies, Ab 2817 and Ab 2818, were generated. Ab 2817 showed superior reactivity (data not shown), and it was used for all the described experiments. Specificity of the generated Abs was confirmed by mixing the Ab (1:500 dilution) with or without 200 μg/ml Reg peptide for 30 min before immunoblotting. Preimmune sera were also obtained from the rabbits before immunization and used as a negative control.
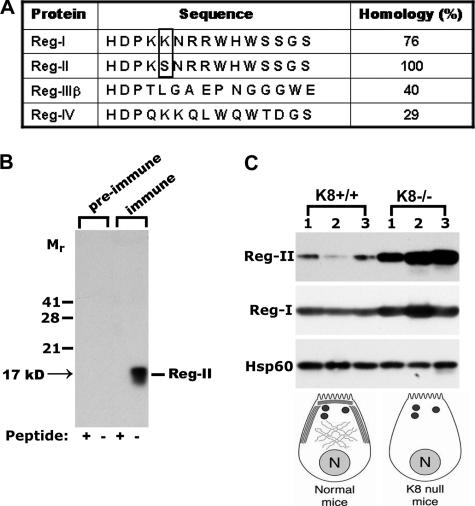
Figure 3.
Reg-II antibody generation and Reg-II/Reg-I expression in K8-WT and K8-null mouse pancreas. (A) The Reg-II peptide sequence (amino acids 109-123) was selected to generate rabbit anti-Reg-II antibodies. This peptide differs by one amino acid (boxed) compared with the Reg-I sequence, but it differs substantially from other Reg sequences. (B) The anti-Reg-II Ab (immune) and preimmune sera were used to blot total pancreas homogenates isolated from K8-null mice. Before immunoblotting, the tested antibodies were incubated with (+) or without (−) the peptide used for immunization. Note that the immune Ab recognizes a 17-kDa protein that corresponds to the predicted size of Reg-II. (C) Reg-II is dramatically up-regulated in K8-null pancreata, whereas Reg-I exhibits only a modest increase. Equal amounts of total pancreas homogenates were loaded in each lane and a blot of Hsp60 was used as a loading control. Schematic acinar cells highlight the phenotype of the keratin filament network in K8-WT and K8-null mice by displaying cytoplasmic filaments and apicolateral membrane-proximal filaments (N, nucleus; large dots, zymogen granules).
Microarray Analysis
Mouse cDNA microarrays were produced at the Stanford Functional Genomics Facility (Stanford, CA). The microarray experiments and data analysis were performed as described previously (Eisen and Brown, 1999; Zhou et al., 2005). Total RNA (80–100 μg) was extracted from pancreatic tissue using RNase midi kit (QIAGEN, Valencia, CA) and reverse transcribed into cDNA by using SuperScript II (Invitrogen, Carlsbad, CA) and random hexamer primers. K8-WT and K8-null derived cDNA were labeled with Cy3- and Cy5-deoxyuridine triphosphate, respectively (GE Healthcare, Chalfont St. Giles, United Kingdom). Twelve independent pairs of K8-null and K8-WT mouse cDNA probes were hybridized with a mouse 42K cDNA array. Fluorescence signals were acquired with a GenePix 4000b microarray scanner, and the images were processed with the GenePix Pro3.0 software (Axon Instruments, Foster City, CA). The original data files were entered into the Stanford Microarray Database and normalized (http://genome-www5.stanford.edu//), and they can be accessed by choosing “public login” and then selecting experimenter “XHJI” and experiments BJI1–12.
A filter was set to select array elements with a regression correlation of r > 0.6. To identify the up-regulated or down-regulated genes in K8-WT versus K8-null mouse pancreata, 1-class significance analysis of microarray (SAM) was performed. This analysis generated a list of genes with an average Cy5/Cy3 ratio significantly different from 1.0, together with an estimate of how many of these genes are false positive (at 90% confidence). The percentage of false-positive genes (i.e., false discovery rate [FDR]) is based on permutations of repeat measurements, and in our analysis we only included genes with an FDR of <1%. Genes were assigned manually to a functional pathway based on information retrieved from the Stanford Online Universal Resource for Clones and Expressed sequence tags (http://genome-www5.stanford.edu.laneproxy.stanford.edu/cgi-bin/SMD/source/sourceSearch).
Reverse Transcription PCR and Immunoblotting
Real-time reverse transcription-polymerase chain reaction (RT-PCR) was performed as described previously (Zhong et al., 2004) by using an ABI Prism 7900 Sequence Detection System (Applied Biosystems). Briefly, total RNA was isolated from pancreatic tissue using an RNeasy midi kit (QIAGEN) and reverse transcribed into cDNA using the SuperScript II reverse transcriptase kit (Invitrogen, Carlsbad, CA) and either random hexamer or oligo(dT) amplification. The selected target genes were amplified using intron-spanning primers (Table 1), and their expression levels were compared with L7 ribosomal protein (used as an internal control reference). Samples were analyzed in quadruplicates, and each experiment was performed at least three times. For immunoblotting, tissues were homogenized using a Teflon homogenizer in a buffer containing 0.187 M Tris-HCl, 3% SDS, and 5 mM EDTA, and the protein concentration was measured using the BCA protein assay (Pierce Chemical, Rockford, IL) (Ku et al., 2004). Equal amounts of proteins were separated by SDS-polyacrylamide gel electrophoresis, transferred to polyvinylidene difluoride membranes, and then immunoblotted and visualized by enhanced chemiluminescence (PerkinElmer Life and Analytical Sciences, Boston, MA). Quantification of the increase in Reg-II protein was carried out by immunoblotting of pancreatic total tissue lysates from two K8 WT and two K8-null mice, analyzed on the same gel with serial dilution of pancreatic lysates from one of the two K8-null mice. Sequential fractionation of a detergent-free, Nonidet P-40–soluble, Empigen-soluble, and remaining pellet solubilized in 2% SDS-containing sample buffer was carried out as described previously (Ku et al., 2004).
Table 1.
Primers used for real time RT-PCR and ISH
| Genea | Primer | Accession no. | Size (bp) |
|---|---|---|---|
| Real-time PCR | |||
| L7 | 5′-GAAAGGCAAGGAGGAAGCTCATCT | NM_011291 | 80 |
| 5′-AATCTCAGTGCGGTACATCTGCCT | |||
| Reg-I | 5′-GAACGCCTACTTCATCCTTGC | NM_009042 | 81 |
| 5′-GATGGCAGGTCTTCTTCAGC | |||
| Reg-II | 5′-GATCAGCATGGCTCAGAACA | NM_009043 | 87 |
| 5′-TCTTCAGCTACCTGGCCTTG | |||
| Reg-IIIδ | 5′-TGGAACCACAGACCTGGGCTA | NM_013893 | 74 |
| 5′-GAGCAGAAATGCCAGGTGTCC | |||
| Ywhae | 5′-TGAGTTTGCCACAGGAAATG | NM_009536 | 101 |
| 5′-GCGTTGGAGGAAGTTCTGTC | |||
| Ppm1b | 5′-GCGAGACCTCCCTTTCTACC | NM_011151 | 116 |
| 5′-ATAACGCAGACCATTCCCAG | |||
| Trap1a | 5′-TCAGAAGATGAGGCCGAAGA | NM_011635 | 101 |
| 5′-AAATGATGGCCAGGAACACA | |||
| Dna1c4 | 5′-TGCTGCAAAGATGATCAAGG | NM_017470 | 99 |
| 5′-ACCTCGTGGGTGATCTCAAA | |||
| AnnexA | 5′-GGGTGACCGTTGTCAGGACT | BC004594 | 100 |
| 5′-CACGTCTGTCCCCTTTCTCC | |||
| MT-AP | 5′-GCAAGTTCTTGCTGGACCTC | BC064444 | 101 |
| 5′-GAGCAGTGACCAGCAGACAG | |||
| Nubp1 | 5′-GTGATGTCAGTGGGCTTCCT | NM_011955 | 98 |
| 5′-ACATCACGGAGGAACTGCTT | |||
| Glg1 | 5′-GAAGCCAGGCTCTCCTACCT | NM_009149 | 94 |
| 5′-AATCCAGCATCTCACCTTGG | |||
| Snrpb2 | 5′-TACCGGGGAGACATGACATT | NM_021335 | 103 |
| 5′-GCATGGGACGGTGTAATCTT | |||
| ISH | |||
| Reg II | 5′-CTTATGCCTGATGTTCCTGTCA | NM_009043 | 489 |
| 5′-TAGGCTCTGAACTTGCAGACAA | |||
| Insulin | 5′-AGACCATCAGCAAGCAGGTC | NM_008386 | 366 |
| 5′-GGTGGGCCTTAGTTGCAGTA |
a L7, ribosomal protein L7; Reg-I, regenerating islet-derived protein-1; Reg-II, regenerating islet-derived protein-2; Reg-IIIδ (Ingaprp), regenerating islet-derived protein-3Δ (also termed islet neogenesis associated protein-related protein); Ywhae, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, ε polypeptide; Ppm1b, protein phosphatase 1B, magnesium dependent, β isoform; Dnalc4, dynein, axonemal, light chain 4; AnnexA, annexin A1; MT-AP, microtubule-associated protein 1; Nubp1, nucleotide binding protein 1; Glg1, Golgi apparatus protein 1; Snrpb2, U2 small nuclear ribonucleoprotein B.
Histological and Immunostaining, and In Situ Hybridization
For histological staining, pancreatic tissues were fixed in Formalin, embedded in paraffin, sectioned, and then stained with hematoxylin and eosin (H&E; Histo-Tec Laboratory, Hayward, CA). Immunohistochemistry staining was performed using the Envision horseradish peroxidase system (Dako North America, Carpinteria, CA) as recommended by the supplier. Immunofluorescence staining was performed as described previously (Ku et al., 2004). Images were analyzed with a Nikon Eclipse TE300 fluorescence microscope (Nikon, Melville, NY) combined with a Bio-Rad confocal laser scanner (Bio-Rad, Hercules, CA). Nonradioactive in situ hybridization (ISH) of paraffin-embedded sections was performed as described previously (St Croix et al., 2000; Iacobuzio-Donahue et al., 2002). Briefly, digoxigenin (DIG)-labeled sense and antisense probes were PCR amplified using primers with incorporated T7 promoter (Table 1). In vitro transcription was performed with T7 RNA polymerase and DIG RNA-labeling reagents (Roche Diagnostics, Indianapolis, IN). Sections (5 μm in thickness) were deparaffinized in xylene followed by dehydration using a graded ethanol/water mixture, incubated with 1% hydrogen peroxide, digested with a 10 μg/ml proteinase K (37°C; 30 min), and hybridized overnight at 55°C in mRNA hybridization buffer (Dako North America) with 200 ng/ml antisense or sense probes, washed in 2× standard saline citrate at 45°C, and incubated with a 1/35 dilution of RNase A cocktail (Ambion, Austin, TX) in 2× standard saline citrate (30 min; 37°C). Slides were then washed twice in 2× standard saline citrate/50% formamide, once in 0.08× standard saline citrate (both at 55°C) and biotin-blocking reagents (Dako North America) were applied to block the endogenous biotin. For signal amplification, a horseradish peroxidase-conjugated rabbit anti-digoxigenin antibody was used followed by incubation with a secondary streptavidin complex (GenPoint kit; Dako North America). The final signal was developed with diaminobenzidine, and the tissues were counterstained with hematoxylin.
RESULTS
Reg Genes Represent the Most Prominently Up-Regulated Gene Group in K8-Null Mouse Pancreata
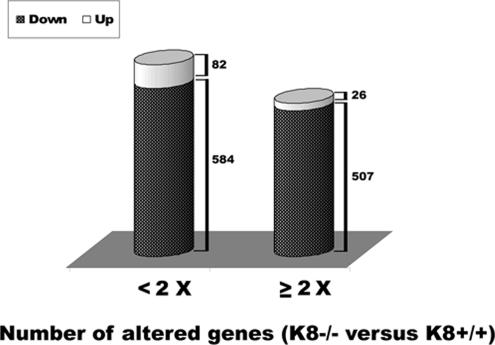
To search for candidate genes that may compensate for keratin cytoprotective functions, the pancreatic gene expression profile was compared in K8-null and K8-WT mice. In total, 8936 genes, which were well identified in at least 80% (10/12) of the arrays, were retrieved from the Stanford Microarray Database. When selecting 1-class SAM with FDR <1% (median FDR), 1199 genes (108 up- and 1091 down-regulated) were identified to be significantly altered (Figure 1). Not included were the 30 invalid spots, i.e., genes without name or accession number (data not shown). Five hundred and seven of 1091 down-regulated and 26 of 108 up-regulated genes were changed at least twofold (Supplemental Table 1). Among them, several Reg genes, including Reg-I, Reg-II, and islet neogenesis associated protein-related protein (Reg-IIIδ), were up-regulated in K8-null mouse pancreas, and of these Regs, Reg-II had the highest overall change (∼15-fold by array analysis; Table 2). Tumor rejection antigen P1A (Trap1a) was the most down-regulated gene (∼8-fold). We then tested the microarray results of the most altered genes, and we confirmed the changes in the Reg genes, particularly Reg-II, by using real-time RT-PCR (Table 2). Not all the changes observed by microarray analysis could be confirmed by real-time RT-PCR (e.g., Trap1a; Table 2).
Figure 1.
Gene expression profile in K8-null versus K8-WT mouse pancreas. Total RNA was extracted from pancreatic tissues of 12 pairs of K8-null and K8-WT mice followed by expression profiling analysis as described in Materials and Methods. Each pair was analyzed separately. The histograms depict the number of genes that are significantly altered in K8-null versus K8-WT mouse pancreata. The selection was performed by 1-class SAM analysis at an FDR of <1% and a -fold change of < or ≥2, respectively.
Table 2.
Expression of selected genes in K8-null versus K8-WT mouse pancreata as determined by microarray analysis and real-time RT-PCR
| Genbank accession no. | Producta | Fold change (microarray) | Fold change (real-time PCR) |
|---|---|---|---|
| K8-null vs. K8 WT up-regulated genes | |||
| AV078371 | Reg-2 | 14.7 | 17.9 |
| AV061702 | Reg-IIIδ | 4.1 | 3.3 |
| AV058239 | Reg-1 | 3.4 | 1.7 |
| AV006208 | Ywhae | 2.3 | 1.0 |
| AI324040 | Ppm1b | 2.0 | 1.3 |
| K8-null vs. K8 WT down-regulated genes | |||
| BG064272 | Trap1a | 8.2 | 0.7 |
| BG067281 | Dnalc4 | 5.5 | 1.1 |
| BG063567 | Annex-A | 4.9 | 1.0 |
| BG064420 | MT-AP | 4.8 | 1.1 |
| BG067587 | Nubp1 | 4.6 | 0.9 |
| BG067283 | Glg1 | 4.1 | 1.1 |
| BG067954 | Snrpb2 | 4.1 | 1.3 |
a Full versions of product abbreviations are given in the footnote to Table 1.
Reg-II Is a Product of Acinar Cells and Is Up-Regulated in K8-Null Pancreata
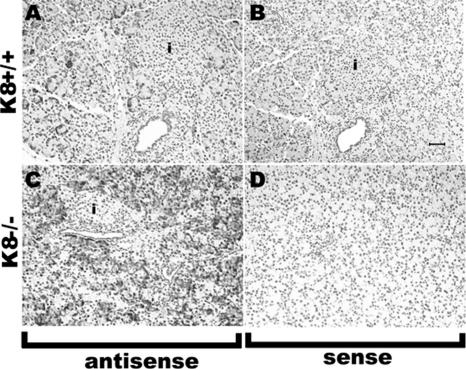
The distribution of Reg-II in K8-null and K8-WT mouse pancreata was investigated using ISH. Reg-II mRNA was localized exclusively in pancreatic acini but not in islets or ducts, and it was much more prominent in K8-null compared with K8-WT pancreata (compare Figure 2, C and G). As a control for islet staining, we used an insulin antisense probe that led to exclusive staining of pancreatic islets (Figure 2, B and F, arrows). The sense probe of Reg-II, which was used as another control, did not provide an appreciable signal (Figure 2, D and H), thereby providing support for the specificity of the findings by using the Reg-II antisense probe.
Figure 2.
Comparison of Reg-II mRNA distribution in K8-WT and K8-null mouse pancreas. Pancreatic tissues isolated from K8-null and K8-WT mice were analyzed by H&E staining (A and E) or by ISH (B–D and F–H) by using antisense probes to insulin (B and F) or Reg-II (C and G). As a control, ISH using a Reg-II sense probe was carried out on sections similar to those shown in C and G (D and H). Note the prominent Reg-II mRNA up-regulation in K8-null exocrine pancreas (G vs. C), whereas the insulin mRNA signal (highlighted by arrows in B and F) is present exclusively in islets as would be predicted. i, islets. Bar (in D, applies to all panels), 50 μm.
Next, we examined the expression of Reg proteins in K8-null pancreas. For this purpose, we generated an anti-peptide Ab 2817 to Reg-II amino acids 109-123. This motif is highly similar among Reg-I and II, differing only by one amino acid (Figure 3A), whereas other Reg family members exhibit a clearly divergent sequence. The generated Ab recognizes a major band of ∼17 kDa in K8-null pancreatic homogenates, which is in good accordance with the predicted size of Reg proteins (Unno et al., 1993). The observed antibody signal is specific, because it is not present in preimmune rabbit sera, and it is completely blocked by preincubation with the synthetic Reg-II peptide used for immunization (Figure 3B). Ab 2817 demonstrated a clearly different signal pattern upon two-dimensional immunoblotting compared with a commercially available Reg-I Ab, suggesting that it is specific to Reg-II (Reg-I and Reg-II were not well resolved using 1-dimensional gels; data not shown). Furthermore, the Reg-II signal is markedly up-regulated in K8-null mice, whereas Reg-I levels are only moderately increased (Figure 3C). Despite some variability between individual mice, 14 K8-null pancreata were tested, and all showed increased Reg-II levels compared with WT pancreata (data not shown). This variability is likely related to Reg-II being an exocrine secreted product that is similar to Reg-1 (De Reggi and Gharib, 2001). Quantification of the increase in protein level in two randomly selected pancreata showed a threefold increase in Reg-II protein levels (Supplemental Figure 1A). Sequential fractionation of WT and K8-null total pancreas homogenates into a detergent-free soluble fraction, a nonionic detergent-soluble fraction, a zwitterionic detergent-soluble pool, and the remaining pellet showed that Reg-II and Reg-I are found primarily in the detergent-free and NP-40 fractions without redistribution as a consequence of keratin absence (Supplemental Figure 1B). Therefore, the microarray findings were confirmed in that the lack of pancreatic keratin filaments in acinar cells is associated with a prominent Reg-II up-regulation at the mRNA and protein levels.
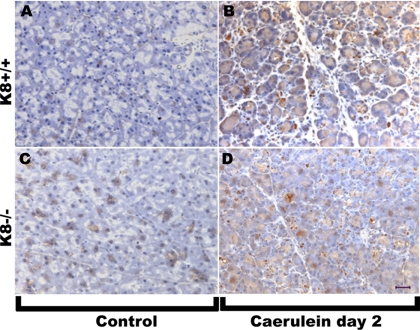
Immunohistochemistry staining of pancreatic tissues from K8+/+ and K8−/− mice confirmed the increase in Reg-II protein in K8-null pancreata (Figure 4, compare A and C). The fixation method afforded different reactivity profiles that may be related to Reg-II extractability or epitope recognition (data not shown). For example, acetone fixation (instead of the 10% Formalin used in Figure 4) did not show the clear increase in Reg-II, but it showed that Reg-II is located in the lumen of pancreatic acini, just outside of the apicolateral K19 fluorescence signal (Supplemental Figure 2).
Figure 4.
Reg-II immune staining in K8 WT and K8-null mouse pancreata before and after caerulein. Pancreata were removed from age and sex matched mice before (A and C) or 2 d after seven hourly injections of caerulein (B and D). After fixation in 10% Formalin and sectioning, immunohistochemistry of Reg-II was performed as described in Materials and Methods. Bar (D), 50 μm.
Effect of Keratin Phosphorylation and Cytoplasmic Organization on Reg-II Expression
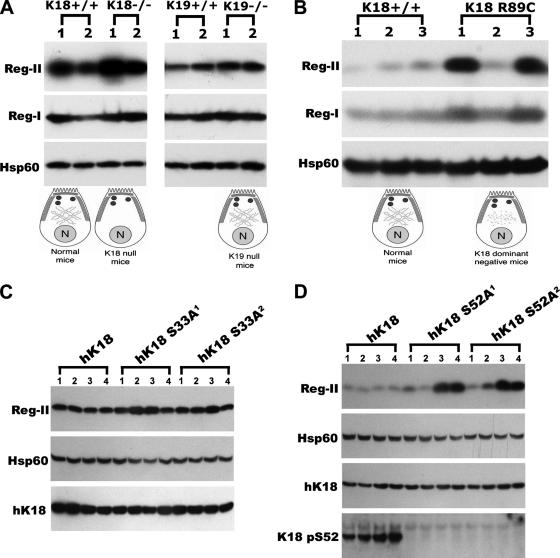
We took advantage of the availability of K18- and K19-null mice to study whether Reg-II up-regulation in K8-null pancreata is specific to K8 absence. K18 and K19 null pancreata have either no acinar cell cytoplasmic filaments (but a normal-looking apicolateral distribution) or a normal-looking acinar cell keratin filament distribution with both cytoplasmic and apicolateral filaments, respectively (Toivola et al., 2000a) (see Figure 5A for schematic representation). This keratin filament distribution contrasts with K8-null acinar cells that have no cytoplasmic or apicolateral filaments (Toivola et al., 2000a) (see Figure 3C for schematic representation). Absence of K18 but not K19 results in increased expression of Reg-II without significantly affecting Reg-I (Figure 5A). Similarly, disruption of the acinar cell keratin cytoskeleton as a consequence of the K18 R89C mutation also induces up-regulation of Reg-II and to a lesser extent Reg-I (Figure 5B). There is some variability between animals (e.g., K18 R89C pancreas 2; Figure 5B), which we attribute to Reg-II being a secreted protein. In support of this, Reg-II mRNA levels in K18 R89C pancreata is typically elevated two- to threefold (not shown but tested in 5 WT and 5 K18 R89C pancreata), whereas Reg-II protein from the same mice can be variable (as exemplified by Figure 5B). Fasting did not seem to affect the variability in Reg-II levels, and there were no obvious sex-related effect on the expression levels (data not shown).
Figure 5.
Reg-II and Reg-I expression in mouse genotypes with different keratin filament distribution and phosphorylation. Total mouse pancreas homogenates were analyzed by immunoblotting by using antibodies to the indicated proteins. (A) Reg-II protein levels are increased in K18-null mice, whose acinar cells lack cytoplasmic keratin filaments, but not in K19-null mice, which exhibit a normal-looking keratin filament network (represented by the schematic; see legend of Figure 3 for schematic descriptors). (B) Disruption of acinar cell cytoplasmic keratin filaments (schematic) as a consequence of K18 R89C mutation is associated with a prominent increase in Reg-II protein levels. In contrast, Reg-I levels are minimally altered. (C and D) Pancreata from mice that express the K18 phosphorylation mutants (two independent transgenic mouse lines per phosphomutant: S33A1, S33A2, S52A1, and S52A2) were analyzed by immunoblotting as carried out in A and B. K18 S52 but not S33 mutation is associated with elevated Reg-II protein levels. Presence of the human (h) K18 transgene product was confirmed in all cases using a human-K18–specific antibody. In addition, an anti-K18 phospho (p)-S52 was also used to confirm expression of the phosphomutant K18. Hsp60 was used as a loading control.
We also examined the role of keratin phosphorylation on Reg-II expression, given that keratin phosphorylation can play a significant role in keratin filament organization and dynamics. Reg-II expression levels are minimally altered in K18 S33A mice (Figure 5C), which exhibit a retraction of the keratin cytoplasmic filaments from the basal supranuclear compartment toward the apical domain (Ku et al., 2002). In contrast, blocking K18 S52 phosphorylation results in elevated Reg-II protein levels in two of four tested mice for each genotype (Figure 5D). Two independent transgenic mouse lines (S33A1 and S33A2; S52A1 and S52A2) were used for each keratin phosphomutant that had similar human K18 transgene protein levels (Figure 5, C and D). Therefore, Reg-II expression levels in the various transgenic lines can be summarized as follows: K8-null > K18-null ≈ K18 R89C ≈ K18 S52A > K18 S33A ≈ K19-null ≈WT, whereas Reg-I is much less affected.
Reg-II, but Not Reg-I, Is up-Regulated during Experimental Mouse Acute Pancreatitis
Because Reg family members are established stress-inducible proteins, we examined their expression during mouse acute pancreatitis. Reg-II protein levels increased dramatically in two different pancreatitis models (Figure 6), whereas Reg-I showed limited induction after CDD feeding (Figure 6A) or only moderate up-regulation after caerulein-induced pancreatitis (Figure 6B). Similar findings were noted after immune staining of pancreata before and after caerulein (Figure 4). The increased Reg-II protein is triggered by the pancreatic injury and remains sustained during the recovery phase (Figure 6). Comparison of Reg-II expression by ISH 2 d after caerulein-induced pancreatitis in K8-null and WT mouse pancreata still shows abundant Reg-II mRNA in the K8-null mice (Figure 7), which is consistent with the Reg-II protein levels. The distribution of Reg-II mRNA after injury is found primarily in acinar cells (Figure 7) as noted under basal conditions (Figure 2).
Figure 6.
Reg-II, but less so Reg-I, is up-regulated during acute pancreatitis. (A) BALB/c mice were fed CDD (an established mouse pancreatitis model) or normal chow (control [C] mice) for 3 d and then switched to normal diet to recover. Mice were euthanized at the indicated times after the beginning of CDD feeding, followed by analysis of total pancreas homogenates by immunoblotting using antibodies to the indicated proteins. Homogenates from two independent mice (1, 2) per time point were analyzed. Note that Reg-II, but not Reg-I, is markedly up-regulated and that the up-regulation begins during the injury and continues through the recovery period. (B) Caerulein was injected intraperitoneally to induce pancreatitis as described in Materials and Methods. The 1-h time point reflects analysis of mice 1 h after receiving a single caerulein injection. The remaining time points reflect mice that were injected hourly seven times then followed for the indicated times (12 h to 10 d from the time of the first injection). Control (C) mice were injected seven times with the carrier followed by isolation of the pancreata 6 h later (i.e., equivalent to the 12-h time point shown in lanes 5 and 6). Homogenates from two independent mice (1, 2) per time point were analyzed.
Figure 7.
Effect of caerulein-induced pancreatitis on Reg-II mRNA expression. In situ hybridization was carried out as described in Figure 2, by using pancreatic tissues isolated from K8-WT (A and B) and K8-null mice (C and D) 2 d after seven hourly caerulein injections. ISH was performed using Reg-II antisense (A and C) and sense probes (B and D). Note the more intense and broadly distributed Reg-II signal in the exocrine pancreas of K8-null compared with K8-WT pancreas. Bar (in B, applies to all panels), 50 μm. i, islets.
DISCUSSION
Overview of Findings
The major conclusions of our study are as follows: 1) K8-null pancreata show compensatory expression profile changes compared with K8-WT tissues, with Regs and particularly Reg-II being the most prominently overexpressed genes. 2) Induced Reg-II is primarily an acinar cell product, and its protein level is enhanced nearly threefold in K8-null pancreas. 3) Reg-II induction is consequent not only to K8 absence but also relates to absence (K18-null) or disruption (K18 R89C) of keratin cytoplasmic filaments and to site-specific K18 Ser52 but not S33 phosphorylation. 4) Reg-II is up-regulated in two acute pancreatitis mouse models. 5) Among the Reg family members, Reg-II inducibility is selective in that Reg-I levels are not as affected as Reg-II.
The Reg Family Members and Response to Injury
The Reg family comprises a heterogeneous group that is divided into four subfamilies (Reg-I–IV) based on structure homology (De Reggi and Gharib, 2001; Zhang et al., 2004). The composition of the Reg family varies among species (De Reggi and Gharib, 2001; Zhang et al., 2004). For example, mice express one type I and one type II members (termed Reg-I and Reg-II, respectively), whereas humans express two type I Regs (Reg-Iα and Reg-Iβ), but no type II (i.e., Reg-II) counterpart (De Reggi and Gharib, 2001; Zhang et al., 2004). The literature pertaining to the nomenclature of the Reg family members is not uniform, because some published work refers to mouse Reg-II when in fact the study pertains to Reg-IIIβ, which is also termed pancreatitis-associated protein I (Unno et al., 1993; Graf et al., 2006; Lieu et al., 2006). Reg-I and Reg-II are independent gene products but highly homologous proteins sharing 76% amino acid sequence identity, and they are found predominantly in the exocrine pancreas under basal conditions (Unno et al., 1993). Changes in Reg-II and its role in pancreatitis are not known, and its function remains elusive with few studies to date focused on Reg-II (Unno et al., 1993; Perfetti et al., 1996; Baeza et al., 1997). In contrast, there is significant evidence supporting an important role for Reg-I in the endocrine and exocrine pancreas. Reg-I is a luminally secreted acinar cell product that is found in pancreatic stones, and it is mitogenic to ductal and islet cells (Zenilman et al., 1996; Unno et al., 2002; Patard et al., 2003). It is also induced in regenerating islets (Terazono et al., 1988) (hence, the name Reg), and it can ameliorate surgical diabetes (Okamoto, 1999). Reg-I is up-regulated during rat pancreatitis, although the extent of induction varies among different studies (Zenilman et al., 2000; Graf et al., 2002). With regard to other Regs, members of the Reg-III subfamily are also up-regulated during mouse pancreatitis, and the use of antisense oligonucleotides directed toward Reg-III exacerbates the severity of rat pancreatitis (Zenilman et al., 2000; Graf et al., 2002; Zhang et al., 2004; Norkina et al., 2006).
As reported herein, Reg-II, but less so Reg-I, is markedly up-regulated in several keratin-deficient or keratin-mutant mouse lines under basal conditions, and in normal mice subjected to two models of experimental pancreatitis. Therefore, Reg-I represents more of a constitutive gene in the context of exocrine pancreatitis, whereas Reg-II is highly inducible. Differential expression of both genes was found in two other independent studies, which analyzed Reg levels during aging and in nonobese diabetic mice (Perfetti et al., 1996; Baeza et al., 1997), and it is supported by the lower homology in 5′-flanking sequence and no significant homology in intervening sequences of both genes (Unno et al., 1993). Furthermore, both Reg genes exhibit slightly different expression patterns. For example, Reg-I is expressed in the gallbladder, but Reg-II is not (Unno et al., 1993). Therefore, despite the striking amino acid homology among them, Reg-I and Reg-II posses several unique characteristics, and they may serve different roles.
Reg-II as a Potential Compensatory Pancreatic Stress Protein in Response to Keratin Absence or Mutation
Reg-II gene expression is markedly up-regulated in K8-null pancreata under basal conditions, and in normal nontransgenic mice in response to pancreatic injury. Reg-II is also up-regulated in K8-null mice after pancreatitis, but the level of induction is less than that noted in WT controls (data not shown). In mouse liver, mRNA for Reg-II was detected at low levels (Unno et al., 1993), but we do not detect Reg-II protein after immunoblotting of WT or K8-null mouse livers (data not shown). However, Reg-IIIβ is expressed in the liver and Reg-IIIβ-null mice have an increased susceptibility to Fas-mediated liver injury and impairment in liver regeneration after partial hepatectomy (Lieu et al., 2006). Together, with the already known effects of other Regs, our results suggest that Reg-II overexpression is likely to compensate for the lack of K8 and might account, at least in part, to the differences in susceptibility of keratin-deficient mice to liver and pancreatic injury. The mechanism for a potential Reg-II-mediated protective effect remains to be defined, because there is no clearly established Reg-II function. By analogy with other Regs, Reg-II may function at one or more levels by enhancing the regenerative capacity (Okamoto, 1999; Patard et al., 2003); or by imparting anti-inflammatory, antiapoptotic, and/or mitogenic effects (Malka et al., 2000; Vasseur et al., 2004; Gironella et al., 2005). It will be important to address whether the effect of Reg-II is paracrine, endocrine, or autocrine. Under normal conditions, Reg-II is secreted into the pancreatic ductal system; however, its fate during and after injury remain to be investigated.
In addition to Reg-II, other factors may also contribute to the dispensability of keratins in terms of cytoprotection in the pancreas. For example, actin is more prominently induced upon pancreatic injury in K8-null mice versus control mice, which raises the possibility that other cytoskeletal proteins may also substitute for the lack of keratins (Zhong and Omary, 2004). However, actin levels under basal conditions are similar in WT and K8-null pancreata (Zhong and Omary, 2004).
Potential Signaling Pathways Regulating Reg-II Expression in Keratin Mutant Animals and during Pancreatitis
One important finding herein is that not only K8 absence but also keratin cytoplasmic filament disruption (in mice that express K18 R89C), cytoplasmic keratin filament absence (in mice that lack K18, with retained apicolateral filaments), and site-specific phosphomutant K18 S52A but not S33A result in pancreas up-regulation of Reg-II under basal conditions. The K18 S52A mutation does not affect keratin filament organization in the pancreas (or the liver) under basal conditions, whereas the K18 S33A mutation maintains acinar cell cytoplasmic filaments, although they become retracted toward the apical pole (Ku et al., 2002). These findings suggest that a signaling event that involves K18 Ser52 is likely to be important in modulating downstream up-regulation of Reg-II expression. Because filament disruption due to K18 R89 mutation is associated with increased K18 S52 phosphorylation in the liver under basal conditions (Ku et al., 2003), the roles of K18 Ser52 phosphorylation and an intact cytoplasmic filament network are likely to be independent in modulating Reg-II expression. Based on available data, K18 S33 and S52 phosphorylation have different cell response behaviors and functions. S33 phosphorylation mediates K18 binding with 14-3-3 proteins, and S33 phosphorylation decreases during apoptosis (Ku et al., 2002, 2003). In contrast, K18 S52 phosphorylation increases during apoptosis (Ku et al., 2003), and its mutation predisposes mice to hepatotoxic injury but the role of these two phosphorylation sites in pancreatic injury has not been studied.
Several emerging lines of evidence support a role for keratins in cell signaling events (Ku and Omary 2006; Pallari and Eriksson 2006), which provide indirect support for the observed involvement of keratin filament organization and site-specific phosphorylation in the up-regulation of Reg-II. For example, K8/K18 bind to tumor necrosis factor (TNF) receptor 2 and Raf-1 kinase (Omary et al., 1992; Liao and Omary, 1996; Caulin et al., 2000; Ku et al., 2004a). In addition, keratin-deficient endodermal cells exhibit increased nuclear factor (NF)-κB and c-Jun NH2-terminal kinase activation in response to TNF-α (Caulin et al., 2000). TNF-α is induced in acute pancreatitis (Granger and Remick, 2005), and increased TNF signaling could be involved in Reg-II overexpression seen in both keratin-deficient animals and in the pancreatitis models, because it modulates the overexpression of other Regs, including Reg-I and Reg-III (Dusetti et al., 1996; Malka et al., 2000). NF-κB signaling may play a role because it occurs during pancreatitis and in keratin-deficient mouse models (Caulin et al., 2000; Algul et al., 2002; Zhong et al., 2004), and it associates with Reg-III (Graf et al., 2006) and keratin induction in response to injury (Ma et al., 1997; Komine et al., 2000; Zhong and Omary, 2004). Another potential candidate modulator is IL-6, given that the Reg promoter contains interleukin (IL)-6 consensus sequences (Unno et al., 1993) that can be activated by IL-6 stimulation (Dusetti et al., 1996; Okamoto, 1999), and the observation that IL-6 also induces K8 expression in intestinal epithelial cells (Wang et al., 2007).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful for the generous sharing of the K8-null (Robert Oshima, The Burnham Institute), K18-null (Thomas Magin, University of Bonn) and K19-null (Makoto Taketo, Kyoto University and Yoshitaka Tamai, Banyu Tsukuba Research Institute) mice. We also thank Evelyn Resurrection for assistance with immunostaining and Pauline Chu for assistance with histochemical staining. This work was supported by National Institutes of Health (NIH) grant DK-47918 and the Department of Veterans Affairs (to M.B.O.), NIH Center Grant DK-56339 to Stanford University, and a Guangdong province, People's Republic of China International Cooperation grant 2004B50301015 (to B.Z.).
Nonstandard abbreviations:
- Ab
antibody
- CDD
choline-deficient diet
- FDR
false discovery rate
- H&E
hematoxylin and eosin
- Hsp
heat-shock protein
- h
human
- IF
intermediate filament
- ISH
in situ hybridization
- K
keratin
- Reg
regenerating islet-derived
- SAM
significance analysis of microarray
- WT
wild type.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-02-0180) on September 26, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Algul H., Tando Y., Schneider G., Weidenbach H., Adler G., Schmid R. M. Acute experimental pancreatitis and NF-kappaB/Rel activation. Pancreatology. 2002;2:503–509. doi: 10.1159/000066090. [DOI] [PubMed] [Google Scholar]
- Baeza N., Sanchez D., Vialettes B., Figarella C. Specific reg II gene overexpression in the non-obese diabetic mouse pancreas during active diabetogenesis. FEBS Lett. 1997;416:364–368. doi: 10.1016/s0014-5793(97)01241-6. [DOI] [PubMed] [Google Scholar]
- Baribault H., Penner J., Iozzo R. V., Wilson-Heiner M. Colorectal hyperplasia and inflammation in keratin 8-deficient FVB/N mice. Genes Dev. 1994;8:2964–2973. doi: 10.1101/gad.8.24.2964. [DOI] [PubMed] [Google Scholar]
- Bershadsky A. D., Vasiliev J. M. New York: Plenum Press; 1988. Cytoskeleton; pp. 133–154. [Google Scholar]
- Bimmler D., Schiesser M., Perren A., Scheele G., Angst E., Meili S., Ammann R., Graf R. Coordinate regulation of PSP/reg and PAP isoforms as a family of secretory stress proteins in an animal model of chronic pancreatitis. J. Surg. Res. 2004;118:122–135. doi: 10.1016/S0022-4804(03)00342-1. [DOI] [PubMed] [Google Scholar]
- Caulin C., Ware C. F., Magin T. M., Oshima R. G. Keratin-dependent, epithelial resistance to tumor necrosis factor-induced apoptosis. J. Cell Biol. 2000;149:17–22. doi: 10.1083/jcb.149.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe P. A., Hutton M. E., Letai A., Hebert A., Paller A. S., Fuchs E. Point mutations in human keratin 14 genes of epidermolysis bullosa simplex patients: genetic and functional analyses. Cell. 1991;66:1301–1311. doi: 10.1016/0092-8674(91)90051-y. [DOI] [PubMed] [Google Scholar]
- Coulombe P. A., Omary M. B. ‘Hard’ and ‘soft’ principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 2002;14:110–122. doi: 10.1016/s0955-0674(01)00301-5. [DOI] [PubMed] [Google Scholar]
- Coulombe P. A., Wong P. Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat. Cell Biol. 2004;6:699–706. doi: 10.1038/ncb0804-699. [DOI] [PubMed] [Google Scholar]
- De Reggi M., Gharib B. Protein-X, pancreatic stone-, pancreatic thread-, Reg-protein, P19, lithostathine, and now what? Characterization, structural analysis and putative function(s) of the major non-enzymatic protein of pancreatic secretions. Curr. Protein Pept. Sci. 2001;2:19–42. doi: 10.2174/1389203013381233. [DOI] [PubMed] [Google Scholar]
- Dusetti N. J., Mallo G. V., Ortiz E. M., Keim V., Dagorn J. C., Iovanna J. L. Induction of lithostathine/reg mRNA expression by serum from rats with acute pancreatitis and cytokines in pancreatic acinar AR-42J cells. Arch. Biochem. Biophys. 1996;330:129–132. doi: 10.1006/abbi.1996.0234. [DOI] [PubMed] [Google Scholar]
- Eisen M. B., Brown P. O. DNA arrays for analysis of gene expression. Methods Enzymol. 1999;303:179–205. doi: 10.1016/s0076-6879(99)03014-1. [DOI] [PubMed] [Google Scholar]
- Fuchs E., Cleveland D. W. A structural scaffolding of intermediate filaments in health and disease. Science. 1998;279:514–519. doi: 10.1126/science.279.5350.514. [DOI] [PubMed] [Google Scholar]
- Gironella M., Iovanna J. L., Sans M., Gil F., Penalva M., Closa D., Miquel R., Pique J. M., Panes J. Anti-inflammatory effects of pancreatitis associated protein in inflammatory bowel disease. Gut. 2005;54:1244–1253. doi: 10.1136/gut.2004.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf R., Schiesser M., Lussi A., Went P., Scheele G. A., Bimmler D. Coordinate regulation of secretory stress proteins (PSP/reg, PAP I, PAP II, and PAP III) in the rat exocrine pancreas during experimental acute pancreatitis. J. Surg. Res. 2002;105:136–144. doi: 10.1006/jsre.2002.6387. [DOI] [PubMed] [Google Scholar]
- Graf R., Schiesser M., Reding T., Appenzeller P., Sun L. K., Fortunato F., Perren A., Bimmler D. Exocrine meets endocrine: pancreatic stone protein and regenerating protein—two sides of the same coin. J. Surg. Res. 2006;133:113–120. doi: 10.1016/j.jss.2005.09.030. [DOI] [PubMed] [Google Scholar]
- Granger J., Remick D. Acute pancreatitis: models, markers, and mediators. Shock. 2005;24(suppl 1):45–51. doi: 10.1097/01.shk.0000191413.94461.b0. [DOI] [PubMed] [Google Scholar]
- Herrmann H., Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular scaffolds. Annu. Rev. Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- Herrmann H., Hesse M., Reichenzeller M., Aebi U., Magin T. M. Functional complexity of intermediate filament cytoskeletons: from structure to assembly to gene ablation. Int. Rev. Cytol. 2003;223:83–175. doi: 10.1016/s0074-7696(05)23003-6. [DOI] [PubMed] [Google Scholar]
- Hesse M., Franz T., Tamai Y., Taketo M. M., Magin T. M. Targeted deletion of keratins 18 and 19 leads to trophoblast fragility and early embryonic lethality. EMBO J. 2000;19:5060–5070. doi: 10.1093/emboj/19.19.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacobuzio-Donahue C. A., Ryu B., Hruban R. H., Kern S. E. Exploring the host desmoplastic response to pancreatic carcinoma: gene expression of stromal and neoplastic cells at the site of primary invasion. Am. J. Pathol. 2002;160:91–99. doi: 10.1016/S0002-9440(10)64353-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. T., Lemp G. F., Gardner J. D. Interaction of cholecystokinin with specific membrane receptors on pancreatic acinar cells. Proc. Natl. Acad. Sci. USA. 1980;77:2079–2083. doi: 10.1073/pnas.77.4.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine M., Rao L. S., Kaneko T., Tomic-Canic M., Tamaki K., Freedberg I. M., Blumenberg M. Inflammatory versus proliferative processes in epidermis. Tumor necrosis factor alpha induces K6b keratin synthesis through a transcriptional complex containing NFkappa B and C/EBPbeta. J. Biol. Chem. 2000;275:32077–32088. doi: 10.1074/jbc.M001253200. [DOI] [PubMed] [Google Scholar]
- Ku N. O., Fu H., Omary M. B. Raf-1 activation disrupts its binding to keratins during cell stress. J. Cell Biol. 2004a;166:479–485. doi: 10.1083/jcb.200402051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N. O., Gish R., Wright T. L., Omary M. B. Keratin 8 mutations in patients with cryptogenic liver disease. N. Engl. J. Med. 2001;344:1580–1587. doi: 10.1056/NEJM200105243442103. [DOI] [PubMed] [Google Scholar]
- Ku N. O., Lim J. K., Krams S. M., Esquivel C. O., Keeffe E. B., Wright T. L., Parry D. A., Omary M. B. Keratins as susceptibility genes for end-stage liver disease. Gastroenterology. 2005;129:885–893. doi: 10.1053/j.gastro.2005.06.065. [DOI] [PubMed] [Google Scholar]
- Ku N. O., Michie S., Oshima R. G., Omary M. B. Chronic hepatitis, hepatocyte fragility, and increased soluble phosphoglycokeratins in transgenic mice expressing a keratin 18 conserved arginine mutant. J. Cell Biol. 1995;131:1303–1314. doi: 10.1083/jcb.131.5.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N. O., Michie S., Resurreccion E. Z., Broome R. L., Omary M. B. Keratin binding to 14-3-3 proteins modulates keratin filaments and hepatocyte mitotic progression. Proc. Natl. Acad. Sci. USA. 2002;99:4373–4378. doi: 10.1073/pnas.072624299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N. O., Michie S. A., Soetikno R. M., Resurreccion E. Z., Broome R. L., Omary M. B. Mutation of a major keratin phosphorylation site predisposes to hepatotoxic injury in transgenic mice. J. Cell Biol. 1998;143:2023–2032. doi: 10.1083/jcb.143.7.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N. O., Michie S. A., Soetikno R. M., Resurreccion E. Z., Broome R. L., Oshima R. G., Omary M. B. Susceptibility to hepatotoxicity in transgenic mice that express a dominant-negative human keratin 18 mutant. J. Clin. Invest. 1996;98:1034–1046. doi: 10.1172/JCI118864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N. O., Omary M. B. A disease- and phosphorylation-related nonmechanical function for keratin 8. J. Cell Biol. 2006;174:115–125. doi: 10.1083/jcb.200602146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku N. O., Soetikno R. M., Omary M. B. Keratin mutation in transgenic mice predisposes to Fas but not TNF-induced apoptosis and massive liver injury. Hepatology. 2003;37:1006–1014. doi: 10.1053/jhep.2003.50181. [DOI] [PubMed] [Google Scholar]
- Ku N. O., Toivola D. M., Zhou Q., Tao G. Z., Zhong B., Omary M. B. Studying simple epithelial keratins in cells and tissues. Methods Cell Biol. 2004;78:489–517. doi: 10.1016/s0091-679x(04)78017-6. [DOI] [PubMed] [Google Scholar]
- Ku N. O., Zhou X., Toivola D. M., Omary M. B. The cytoskeleton of digestive epithelia in health and disease. Am. J. Physiol. 1999;277:G1108–G1137. doi: 10.1152/ajpgi.1999.277.6.G1108. [DOI] [PubMed] [Google Scholar]
- Liao J., Lowthert L. A., Ku N. O., Fernandez R., Omary M. B. Dynamics of human keratin 18 phosphorylation: polarized distribution of phosphorylated keratins in simple epithelial tissues. J. Cell Biol. 1995;131:1291–1301. doi: 10.1083/jcb.131.5.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J., Omary M. B. 14–3-3 proteins associate with phosphorylated simple epithelial keratins during cell cycle progression and act as a solubility cofactor. J. Cell Biol. 1996;133:345–357. doi: 10.1083/jcb.133.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieu H. T., et al. Reg2 inactivation increases sensitivity to Fas hepatotoxicity and delays liver regeneration post-hepatectomy in mice. Hepatology. 2006;44:1452–1464. doi: 10.1002/hep.21434. [DOI] [PubMed] [Google Scholar]
- Lombardi B., Estes L. W., Longnecker D. S. Acute hemorrhagic pancreatitis (massive necrosis) with fat necrosis induced in mice by DL-ethionine fed with a choline-deficient diet. Am. J. Pathol. 1975;79:465–480. [PMC free article] [PubMed] [Google Scholar]
- Loranger A., Duclos S., Grenier A., Price J., Wilson-Heiner M., Baribault H., Marceau N. Simple epithelium keratins are required for maintenance of hepatocyte integrity. Am. J. Pathol. 1997;151:1673–1683. [PMC free article] [PubMed] [Google Scholar]
- Ma S., Rao L., Freedberg I. M., Blumenberg M. Transcriptional control of K5, K6, K14, and K17 keratin genes by AP-1 and NF-kappaB family members. Gene Expr. 1997;6:361–370. [PMC free article] [PubMed] [Google Scholar]
- Magin T. M., Schroder R., Leitgeb S., Wanninger F., Zatloukal K., Grund C., Melton D. W. Lessons from keratin 18 knockout mice: formation of novel keratin filaments, secondary loss of keratin 7 and accumulation of liver-specific keratin 8-positive aggregates. J. Cell Biol. 1998;140:1441–1451. doi: 10.1083/jcb.140.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malka D., Vasseur S., Bodeker H., Ortiz E. M., Dusetti N. J., Verrando P., Dagorn J. C., Iovanna J. L. Tumor necrosis factor alpha triggers antiapoptotic mechanisms in rat pancreatic cells through pancreatitis-associated protein I activation. Gastroenterology. 2000;119:816–828. doi: 10.1053/gast.2000.16491. [DOI] [PubMed] [Google Scholar]
- Moll R., Franke W. W., Schiller D. L., Geiger B., Krepler R. The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell. 1982;31:11–24. doi: 10.1016/0092-8674(82)90400-7. [DOI] [PubMed] [Google Scholar]
- Norkina O., Graf R., Appenzeller P., De Lisle R. C. Caerulein-induced acute pancreatitis in mice that constitutively overexpress Reg/PAP genes. BMC Gastroenterol. 2006;6:16. doi: 10.1186/1471-230X-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H. The Reg gene family and Reg proteins: with special attention to the regeneration of pancreatic beta-cells. J. Hepatobiliary Pancreat. Surg. 1999;6:254–262. doi: 10.1007/s005340050115. [DOI] [PubMed] [Google Scholar]
- Omary M. B., Baxter G. T., Chou C. F., Riopel C. L., Lin W. Y., Strulovici B. PKC epsilon-related kinase associates with and phosphorylates cytokeratin 8 and 18. J. Cell Biol. 1992;117:583–593. doi: 10.1083/jcb.117.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omary M. B., Coulombe P. A., McLean W. H. Intermediate filament proteins and their associated diseases. N. Engl. J. Med. 2004;351:2087–2100. doi: 10.1056/NEJMra040319. [DOI] [PubMed] [Google Scholar]
- Omary M. B., Ku N. O., Liao J., Price D. Keratin modifications and solubility properties in epithelial cells and in vitro. Subcell. Biochem. 1998;31:105–140. [PubMed] [Google Scholar]
- Omary M. B., Ku N. O., Tao G. Z., Toivola D. M., Liao J. “Heads and tails” of intermediate filament phosphorylation: multiple sites and functional insights. Trends Biochem. Sci. 2006;31:383–394. doi: 10.1016/j.tibs.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Pallari H. M., Eriksson J. E. Intermediate filaments as signaling platforms. Sci. STKE. 2006;2006:pe53. doi: 10.1126/stke.3662006pe53. [DOI] [PubMed] [Google Scholar]
- Patard L., Lallemand J. Y., Stoven V. An insight into the role of human pancreatic lithostathine. JOP. 2003;4:92–103. [PubMed] [Google Scholar]
- Perfetti R., Egan J. M., Zenilman M. E., Shuldiner A. R. Differential expression of reg-I and reg-II genes during aging in the normal mouse. J. Gerontol. A Biol. Sci. Med. Sci. 1996;51:B308–B315. doi: 10.1093/gerona/51a.5.b308. [DOI] [PubMed] [Google Scholar]
- Schweizer J., et al. New consensus nomenclature for mammalian keratins. J. Cell Biol. 2006;174:169–174. doi: 10.1083/jcb.200603161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croix St, et al. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- Strnad P., Lienau T. C., Tao G. Z., Lazzeroni L. C., Stickel F., Schuppan D., Omary M. B. Keratin variants associate with progression of fibrosis during chronic hepatitis C infection. Hepatology. 2006;43:1354–1363. doi: 10.1002/hep.21211. [DOI] [PubMed] [Google Scholar]
- Tamai Y., Ishikawa T., Bosl M. R., Mori M., Nozaki M., Baribault H., Oshima R. G., Taketo M. M. Cytokeratins 8 and 19 in the mouse placental development. J. Cell Biol. 2000;151:563–572. doi: 10.1083/jcb.151.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terazono K., Yamamoto H., Takasawa S., Shiga K., Yonemura Y., Tochino Y., Okamoto H. A novel gene activated in regenerating islets. J. Biol. Chem. 1988;263:2111–2114. [PubMed] [Google Scholar]
- Toivola D. M., Baribault H., Magin T., Michie S. A., Omary M. B. Simple epithelial keratins are dispensable for cytoprotection in two pancreatitis models. Am. J. Physiol. 2000a;279:G1343–G1354. doi: 10.1152/ajpgi.2000.279.6.G1343. [DOI] [PubMed] [Google Scholar]
- Toivola D. M., Ku N. O., Ghori N., Lowe A. W., Michie S. A., Omary M. B. Effects of keratin filament disruption on exocrine pancreas-stimulated secretion and susceptibility to injury. Exp. Cell Res. 2000b;255:156–170. doi: 10.1006/excr.1999.4787. [DOI] [PubMed] [Google Scholar]
- Toivola D. M., Ku N. O., Resurreccion E. Z., Nelson D. R., Wright T. L., Omary M. B. Keratin 8 and 18 hyperphosphorylation is a marker of progression of human liver disease. Hepatology. 2004;40:459–466. doi: 10.1002/hep.20277. [DOI] [PubMed] [Google Scholar]
- Toivola D. M., Omary M. B., Ku N. O., Peltola O., Baribault H., Eriksson J. E. Protein phosphatase inhibition in normal and keratin 8/18 assembly-incompetent mouse strains supports a functional role of keratin intermediate filaments in preserving hepatocyte integrity. Hepatology. 1998;28:116–128. doi: 10.1002/hep.510280117. [DOI] [PubMed] [Google Scholar]
- Unno M., Nata K., Noguchi N., Narushima Y., Akiyama T., Ikeda T., Nakagawa K., Takasawa S., Okamoto H. Production and characterization of Reg knockout mice: reduced proliferation of pancreatic beta-cells in Reg knockout mice. Diabetes. 2002;51(suppl 3):S478–S483. doi: 10.2337/diabetes.51.2007.s478. [DOI] [PubMed] [Google Scholar]
- Unno M., Yonekura H., Nakagawara K., Watanabe T., Miyashita H., Moriizumi S., Okamoto H., Itoh T., Teraoka H. Structure, chromosomal localization, and expression of mouse reg genes, reg I and reg II. A novel type of reg gene, reg II, exists in the mouse genome. J. Biol. Chem. 1993;268:15974–15982. [PubMed] [Google Scholar]
- Vassar R., Coulombe P. A., Degenstein L., Albers K., Fuchs E. Mutant keratin expression in transgenic mice causes marked abnormalities resembling a human genetic skin disease. Cell. 1991;64:365–380. doi: 10.1016/0092-8674(91)90645-f. [DOI] [PubMed] [Google Scholar]
- Vasseur S., Folch-Puy E., Hlouschek V., Garcia S., Fiedler F., Lerch M. M., Dagorn J. C., Closa D., Iovanna J. L. p8 improves pancreatic response to acute pancreatitis by enhancing the expression of the anti-inflammatory protein pancreatitis-associated protein I. J. Biol. Chem. 2004;279:7199–7207. doi: 10.1074/jbc.M309152200. [DOI] [PubMed] [Google Scholar]
- Wang L., Srinivasan S., Theiss A., Merlin D., Sitaraman S. V. IL-6 induces keratin expression in intestinal epithelial cells: potential role of keratin-8 in IL-6 induced barrier function alterations. J. Biol. Chem. 2007;282:8219–8227. doi: 10.1074/jbc.M604068200. [DOI] [PubMed] [Google Scholar]
- Zatloukal K., Stumptner C., Lehner M., Denk H., Baribault H., Eshkind L. G., Franke W. W. Cytokeratin 8 protects from hepatotoxicity, and its ratio to cytokeratin 18 determines the ability of hepatocytes to form Mallory bodies. Am. J. Pathol. 2000;156:1263–1274. doi: 10.1016/S0002-9440(10)64997-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenilman M. E., Magnuson T. H., Swinson K., Egan J., Perfetti R., Shuldiner A. R. Pancreatic thread protein is mitogenic to pancreatic-derived cells in culture. Gastroenterology. 1996;110:1208–1214. doi: 10.1053/gast.1996.v110.pm8613011. [DOI] [PubMed] [Google Scholar]
- Zenilman M. E., Tuchman D., Zheng Q., Levine J., Delany H. Comparison of reg I and reg III levels during acute pancreatitis in the rat. Ann. Surg. 2000;232:646–652. doi: 10.1097/00000658-200011000-00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Kandil E., Lin Y. Y., Levi G., Zenilman M. E. Targeted inhibition of gene expression of pancreatitis-associated proteins exacerbates the severity of acute pancreatitis in rats. Scand. J. Gastroenterol. 2004;39:870–881. doi: 10.1080/00365520410006477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B., Omary M. B. Actin overexpression parallels severity of pancreatic injury. Exp. Cell Res. 2004;299:404–414. doi: 10.1016/j.yexcr.2004.05.026. [DOI] [PubMed] [Google Scholar]
- Zhong B., Zhou Q., Toivola D. M., Tao G. Z., Resurreccion E. Z., Omary M. B. Organ-specific stress induces mouse pancreatic keratin overexpression in association with NF-kappaB activation. J. Cell Sci. 2004;117:1709–1719. doi: 10.1242/jcs.01016. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Ji X., Chen L., Greenberg H. B., Lu S. C., Omary M. B. Keratin mutation primes mouse liver to oxidative injury. Hepatology. 2005;41:517–525. doi: 10.1002/hep.20578. [DOI] [PubMed] [Google Scholar]
- Zhou Q., Toivola D. M., Feng N., Greenberg H. B., Franke W. W., Omary M. B. Keratin 20 helps maintain intermediate filament organization in intestinal epithelia. Mol. Biol. Cell. 2003;14:2959–2971. doi: 10.1091/mbc.E03-02-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.