Abstract
The chaperones of the ClpB/HSP100 family play a central role in thermotolerance in bacteria, plants, and fungi by ensuring solubilization of heat-induced protein aggregates. In addition in yeast, Hsp104 was found to be required for prion propagation. Herein, we analyze the role of Podospora anserina Hsp104 (PaHsp104) in the formation and propagation of the [Het-s] prion. We show that ΔPaHsp104 strains propagate [Het-s], making [Het-s] the first native fungal prion to be propagated in the absence of Hsp104. Nevertheless, we found that [Het-s]-propagon numbers, propagation rate, and spontaneous emergence are reduced in a ΔPaHsp104 background. In addition, inactivation of PaHsp104 leads to severe meiotic instability of [Het-s] and abolishes its meiotic drive activity. Finally, we show that ΔPaHSP104 strains are less susceptible than wild type to infection by exogenous recombinant HET-s(218–289) prion amyloids. Like [URE3] and [PIN+] in yeast but unlike [PSI+], [Het-s] is not cured by constitutive PaHsp104 overexpression. The observed effects of PaHsp104 inactivation are consistent with the described role of Hsp104 in prion aggregate shearing in yeast. However, Hsp104-dependency appears less stringent in P. anserina than in yeast; presumably because in Podospora prion propagation occurs in a syncitium.
INTRODUCTION
The prion concept originates from the study of mammalian neurodegenerative diseases (Prusiner, 1982). Prions are defined as proteinaceous infectious particles. In 1994, the boundaries of the prion concept were expanded when a non-Mendelian genetic element of yeast was shown to be a prion (Wickner, 1994). The [URE3], [PSI+], and [PIN+] yeast prions correspond to self-perpetuating aggregates of the Ure2p, Sup35, and Rnq1p proteins, respectively (Wickner et al., 2004). Currently the cycle of yeast prion propagation is best understood as an amyloid nucleation/polymerization process followed by a step of aggregate fragmentation that is conceptually analogous to the replication step in nucleic acid–based genetic elements (Tuite and Koloteva-Levin, 2004). Obviously, in order to be maintained in a dividing cell population, a prion aggregate must replicate at least as rapidly as the host cell.
Genetic studies on yeast prions have revealed the existence of cellular cofactors involved in prion maintenance. Among them, the Hsp104 chaperone has the most critical and universal role in yeast prion maintenance (Chernoff et al., 1995; True, 2006). Yeast Hsp104 is strictly required for maintenance of all known native yeast prions (Chernoff et al., 1995; Derkatch et al., 1997; Moriyama et al., 2000; Sondheimer and Lindquist, 2000). Intriguingly, not only lack of Hsp104 activity but also an excess of Hsp104 can lead to prion elimination. Overexpression of Hsp104 cures [PSI+] but not [URE3] and [PIN+] (Chernoff et al., 1995; Moriyama et al., 2000; Derkatch et al., 1997; Sondheimer et al., 2001).
Hsp104 belongs to the Hsp100/ClpB family of heat-inducible chaperones (Lee et al., 2004). Hsp100/ClpB family members are found in bacteria, plants, and fungi (but not in animals). These chaperones function to resolubilize protein aggregates formed after various stresses. Hsp100s are AAA+ ATPase with two nucleotide-binding domains per subunit and form an hexameric ring-shaped structure with a central pore (Lee et al., 2003a,b). ATPase activity and conformational changes are required for the protein-remodeling activity of Hsp104. It is believed that loops located in the central pore bind the substrate and then translocate the substrate through the central pore upon ATP hydrolysis by repeated rounds of pulling action (Horwich, 2004; Weibezahn et al., 2004; Bukau et al., 2006). The mechanism by which Hsp104 participates in prion replication has been intensively studied in yeast, in particular using the [PSI+] model (True, 2006). It has been initially proposed that Hsp104 might be required to allow Sup35 to reach a prion-competent stage (Chernoff et al., 1995). In support of this model is the fact that, in vitro, Hsp104 favors nucleation of Sup35 (and Ure2p) prion aggregates (Shorter and Lindquist, 2004, 2006). However, the observations that 1) Hsp104 is not required in vivo for newly synthesized Sup35 to enter existing aggregates (Ness et al., 2002), 2) [PSI+] aggregates can form de novo in the absence of Hsp104 (Zhou et al., 2001; Osherovich et al., 2004), and 3) formation of infectious Sup35, Ure2p, and Rnq1 amyloids occurs spontaneously in vitro (in the absence of Hsp104; Sparrer et al., 2000; Brachmann et al., 2005; Patel and Liebman, 2006), challenge this model. Ter-Avanesyan and coworkers proposed an alternative view by suggesting that Hsp104 might be required for fragmentation of prion aggregates (Paushkin et al., 1996). In this model, Hsp104 severs existing prion aggregates and increases the number of prion seeds, thus ensuring efficient prion propagation and mitotic stability of the prion template. This model has received ample experimental support. Chemical inactivation of Hsp104 (by low concentration of GuHCl) leads to prion loss in dividing cells by progressive dilution of the propagons, the genetic entity that allows [PSI+] maintenance (Eaglestone et al., 2000; Byrne et al., 2008). In addition, it appears that Sup35 aggregate size is increased in a Δhsp104 background (Kryndushkin et al., 2003). A recent study further supports and elaborates this model and indicates that inactivation of Hsp104 reduces mobility of Sup35-GFP aggregates, which impacts their mitotic segregation (Satpute-Krishnan et al., 2007). Then, importantly it was found that Hsp104 has the ability to sever Sup35 and Ure2p amyloid aggregates in vitro (Shorter and Lindquist, 2004, 2006). Hsp104 fragments Sup35 fibrils into noninfectious entities, whereas at fragmentation end points Hsp104-treated Ure2p fibrils are still infectious (Shorter and Lindquist, 2006), which readily explains why [PSI+] but not [URE3] is cured by Hsp104 overexpression. Considering the current threading mechanistic model of Hsp100 activity, this shearing action could result from extraction of protein monomers from the fibril, leading to the formation of gaps in the prion fibril.
Although the [URE3], [PSI+], and [PIN+] native yeast prions are strictly dependent on Hsp104 for propagation, it was reported that certain mutants of Sup35 and Ure2p can form Hsp104-independent prions in yeast (Liu et al., 2002; Crist et al., 2003; Ripaud et al., 2003).
A prion has also been identified in the filamentous fungus Podospora anserina (Coustou et al., 1997). Like Saccharomyces cerevisiae, P. anserina is also an ascomycete, but it differs markedly from yeast in terms of cellular organization. Although S. cerevisiae is essentially an unicellular organism, filamentous fungi grow as an interconnected network of filaments. There is a cytoplasmic continuity throughout most of the mycelium because cross-walls that compartmentalize the filaments into articles are incomplete. In addition, cell fusion events (anastomoses) spontaneously occur within and between colonies. Prion propagation in such a coenocytic structure could be inherently different from prion propagation in a population of dividing individualized cells.
[Het-s] controls a fungal somatic allorecognition process termed heterokaryon incompatibility (Rizet, 1952). Organisms that form somatic chimeras display genetic systems, allowing them to recognize conspecific self from nonself. It is believed that these genetic systems have been selected to prevent different forms of conspecific parasitism (Buss, 1982). Filamentous fungi, which spontaneously form somatic heterokaryons, possess a set of genes devoted to non-self-recognition (Glass and Dementhon, 2006). These genes are termed het genes (for heterokaryon incompatibility). The het-s locus is one member of this het-gene set and exists as two incompatible polymorphic variants termed het-s and het-S. Cell fusion between a het-s and a het-S strain leads to death of the mixed fusion cell. The HET-s protein is a prion, and the incompatibility reaction only occurs when the protein is in the prion state (Coustou et al., 1997). Strains that bear the HET-s protein in its nonprion state are designated [Het-s*] and are compatible with het-S strains. Strains that bear HET-s in its prion state are designated [Het-s] and are incompatible with het-S strains. [Het-s] is transmitted vertically between strains by the somatic cell fusions that spontaneously occur when mycelia are confronted. During the sexual cycle, [Het-s] is inherited as a non-Mendelian genetic element and shows strict maternal inheritance (Rizet, 1952; Beisson-Schecroun, 1962). During this sexual stage, the het-s/het-S antagonism is also expressed and leads to meiotic drive of the het-s allele (Bernet, 1965; Dalstra et al., 2003, 2005). Namely in a [Het-s] × het-S cross (when [Het-s] is the maternal parent), the het-S ascospores specifically abort, thus leading to production of two-spored asci. This spore-killing phenomenon leads to an excess het-s over het-S progeny, thus making the het-s allele/[Het-s] prion a meiotic drive element. Specific aspects of the P. anserina sexual cycle relevant for the [Het-s]-prion propagation process are depicted in Supplementary Figure 1. It should simply be noted here that during the sexual cycle, before meiosis, a transition from a coenocytic to a cellular structure takes place.
Previous studies have identified the C-terminal region of HET-s as the prion-forming domain (Balguerie et al., 2003). The HET-s(218–289) region was found to be both necessary and sufficient for [Het-s] prion propagation in P. anserina but also in an heterologous host such as S. cerevisiae (Taneja et al., 2007). Recombinant full-length and HET-s(218–289) amyloids display prion infectivity in protein transfection assays (Maddelein et al., 2002; Nazabal et al., 2005; Sabate et al., 2007). HET-s differs from yeast prions such as [PSI+] and [URE3] in terms of amino acid composition. Yeast PFDs are N/Q-rich but nearly devoid of charged residues. In contrast, the HET-s PFD is not N/Q-rich but is highly charged. A structure model of the infectious amyloid form of HET-s(218–289) has been proposed and corresponds to a β -roll structure composed of the stacking of two homologous strand-turn-strand motifs connected by an unstructured loop (Ritter et al., 2005; Sen et al., 2007). Current structural models for Sup35 NM and Ure2p propose a parallel in-register β-sheet organization, which is quite different from the organization proposed for HET-s (Kajava et al., 2004; Shewmaker et al., 2006). Thus [Het-s] and yeast prions might differ structurally but also by the cellular context in which they are propagated (syncitium vs. isolated cells). This prompted us to study the role of the P. anserina Hsp104 ortholog on [Het-s] propagation in P. anserina. Herein, we show that [Het-s] can be propagated in somatic cells in the absence of P. anserina Hsp104 (PaHsp104). But we also show that inactivation of PaHsp104 leads to an alteration of somatic [Het-s] propagation and a marked destabilization of the prion during the sexual cycle. Globally, our results are consistent with a described role of Hsp104 prion replication in yeast but indicate that the requirement for Hsp104 is less stringent for [Het-s] than for yeast prions.
MATERIALS AND METHODS
Cloning of PaHsp104
The P. anserina Hsp104 ortholog was isolated using an heterologous hybridization approach from a genomic cosmid library organized as 16 pools of 192 clones each as previously described (Saupe et al., 2000). Two oligonucleotides (NCHSP1045: 5′CAGCCATGGCCAGGGAAGGCAAGATTG3′ and NCHSP1043: 5′TGCGATCGTTGTCTTGGAGCCGCTTC3′) were designed to amplify a 1.8-kb fragment of the putative Hsp104 ortholog of Neurospora crassa corresponding to a highly conserved region of the gene encompassing the region coding for NBD1 and NBD2 (amino acid position 199–798). This fragment was then used as heterologous probe in a low stringency (1× SSC, 50°C) genomic Southern blot on genomic P. anserina DNA digested with a range of different enzymes. This allowed identification of a single 3.5-kb XhoI fragment that specifically hybridized to the probe. XhoI-digested DNA from each of the 16 library pools was then probed by Southern blot in the same condition. The diagnostic 3.5-kb XhoI fragment was identified in three different pools. Individual clones of one such cosmid pool (3C4C) were then probed; then a single cosmid (4CD5) hybridizing with the probe was isolated, and the 3.5-kb XhoI was subcloned and sequenced. As expected, the sequence corresponded to the putative P. anserina ortholog of Hsp104, but the XhoI fragment did not encompass the entire gene. Therefore a 9-kb BamHI fragment containing the entire PaHsp104 gene was isolated from cosmid 4CD5 and cloned in pCB1004 (Carrol et al., 1994). The corresponding plasmid was designated pCB-PaHsp104. The PaHsp104 sequence is deposited at GenBank (accession number bankit1012911 EU128751).
Inactivation and Overexpression of PaHsp104
For construction of the pGPD-PaHSP104 plasmid allowing constitutive overexpression of PaHsp104, the open reading frame (ORF) was amplified by PCR and cloned downstream of the pGPD promotor of Aspergillus nidulans inserted in the pCB1004 plasmid (Punt et al., 1988; Carrol et al., 1994). The corresponding pGPD-PaHsp104 plasmid was introduced into a wild-type het-s strain. DNA-mediated transformation of Podospora strains was performed as described (Bergès and Barreau, 1989).
The gene disruption of PaHsp104 was carried as previously described (Dementhon et al., 2004). In brief, the pCB-PaHsp104 plasmid was used as template in an inverse PCR using primer del-Hsp1045 (5′GCTTAATTAATGTGAATTAGACTTTTGAGACAGAGCG3′) and del-Hsp1043 (5′GCTTAATTAATAGAGGTGTCGTTGAGATGCCCAAGAAG3′) and the “Expand 20 kb plus” kit (Roche, Indianapolis, IN). The corresponding 12-kb PCR fragment was then restricted with PacI, circularized with T4 DNA ligase, and transformed into DH5α E. coli cells. The PacI fragment of the ura5 gene of P. anserina (Dementhon et al., 2004) was then inserted at the single PacI site. The resulting plasmid thus contains the ura5 marker flanked by 2.5 and 3.5 kb of genomic sequence of the PaHsp104 locus and also bears the hygR marker on the plasmid backbone. This plasmid was used to transform a ura5 P. anserina strain, and prototrophic hygS transformants were selected. In 12 of 20 transformants screened by PCR, the ura5 marker replaced the PaHsp104 ORF. Gene disruption was also confirmed by Southern blot.
Prion Propagation, Incompatibility, and Spore-killing Assays
Incompatibility phenotypes were determined by performing barrage tests on corn meal agar medium as previously described (Benkemoun et al., 2006).
[Het-s] prion propagation was assayed as the ability to transmit the [Het-s] prion from a [Het-s]-donor strain to a [Het-s*] prion-free tester strain after confrontation on solid medium. After contact, the tester strains were assayed for their [Het-s] status in barrage tests as described here above.
Spore-killing was studied as described previously (Bernet, 1965; Dalstra et al., 2003, 2005).
To determine the spontaneous emergence rate of [Het-s], four [Het-s*] or [Het-s*] ΔPaHSP104 implants and a central [Het-S] tester were placed on corn meal agar medium and grown for 5 d at 26° (Beisson-Schecroun, 1962). Spontaneous emergence of [Het-s] is detected by formation of a barrage with the central [Het-S] tester strain. A Chi square test was performed (one degree of freedom and a 5% risk) to verify that the observed difference in [Het-s] spontaneous emergence rate between wild-type and ΔPaHSP104 strains is significant.
Estimation of the Numbers of [Het-s]-propagons
To estimate the number of propagons per protoplasts, we then applied the method described by Belcour (1976). This method is based on the assumption that random sampling of the cytoplasm occurs during protoplast formation and that protoplasts containing one or more [Het-s]-propagons yield [Het-s] mycelia, whereas protoplast containing no [Het-s] propagons yield [Het-s*] mycelia. The mean number of propagons per protoplast is deduced by applying the Poisson law. The Poisson distribution is given by the formula:
where P(X) is the probability a having sampled X propagons per protoplast. The value for λ, here the mean number of propagons per protoplast, was deduced from the value of P(0), the probability of having no propagons, which corresponds to frequency of prion loss events observed experimentally. From the value of λ, the probability distribution can be inferred for all values of X.
In practice, protoplasts of a [Het-s], [Het-s] ΔPaHSP104, and [Het-s]::pGPD-Pahsp104 strains were prepared as previously described (Bergès and Barreau, 1989). Protoplasts generated by this procedure have an average volume of ∼35 μm3 and are generally multinucleated. Protoplasts were serially diluted to a final concentration of ∼20 protoplasts per μl. One hundred microliters of protoplasts was mixed with 50 ml preheated synthetic medium and 15 ml of 0.8 M sorbitol was used as an overlay for 10 synthetic-medium Petri dishes. Regenerated mycelia were confronted with a het-S tester strain to assay for the presence of the [Het-s] prion.
Microscopy
For fluorescence microscopy, synthetic medium containing 2% (wt/vol) agarose was poured as two 10-ml layers of medium. P. anserina hyphae were inoculated on this medium and cultivated for 16–24 h at 26°C. The top layer of the medium was then cut out, and the mycelium was examined with a Leica DMRXA microscope equipped with a Micromax CCD (Princeton Scientific Instruments, Monmouth Junction, NJ) controlled by the Metamorph 5.06 software (Roper Scientific, Tucson, AZ). The microscope was fitted with a standard fluorescein isothiocyanate filter set (Leica L4, Deerfield, IL) and a Leica PL APO 100× immersion lens.
Thermotolerance Assay
Mycelia were grown on synthetic medium for 2 d at 26°C before be tested for thermotolerance. Fragments of mycelium were sampled with a Pasteur pipette tip to calibrate their size. Implants were placed on solid corn meal agar medium at 48°C for 2 h and incubate at 26°C in the dark for 24 h. The experiment was repeated three times, and strains were observed after 72 h.
Protein Transfection Assays
Protein transfection experiments were performed using a cell disruptor (Fast-prep FP120, BIO101, Qbiogen, Carlsbad, CA). For each test, ∼0.5 cm3 of [Het-s*] mycelium grown on solid medium was sheared (run time 30 s, speed 6) in 500 μl of STC50 buffer (0.8 M sorbitol, 50 mM CaCl2, 100 mM Tris HCl, pH 7.5), and the sonicated HET-s(218–289) amyloids were assembled at pH 7 (20 μl at a variable concentration) in a 2-ml screw-cap tube. The sheared mycelium was then diluted with 600 μl of STC50 buffer, then plated as serial dilutions onto DO-0.8M sorbitol medium, and incubated at 26°C until being confluent (7–8 d). Then several implants (at least two per mycelium) were checked for the [Het-s] phenotype in barrage tests.
RESULTS
PaHsp104 Is Required for Thermotolerance in P. anserina
The P. anserina Hsp104 ortholog (PaHsp104) was isolated from a genomic cosmid library using the sequence of the putative Hsp104 ortholog of N. crassa as the probe. The PaHsp104 gene encodes a 926-amino acid long protein and is devoid of introns. The PaHsp104 protein displays 50% identity with S. cerevisiae Hsp104 (Supplementary Figure 2). PaHsp104 is a single copy gene and does not display close paralogues in the P. anserina genome. Using antibodies directed to the walker A motif of the NBD1 domain of S. cerevisiae Hsp104 (Parsell et al., 1991), we detected PaHsp104 in crude protein extracts after a 15-min heat shock at 48°C but not in strains grown at 26°C (not shown).
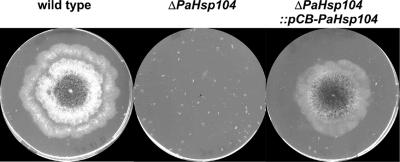
ΔPaHSP104 strains in which the PaHsp104 gene is inactivated were generated. Viability of ΔPaHsp104 strains is greatly affected by a heat shock at 48°C (Figure 1). Ectopic insertion of a pCB-PaHsp104 plasmid bearing a genomic insert encompassing the PaHsp104 gene complements this thermotolerance defect (Figure 1). ΔPaHsp104 strains also displayed a growth retardation upon exit of stationary phase. This growth retardation could either represent a defect in stationary phase exit per se or a reduction of viability in quiescence. In yeast, Hsp104 is strongly induced in stationary phase cells and plays a role in survival of quiescent cells (Sanchez et al., 1992).
Figure 1.
Inactivation of PaHsp104 affects thermotolerance. A wild-type, a ΔPaHsp104, and a ΔPaHsp104 :: pCB-PaHsp104 strain (bearing an ectopic copy of PaHsp104) were inoculated on solid medium, heat-shocked at 48°C for 2 h, and incubated for 24 h at 26°C. The procedure was repeated two more times before imaging the Petri plates.
Constitutive overexpression of PaHsp104 was achieved by driving its expression from the strong constitutive GPD promoter of A. nidulans (Punt et al., 1988). In Western blots, the amount of PaHsp104 protein in transformants expressing this construct was higher than in extract from wild-type strains that had been heat-shocked (not shown). One such transformant was selected and designated het-s::pGPD-PaHsp104.
[Het-s] Somatic Maintenance and Propagation in the Absence of Hsp104
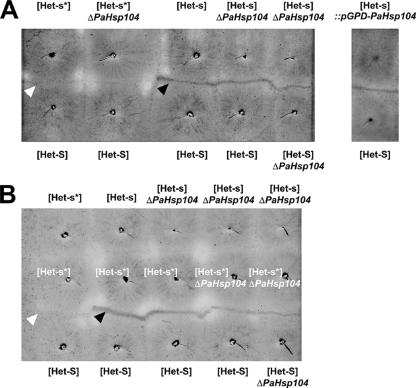
[Het-s*] ΔPaHsp104 strains could readily acquire [Het-s]. After cytoplasmic contact with a wild-type [Het-s] strain, ΔPaHsp104 het-s strains produced a barrage reaction when confronted with het-S or ΔPaHsp104 het-S strains (Figure 2A). ΔPaHsp104 [Het-s] strains were able to infect both wild-type [Het-s*] and ΔPaHsp104 [Het-s*] strains (Figure 2B). As observed for wild type, [Het-s] was never lost in somatic subcultures of ΔPaHsp104 strains. We conclude that [Het-s] can be propagated in a ΔPaHsp104 strain.
Figure 2.
[Het-s] somatic maintenance and propagation in the absence of PaHsp104 and in strains overexpressing PaHsp104. [Het-s] maintenance was assayed by performing barrage tests with wild-type het-s, het-s ΔPaHsp104, and het-s::pGPD-PaHsp104 strains. Presence of [Het-s] is detected by the formation of a dense abnormal contact line in confrontation with het-S tester strains (A). Note that neither inactivation nor overexpression of PaHsp104 abolishes barrage formation with a het-S (or het-S ΔPaHsp104) tester. [Het-s] transmission was assayed by confronting wild-type or ΔPaHsp104 [Het-s] strains with wild-type or ΔPaHsp104 [Het-s*] strains (B). Conversion of the [Het-s*] strains to the [Het-s] phenotype is then detected by formation of a barrage line in confrontation with het-S tester strains. Note that inactivation of PaHsp104 does not prevent conversion of a [Het-s*] strain to the [Het-s] phenotype. The white and black arrows mark, respectively, a normal confrontation and a barrage.
Strains constitutively overexpressing PaHsp104 were not affected in [Het-s] maintenance in the same assays.
Inactivation of PaHsp104 Leads to a Reduction of [Het-s]-Propagon Numbers
We next sought approaches to detect possible subtle alterations of [Het-s] somatic maintenance and propagation in the ΔPaHsp104 background. Léon Belcour developed a method to estimate the number of [Het-s] genetic units per cell (Belcour, 1976). Protoplasts are isolated from a [Het-s] mycelium and plated on solid medium. The average number of infectious [Het-s] units per protoplast is then deduced from the fraction of prion-free regenerating mycelia (Belcour, 1976). The genetic entities characterized by this method are conceptually related to the propagons defined in [PSI+] studies (Eaglestone et al., 2000). For simplicity, herein, [Het-s] units will be thus designated propagons, although we understand that [Het-s] units as defined by Belcour are not totally analogous to [PSI+] propagons. Although the number of propagons deduced by this method should only be considered a rough estimate, this method nonetheless allows a comparative analysis between wild-type and strains lacking and overexpressing PaHsp104.
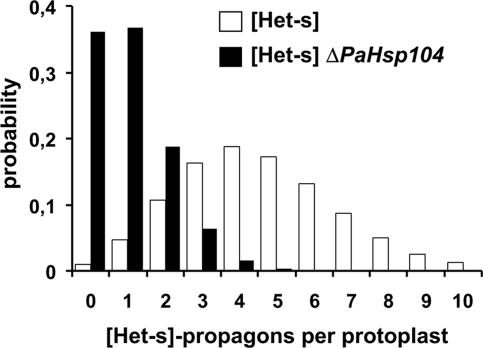
Table 1 gives the fraction of [Het-s*] mycelia obtained after regeneration of wild-type and ΔPaHsp104 protoplasts. The fraction of recovered [Het-s*] prion-free strains is increased from 1 ± 1% in wild-type [Het-s] to 36 ± 14% in a ΔPaHsp104 background. The deduced probability of propagon distribution in wild-type and ΔPaHsp104 protoplasts is given Figure 3. The estimated average number of propagons per protoplast decreases from ∼4.6 in wild type to 1.02 in ΔPaHsp104 (Table 1 and Figure 3). We conclude that inactivation of PaHsp104 strongly decreases the number of [Het-s] propagons per protoplast.
Table 1.
[Het-s]-prion loss after protoplast regeneration and estimated number of propagons in strains lacking or overexpressing PaHsp104
| Strain | [Het-s*] mycelia/total tested mycelia | Mean percentage of prion loss ± SD (%)a | Estimated no. of [Het-s]-propagons per protoplasts |
|---|---|---|---|
| [Het-s] | 4/361 | 1 ± 1 | 4.6 |
| [Het-s] ΔPaHsp104 | 87/241 | 36 ± 14 | 1.0 |
| [Het-s]::pGPD-PaHsp104 | 3/337 | 1 ± 1 | 4.6 |
a SD over three or four independent experiments.
Figure 3.
[Het-s]-propagon numbers per cell in wild-type and ΔPaHsp104. The histogram gives the probability of [Het-s]-propagon numbers per protoplast estimated by applying the Poisson distribution and based on the rate of [Het-s] loss after protoplast regeneration reported in Table 1 (see text for details); □: [Het-s], ■: [Het-s] ΔPaHsp104.
Constitutive overexpression of PaHsp104 did not lead to a significant modification of the fraction of [Het-s*] prion-free mycelia in this assay (Table 1).
Inactivation of PaHsp104 Lowers the Rate of [Het-s] Spreading through the Mycelium
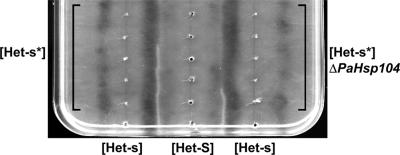
Janine Beisson developed a simple method to follow the spreading of the prion conversion process in a [Het-s*] mycelium (Figure 4) (Beisson-Schecroun, 1962). In this assay, a row of [Het-s*] subcultures is inoculated on solid medium, and the bottom subculture is infected with [Het-s]. This will lead in a domino effect to successive infection and conversion of the entire row. After the initial infection event, one monitors the progression of the prion-conversion process from the bottom to the top of the row. This can conveniently be achieved by confronting the het-s strains with a parallel row het-S tester. A barrage line will form only if prion conversion has occurred before anastomosis of the het-s and het-S strains. The barrage line marks the progression of the prion-conversion process at the time of contact between the het-s and het-S testers. When ΔPaHsp104 strains were compared with wild type in such assays, we found that the barrage lines were systematically shorter for ΔPaHsp104 (Figure 4). We conclude that the rate of spreading of the [Het-s]-infection in a [Het-s*] mycelium is reduced in the absence of PaHsp104.
Figure 4.
Inactivation of PaHsp104 reduces the rate of spreading of [Het-s]-infection in somatic mycelium. A series of five [Het-s*] strains (left vertical row) and a series of five [Het-s*] ΔPaHsp104 strains (right vertical row) were inoculated on solid medium together with a central row of [Het-S] testers. At the bottom of the plate prion-infected [Het-s] was used to infect the [Het-s*] strains. In a barrage test, the abnormal contact line only forms if the het-s is prion-infected at the time of initial contact with the [Het-S] tester. In the example depicted here, a barrage reaction is detected for the three lower wild-type het-s mycelia, indicating that the prion infection has invaded the three lower [Het-s*] strains at the time of contact with the [Het-S] tester. In ΔPaHsp104, only the first mycelium produces a barrage reaction, indicating that only the first [Het-s*] ΔPaHsp104 mycelium has been converted to the [Het-s] phenotype at the time of contact with the [Het-S] tester.
Inactivation of Hsp104 Reduces the Frequency of Spontaneous [Het-s] Emergence
Spontaneous emergence rate of [Het-s] was measured as previously described (Beisson-Schecroun, 1962). In wild type, we counted seven events of spontaneous emergence of [Het-s] in a total of 600 cultures grown for 5 d. In pGPD-PaHsp104, 5 spontaneous emergence events occurred in 400 analyzed cultures. In contrast, no spontaneous emergence events were detected in 1200 tested ΔPaHsp104 cultures (Table 2). Statistical analysis indicates that based on these results, it can be stated that the rate of spontaneous [Het-s] emergence is decreased in a ΔPaHsp104 background. Practically speaking, during the course of this study we have never detected spontaneous [Het-s] emergence events in the ΔPaHsp104 het-s background. This could indicate that spontaneous [Het-s] emergence cannot be achieved in a ΔPaHsp104 background. We have therefore analyzed a situation where spontaneous emergence of [Het-s] is favored, namely in transformants overexpressing HET-s-GFP (Coustou et al., 1997; Coustou-Linares et al., 2001). In this situation, [Het-s] emergence readily occurred in a ΔPaHsp104 background, and the rate was not significantly different from that in a wild-type control (not shown), indicating that there is not an absolute requirement for PaHsp104 for spontaneous [Het-s] emergence, at least when HET-s is overexpressed.
Table 2.
Spontaneous emergence of [Het-s] in strains lacking or overexpressing PaHsp104
| Strain | Total no. of tested mycelia | Events of spontaneous [Het-s] emergence |
|---|---|---|
| [Het-s*] | 600 | 7 |
| [Het-s*] ΔPaHsp104 | 1200 | 0a |
| [Het-s*]::pGPD-PaHsp104 | 400 | 5 |
a Based on a Chi square test, frequency of spontaneous emergence of [Het-s] is significantly lower in ΔPaHsp104 than in wild-type [Het-s*] or [Het-s*]::pGPD-PaHsp104.
Inactivation of Hsp104 Modifies HET-s-GFP Aggregate Morphology
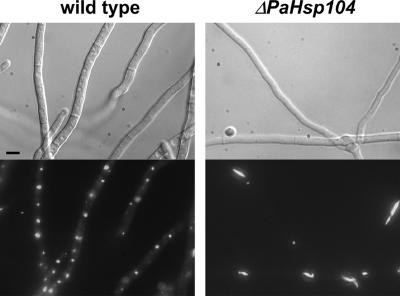
It was previously shown that in a strain expressing a HET-s-GFP fusion the transition to the prion state leads to formation of dot-like fluorescent foci (Coustou-Linares et al., 2001). We analyzed HET-s-GFP aggregate formation and morphology in a ΔPaHsp104 background. On transition to the [Het-s] prion state, we observed the formation of fluorescent foci in ΔPaHsp104 strains. The morphology of these foci was however slightly different from wild-type. Fluorescent foci were often elongated, whereas they are invariably dot-like in wild type (Figure 5).
Figure 5.
Inactivation of PaHsp104 slightly alters morphology of HET-s-GFP aggregates. A wild-type [Het-s] (left panel) and a ΔPaHsp104 [Het-s] strain (right panel) expressing a HET-s-GFP fusion protein were imaged by fluorescence microscopy. Scale bar, 4 μm.
Inactivation of Hsp104 Leads to Meiotic [Het-s] Instability and Abolishes [Het-s]-induced Meiotic Drive
Because [Het-s] is maintained at the vegetative stage, we had the opportunity to study meiotic prion inheritance in the absence of PaHsp104. Although meiotic loss of [Het-s] is never observed in wild type, only a minute fraction (∼3%) of the progeny in an homozygous ΔPaHsp104 cross retained [Het-s] (Table 3). No increase in meiotic loss was detected in crosses homozygous for the PaHsp104 overexpression (Table 3). In crosses heterozygous for ΔPaHsp104 meiotic stability of [Het-s] was partially restored (Table 3).
Table 3.
Effect of PaHsp104 overexpression and inactivation on meiotic stability of [Het-s]
| Crossa | [Het-s] spores/total | Percentage (%) |
|---|---|---|
| [Het-s] × [Het-s] | 192/192 | 100 |
| [Het-s] ΔPaHsp104 × [Het-s] ΔPaHsp104 | 8/242 | 3 |
| [Het-s] ΔPaHsp104 × [Het-s] | 94/99 | 95 |
| [Het-s] × [Het-s] ΔPaHsp104 | 41/47 | 87 |
| [Het-s]::pGPD-PaHsp104 × [Het-s] ::pGPD-PaHsp104 | 48/48 | 100 |
a In each cross the maternal parent is given first.
Next, we analyzed the spore-killing activity of [Het-s] in a ΔPaHsp104 background. (Table 4). In the wild-type [Het-s] female × [Het-S] male control cross, in agreement with previous reports (Bernet, 1965; Dalstra et al., 2003, 2005), we counted 22% of two-spored asci. In contrast, in crosses homozygous for ΔPaHsp104, the percentage of two-spored asci was very low and was at the level of a control het-S× het-S cross showing no killing. We conclude that PaHsp104 is required for the spore-killing activity of [Het-s].
Table 4.
[Het-s]-spore–killing activity in crosses performed at 18°DC with strains lacking PaHsp104
| Crossa | Two-spored asci/total | Percentage of two-spored asci |
|---|---|---|
| [Het-S] × [Het-S] | 3/313 | 1 |
| [Het-s] × [Het-S] | 375/528 | 22 |
| [Het-s] ΔPaHSP104 ×[Het-S] ΔPaHSP104 | 3/334 | 1 |
| [Het-s] ΔPaHSP104 × [Het-S] | 64/658 | 14 |
| [Het-s] × [Het-S] ΔPaHSP104 | 19/142 | 13 |
a In each cross the maternal parent is given first.
ΔPaHsp104 Strains Are Partially Resistant to Infection by Exogenous Recombinant HET-s(218–289) Amyloids
Because [Het-s] can be propagated both in strains lacking or overexpressing PaHsp104, it is possible to determine if PaHsp104 is capable of modulating infectivity of exogenous HET-s(218–289) amyloids. We have performed protein transfection experiments with HET-s(218–289) amyloids assembled in vitro using either wild-type, ΔPaHsp104, or pGPD-PaHsp104 [Het-s*] strains as recipients. Infectivity assays were carried out as described previously either using a fixed amount of protein and by plating serial dilutions of the reaction mixture (Table 5) or by using a range of protein concentrations (Table 6; Sabate et al., 2007). As previously reported, HET-s(218–289) amyloids assembled in vitro at pH 7 show high [Het-s]-infectivity levels when transfected into wild-type [Het-s*] recipients (Tables 5 and 6). In sharp contrast, a ΔPaHsp104 strain is nearly refractory to infection with recombinant HET-s(218–289) amyloids. Strains overexpressing PaHsp104 were readily infected by HET-s(218–289). However, the fraction of strains converted to the [Het-s] phenotype was decreased compared with the control, (Table 6). In particular, when low amount of HET-s(218–289) protein were used, infectivity dropped off sharply when a recipient overexpressing PaHsp104 was used.
Table 5.
Effect of PaHsp104 deletion and overexpression on infectivity of HET-s(218–289) amyloid fibers
| Amount of fast-prep solution |
||||
|---|---|---|---|---|
| 200 μl | 100 μ | 10 μl | 1 μl | |
| Recipient straina | ||||
| [Het-s*] | 58/75 | 27/42 | 17/42 | 6/45 |
| [Het-s*] ΔPaHSP104 | 3/27 | 0/9 | 0/9 | 0/9 |
| [Het-s*]::pGPD-PaHsp104 | 30/36 | 17/21 | 11/21 | 0/21 |
a Infected strains over total tested strains.
Table 6.
Effect of PaHsp104 deletion and over-expression on infectivity of HET-s(218–289) amyloid fibers
| Amount of HET-s(218–289) amyloid fibers |
No protein | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 2 nmol | 1 nmol | 0.5 nmol | 0.25 nmol | 120 pmol | 60 pmol | 30 pmol | 15 pmol | ||
| Recipient straina | |||||||||
| Experiment 1 | |||||||||
| wt | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 3/3 | 1/3 | 0/6 |
| ΔPaHSP104 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/6 |
| pGPD-PaHsp104 | 1/3 | 1/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/3 | 0/6 |
| Experiment 2 | |||||||||
| wt | ND | 3/3 | 3/3 | ND | 3/3 | ND | ND | 0/3 | 0/6 |
| ΔPaHSP104 | ND | 0/6 | 0/6 | ND | 0/6 | ND | ND | 0/6 | 0/6 |
| pGPD-PaHsp104 | ND | 6/6 | 3/6 | ND | 1/6 | ND | ND | 0/6 | 0/6 |
| Experiment 3 | |||||||||
| wt | ND | 6/6 | 6/6 | ND | 6/6 | ND | ND | 6/6 | 0/6 |
| ΔPaHSP104 | ND | 0/12 | 0/12 | ND | 0/12 | ND | ND | 0/12 | 0/6 |
| pGPD-PaHsp104 | ND | 8/12 | 7/12 | ND | 3/12 | ND | ND | 0/12 | 0/6 |
| Total | |||||||||
| wt | 3/3 | 12/12 | 12/12 | 3/3 | 12/12 | 3/3 | 3/3 | 7/12 | 0/18 |
| ΔPaHSP104 | 0/3 | 0/21 | 0/21 | 0/3 | 0/21 | 0/3 | 0/3 | 0/21 | 0/18 |
| pGPD-PaHsp104 | 1/3 | 15/21 | 10/21 | 0/3 | 4/21 | 0/3 | 0/3 | 0/21 | 0/18 |
a Infected strains over total tested strains.
We conclude that inactivation of PaHsp104 (but also to a lesser extent its overexpression) decreases infectivity of recombinant HET-s(218–289) amyloids assembled in vitro. These results suggest that HET-s(218–289) amyloids assembled in vitro are a substrate of the PaHsp104 activity.
DISCUSSION
Herein we have isolated and characterized PaHsp104, the P. anserina Hsp104 ortholog in order to analysis its role in [Het-s] prion propagation.
Somatic [Het-s] Propagation Can Occur in the Absence of PaHsp104
[Het-s] can be maintained in P. anserina in the absence of Hsp104 activity. Although certain mutants of Sup35 and a Ure2p-GFP fusion protein were shown to be able to propagate as prions in a ΔHsp104 background (Liu et al., 2002; Crist et al., 2003; Ripaud et al., 2003), [Het-s] appears to be the first example of a native (conformation-based) prion that can be maintained in the absence of this chaperone. Nevertheless, in line with the central role of Hsp104 in yeast prion propagation, our results indicate that PaHsp104 also has a role in [Het-s] propagation. Inactivation of PaHsp104 impacts [Het-s] propagon numbers, spontaneous emergence, [Het-s] spreading rate, HET-s-GFP aggregate morphology, meiotic stability, and [Het-s]-spore killing activity. Also, in the absence of PaHsp104, P. anserina becomes nearly immune to infection with recombinant HET-s amyloids. PaHsp104 is not strictly required for the [Het-s] prion replication cycle in vivo. Apparently, this chaperone simply makes the process more efficient.
Why Is the Requirement for Hsp104 Less Stringent for P. anserina [Het-s] than for Native Yeast Prions?
One can envision two hypotheses to explain why [Het-s] propagation can be PaHsp104-independent. First, HET-s prion aggregates might differ from yeast prion aggregates. HET-s and yeast prions markedly differ in amino acid composition and proposed structure. It might be that such structural features allow HET-s amyloids to be fragmented in vivo even in the absence of Hsp104. This first hypothesis is however contradicted by the finding that propagation of [Het-sy], the prion form of a HET-s(218–289)-GFP fusion protein, requires Hsp104 in yeast (Taneja et al., 2007). Thus a second, more likely hypothesis is that the relative Hsp104-independency of [Het-s] propagation in Podospora compared with yeast prions is due to the difference in cellular organization in these two fungi. Somatic tissue of P. anserina is a coenocytic structure, whereas yeast grows as isolated cells. In dividing cells, a prion can obviously only be maintained if the prion replication rate matches mitotic division rates and if prions are correctly segregated between mother and daughter cells. In Podospora, the number of [Het-s]-propagons was estimated to be ∼4–5 per protoplast in wild type. Based on the average cytoplasmic volume of protoplasts and fungal articles (∼35 and 280 μm3, respectively), this leads to an estimate of ∼30–40 propagons per fungal article. In the ΔPaHsp104 background, the number of propagons was estimated to be ∼1 per protoplast, which would still represent ∼10 propagons per article. The severe reduction in propagon number upon PaHsp104 deletion is still compatible with [Het-s]-maintenance in this syncitial cellular structure. Transposed to a yeast cell volume, this would however only represent ∼1–2 propagons per cell, which appears insufficient for stable prion maintenance in a population of dividing cells. It is also important to note that [Het-s] distribution in the somatic mycelium of a ΔPaHsp104 strain could even be discontinuous, meaning that a fraction of the fungal articles could lack [Het-s] propagons altogether. The corresponding mycelium would still be scored as [Het-s], because practically speaking subcultures are always made from mycelial plugs that contain a large number of cells. Finally, prion loss events in one article could be counteracted by reinfection by prion seeds from adjacent articles or brought in by cell fusion events occurring within the colony; again, a situation that cannot occur in yeast.
In summary, because propagation of [Het-sy] is Hsp104-dependent in yeast (Taneja et al., 2007), we think that the ability of [Het-s] to propagate somatically in the absence of PaHsp104 does not reflect a fundamental difference in the mode of prion replication in comparison with native yeast prions. Rather, we content that Hsp104-independancy could be explained by the coenocytic structure of the organism. The fact that [Het-s] is highly destabilized during the sexual cycle—when this coenocytic structure is lost—supports this hypothesis. If this hypothesis is valid, Hsp104-dependent yeast prions should be able to propagate (at least somatically) in a ΔPaHsp104 background in P. anserina.
PaHsp104 Requirement for [Het-s]-Inheritance during the Sexual Cycle
PaHsp104 is dispensable for horizontal transmission of [Het-s] but is required for its vertical transmission. After fertilization, male and female nuclei are entrapped in specialized hyphae termed ascogenous hyphae. It has been proposed that this compartmentation occurs to protect the germ line from invasion by conspecific parasitic nuclei (Buss, 1982). Ascogenous hyphae undergo an intricate developmental program characterized by a series of consecutive divisions, leading to formation of ∼20–50 ascus mother cells (Supplementary Figure 2). Genetically, it is possible to delimit to some extent the timing of sexual [Het-s] loss. Close to normal [Het-s] inheritance can be restored in heterozygous crosses with a wild-type male parent, indicating that ΔPaHsp104 ascogonia still contain [Het-s]. It can thus be stated that in homozygous ΔPaHsp104 crosses, [Het-s] is lost after fertilization and not at the stage of female organ differentiation. The spore-killing activity of [Het-s] is abolished in ΔPaHsp104 crosses. The simplest interpretation of this result is that the [Het-s] prion is lost in ΔPaHsp104 crosses before the developmental stage at which spore-killing occurs (namely after spore delimitation during the ascospore maturation process). On the basis of this genetic evidence, we propose that [Het-s] loss occurs after fertilization and before ascospore delimitation and thus apparently coincides with the series of consecutive cell divisions that leads to formation of the ascus mother cells. The proposed roles of Hsp104 in prion aggregate shearing and segregation are consistent with the observed meiotic loss of [Het-s] in ΔPaHsp104 crosses.
Meiotic stability of [Het-s] and spore-killing activity are not totally restored in crosses heterozygous for ΔPaHsp104. It is possible that the amount of PaHsp104 protein is insufficient for full [Het-s] stabilization when only one functional copy of the gene is present.
In the wild, the somatic mycelium of P. anserina is ephemeral and sexual spores are the sole resistance form. Therefore, the [Het-s] element can presumably only be maintained in wild-type populations if it is stable meiotically. Hence, from the biological point of view, it can be stated that in nature, [Het-s] is in fact PaHsp104 dependent.
Other [Het-s]-related Phenotypic Alterations Caused by Inactivation of PaHsp104
Inactivation of PaHsp104 led to a number of other [Het-s]-related phenotypic alterations that shall now be discussed.
Spreading of the [Het-s]-prion infection within a [Het-s*] mycelium is reduced in a ΔPaHsp104 background. Interestingly, Satpute-Krishnan et al. (2007) have reported that at the cellular level inactivation of Hsp104 in yeast reduces mobility of Sup35[PSI+] aggregates. The reduction in the rate of [Het-s] spreading might be explained both by the observed reduction in propagon numbers and also potentially by such a reduction in their cellular mobility. Experiments aimed at the detection of a potential reduction in HET-s-GFP aggregate mobility have not been performed here. We found that HET-s-GFP[Het-s] morphology was modified in the absence of PaHsp104: a fraction of the aggregates were slightly elongated instead of being invariably dot-like. It is difficult at this stage to propose a convincing interpretation of this observation. But this modification of HET-s-GFP aggregate morphology upon inactivation of PaHsp104 suggests that in wild type, the dot-like shape of the aggregates results from the aggregate remodeling activity of this chaperone. Of note here is the fact that in yeast HET-s(218–289)-GFP dot-like aggregates are formed only in wild type, whereas ring (elongated aggregates) can still form in a ΔHsp104 background (Taneja et al., 2007).
Spontaneous emergence of [Het-s] is decreased in the absence of PaHsp104. Our present results allow us to state that this reduction is statically significant but do not allow quantification of this reduction. Several studies in yeast have suggested that Hsp104 is not required for the initial aggregation of the prion proteins (Zhou et al., 2001; Osherovich et al., 2004), although in vitro Hsp104 appears to catalyze prion aggregate formation in certain conditions (Shorter and Lindquist, 2004, 2006). In yeast, Hsp104 was found to be dispensable for formation of [Het-sy] aggregates (Taneja et al., 2007). The observed effect of PaHsp104 inactivation on spontaneous prion formation in Podospora may therefore seem somewhat surprising. This result could represent an indication that PaHsp104 controls the initial event of [Het-s]-prion formation. However, alternatively, it might be that the measured reduction in spontaneous emergence is only apparent and results in fact from the [Het-s]-propagation defects observed in the ΔPaHsp104 background. It might be that in the ΔPaHsp104 background, a significant fraction of elementary [Het-s]-emergence events are abortive at the macroscopic level; for instance, because spreading of newly emerging [Het-s] is upset by filament compartmentation before the infection has efficiently invaded the whole mycelium. We are therefore reluctant at this stage to consider that our results necessarily reflect an actual reduction of spontaneous [Het-s] emergence. PaHsp104 is not strictly required for [Het-s] emergence since spontaneous [Het-s]-emergence readily occurs in a ΔPaHsp104 strain when HET-s is overexpressed.
PaHsp104 Is Required to Reveal Prion Infectivity of HET-s(218–289) Amyloids Assembled In Vitro
In vitro studies have established that Sup35 and Ure2p prion aggregates are direct substrates of Hsp104 in vitro (Shorter and Lindquist, 2004, 2006). Although similar evidence is yet lacking in the case of HET-s, our observation that inactivation of PaHsp104 dramatically reduces infectivity levels of recombinant HET-s(218–289) amyloids strongly suggests that PaHsp104 participates in the processing of such amyloids formed in vitro. This result also indicates that in our protein transfection experiments amyloid entry is not the sole condition for generation of infectivity. It appears that HET-s(218–289) amyloids are turned into efficient prions only after being processed in vivo by PaHsp104.
This experiment is the only one in which we could assign a [Het-s]-related phenotype to the strain overexpressing PaHsp104. Namely, we found that overexpression of PaHsp104 somewhat reduced infectivity of HET-s(218–289) amyloids. In this experiment (and this experiment only), the paradoxical effects of Hsp104 inactivation and overexpression on the [PSI+] prion originally described by Chernoff et al. (1995) are recapitulated in the [Het-s]-system.
In conclusion, our work is not a fundamental addition to our current understanding of the role of Hsp104 in prion propagation as established by work on yeast prions. The effects on PaHsp104 inactivation we describe here are fully consistent with reported roles and activities of Hsp104 on prions in yeast. This study together with the recent report by Taneja et al. (2007), showing that [Het-s] can be a prion in yeast suggests that in spite of marked differences in terms of primary sequence and probably overall structure, the mode of propagation of Podospora [Het-s] is likely to be essentially comparable to that of N/Q-rich yeast prions. Nevertheless, this work illustrates how the difference in cellular organization in these two fungi can impact prion propagation, in particular the dependency toward the Hsp104 chaperone
Supplementary Material
ACKNOWLEDGMENTS
Martine Sicault-Sabourin is warmly acknowledged for her ample technical assistance in the genetic analyses. We thank Susan Lindquist (Whitehead Institute, MIT) for the anti-Hsp104 antibody. Suzana Dos Reis, Laurent Malato and Laura Benkemoun were recipients of PhD fellowships from the Ministère de La Recherche et de L'Enseignement Supérieur. This work was funded by grants from the Ministèrede La Recherche et de L'Enseignement Supérieur (ACI jeune chercheur), a grant from the Groupement d'interet scientifique infections à Prion to S.J.S., and a grant from the Agence National de La Kecherche (PanPrionDrug).
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-07-0657) on September 19, 2007.
 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Balguerie A., et al. Domain organization and structure-function relationship of the HET-s prion protein of Podospora anserina. EMBO J. 2003;22:2071–2081. doi: 10.1093/emboj/cdg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson-Schecroun J. Incompatibilité cellulaire et interactions nucléocytoplamsiques dans les phénomènes de barrage chez le Podospora anserina. Ann. Genet. 1962;4:3–50. [PubMed] [Google Scholar]
- Belcour L. Loss of a cytoplasmic determinant through formation of protoplasts in Podospora. Neurospora. Newslett. 1976;23:26–27. [Google Scholar]
- Benkemoun L., Sabate R., Malato L., Dos Reis S., Dalstra H., Saupe S. J., Maddelein M. L. Methods for the in vivo and in vitro analysis of [Het-s] prion infectivity. Methods. 2006;39:61–67. doi: 10.1016/j.ymeth.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Bergès T., Barreau C. Heat-shock at elevated temperature improves transformation efficiency of protoplasts from Podospora anserina. J. Gen. Microbiol. 1989;135:601–604. doi: 10.1099/00221287-135-3-601. [DOI] [PubMed] [Google Scholar]
- Bernet J. Mode d'action des gènes de barrage et relation entre l'incompatibilité cellulaire et l'incompatibilité sexuelle chez le Podospora anserina. Ann. Sci. Natl. Bot. 1965;6:611–768. [Google Scholar]
- Brachmann A., Baxa U., Wickner R. B. Prion generation in vitro: amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukau B., Weissman J., Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- Buss L. W. Somatic cell parasitism and the evolution of somatic tissue compatibility. Proc. Natl. Acad. Sci. USA. 1982;79:5337–5341. doi: 10.1073/pnas.79.17.5337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne L. J., Cox B. S., Cole D. J., Ridout M. S., Morgan B. J., Tuite M. F. Cell division is essential for elimination of the yeast [PSI+] prion by guanidine hydrochloride. Proc. Natl. Acad. Sci. USA. 2007;104:11688–11693. doi: 10.1073/pnas.0701392104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrol A. M., Sweigard J. A., Valent-Central B. Improved vectors for selecting resistance to hygromycin. Fungal Genet. Newslett. 1994;41:22. [Google Scholar]
- Chernoff Y. O., Lindquist S. L., Ono B., Inge-Vechtomov S. G., Liebman S. W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- Coustou V., Deleu C., Saupe S., Begueret J. The protein product of the het-s heterokaryon incompatibility gene of the fungus Podospora anserina behaves as a prion analog. Proc. Natl. Acad. Sci. USA. 1997;94:9773–9778. doi: 10.1073/pnas.94.18.9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coustou-Linares V., Maddelein M. L., Begueret J., Saupe S. J. In vivo aggregation of the HET-s prion protein of the fungus Podospora anserina. Mol. Microbiol. 2001;42:1325–1335. doi: 10.1046/j.1365-2958.2001.02707.x. [DOI] [PubMed] [Google Scholar]
- Crist C. G., Nakayashiki T., Kurahashi H., Nakamura Y. [PHI+], a novel Sup35-prion variant propagated with non-Gln/Asn oligopeptide repeats in the absence of the chaperone protein Hsp104. Genes Cells. 2003;8:603–618. doi: 10.1046/j.1365-2443.2003.00661.x. [DOI] [PubMed] [Google Scholar]
- Dalstra H. J., Swart K., Debets A. J., Saupe S. J., Hoekstra R. F. Sexual transmission of the [Het-S] prion leads to meiotic drive in Podospora anserina. Proc. Natl. Acad. Sci. USA. 2003;100:6616–6621. doi: 10.1073/pnas.1030058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalstra H. J., van der Zee R., Swart K., Hoekstra R. F., Saupe S. J., Debets A. J. Non-mendelian inheritance of the HET-s prion or HET-s prion domains determines the het-S spore killing system in Podospora anserina. Fungal Genet. Biol. 2005;42:836–847. doi: 10.1016/j.fgb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Dementhon K., Saupe S. J., Clave C. Characterization of IDI-4, a bZIP transcription factor inducing autophagy and cell death in the fungus Podospora anserina. Mol. Microbiol. 2004;53:1625–1640. doi: 10.1111/j.1365-2958.2004.04235.x. [DOI] [PubMed] [Google Scholar]
- Derkatch I. L., Bradley M. E., Zhou P., Chernoff Y. O., Liebman S. W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaglestone S. S., Ruddock L. W., Cox B. S., Tuite M. F. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 2000;97:240–244. doi: 10.1073/pnas.97.1.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass N. L., Dementhon K. Non-self recognition and programmed cell death in filamentous fungi. Curr. Opin. Microbiol. 2006;9:553–558. doi: 10.1016/j.mib.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Horwich A. L. Chaperoned protein disaggregation—the ClpB ring uses its central channel. Cell. 2004;119:579–581. doi: 10.1016/j.cell.2004.11.018. [DOI] [PubMed] [Google Scholar]
- Kajava A. V., Baxa U., Wickner R. B., Steven A. C. A model for Ure2p prion filaments and other amyloids: the parallel superpleated beta-structure. Proc. Natl. Acad. Sci. USA. 2004;101:7885–7890. doi: 10.1073/pnas.0402427101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryndushkin D. S., Alexandrov I. M., Ter-Avanesyan M. D., Kushnirov V. V. Yeast [PSI+] prion aggregates are formed by small Sup35 polymers fragmented by Hsp104. J. Biol. Chem. 2003;278:49636–49643. doi: 10.1074/jbc.M307996200. [DOI] [PubMed] [Google Scholar]
- Lee S., Hisayoshi M., Yoshida M., Tsai F. T. Crystallization and preliminary X-ray crystallographic analysis of the Hsp100 chaperone ClpB from Thermus thermophilus. Acta. Crystallogr. D Biol. Crystallogr. 2003a;59:2334–2336. doi: 10.1107/s0907444903023266. [DOI] [PubMed] [Google Scholar]
- Lee S., Sowa M. E., Choi J. M., Tsai F. T. The ClpB/Hsp104 molecular chaperone-a protein disaggregating machine. J. Struct. Biol. 2004;146:99–105. doi: 10.1016/j.jsb.2003.11.016. [DOI] [PubMed] [Google Scholar]
- Lee S., Sowa M. E., Watanabe Y. H., Sigler P. B., Chiu W., Yoshida M., Tsai F. T. The structure of ClpB: a molecular chaperone that rescues proteins from an aggregated state. Cell. 2003b;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- Liu J. J., Sondheimer N., Lindquist S. L. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+] Proc. Natl. Acad. Sci. USA. 2002;99(Suppl 4):16446–16453. doi: 10.1073/pnas.252652099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddelein M. L., Dos Reis S., Duvezin-Caubet S., Coulary-Salin B., Saupe S. J. Amyloid aggregates of the HET-s prion protein are infectious. Proc. Natl. Acad. Sci. USA. 2002;99:7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama H., Edskes H. K., Wickner R. B. [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone hsp104 and curing by overexpressed chaperone ydj1p. Mol. Cell. Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazabal A., Maddelein M. L., Bonneu M., Saupe S. J., Schmitter J. M. Probing the structure of the infectious amyloid form of the prion-forming domain of HET-s using high resolution hydrogen/deuterium exchange monitored by mass spectrometry. J. Biol. Chem. 2005;280:13220–13228. doi: 10.1074/jbc.M413185200. [DOI] [PubMed] [Google Scholar]
- Ness F., Ferreira P., Cox B. S., Tuite M. F. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 2002;22:5593–5605. doi: 10.1128/MCB.22.15.5593-5605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osherovich L. Z., Cox B. S., Tuite M. F., Weissman J. S. Dissection and design of yeast prions. PLoS Biol. 2004;2:E86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell D. A., Sanchez Y., Stitzel J. D., Lindquist S. Hsp104 is a highly conserved protein with two essential nucleotide-binding sites. Nature. 1991;353:270–273. doi: 10.1038/353270a0. [DOI] [PubMed] [Google Scholar]
- Patel B. K., Liebman S. W. “Prion-proof” for [PIN(+)]: Infection with in vitro-made amyloid aggregates of Rnq1p-(132–405) induces [PIN(+)] J. Mol. Biol. 2006;365:773–782. doi: 10.1016/j.jmb.2006.10.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S. V., Kushnirov V. V., Smirnov V. N., Ter-Avanesyan M. D. Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. B. Novel proteinaceous infectious particles cause scrapie. Science. 1982;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- Punt P. J., Dingemanse M. A., Jacobs-Meijsing B. J., Pouwels P. H., van den Hondel C. A. Isolation and characterization of the glyceraldehyde-3-phosphate dehydrogenase gene of Aspergillus nidulans. Gene. 1988;69:49–57. doi: 10.1016/0378-1119(88)90377-0. [DOI] [PubMed] [Google Scholar]
- Ripaud L., Maillet L., Cullin C. The mechanisms of [URE3] prion elimination demonstrate that large aggregates of Ure2p are dead-end products. EMBO J. 2003;22:5251–5259. doi: 10.1093/emboj/cdg488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter C., Maddelein M. L., Siemer A. B., Luhrs T., Ernst M., Meier B. H., Saupe S. J., Riek R. Correlation of structural elements and infectivity of the HET-s prion. Nature. 2005;435:844–848. doi: 10.1038/nature03793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizet G. Les phénomènes de barrage chez Podospora anserina. I. Analyse de barrage entre les souches s et S. Rev. Cytol. Biol. Veg. 1952;13:51–92. [Google Scholar]
- Sabate R., et al. Prion and non-prion amyloids of the HET-s prion forming domain. J. Mol. Biol. 2007;370:768–783. doi: 10.1016/j.jmb.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Sanchez Y., Taulien J., Borkovich K. A., Lindquist S. Hsp104 is required for tolerance to many forms of stress. EMBO J. 1992;11:2357–2364. doi: 10.1002/j.1460-2075.1992.tb05295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpute-Krishnan P., Langseth S. X., Serio T. R. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 2007;5:e24. doi: 10.1371/journal.pbio.0050024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saupe S. J., Clave C., Sabourin M., Begueret J. Characterization of hch, the Podospora anserina homolog of the het-c heterokaryon incompatibility gene of Neurospora crassa. Curr. Genet. 2000;38:39–47. doi: 10.1007/s002940000130. [DOI] [PubMed] [Google Scholar]
- Sen A., Baxa U., Simon M. N., Wall J. S., Sabate R., Saupe S. J., Steven A. C. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked beta-solenoid model of HET-s prion fibrils. J. Biol. Chem. 2007;282:5545–5550. doi: 10.1074/jbc.M611464200. [DOI] [PubMed] [Google Scholar]
- Shewmaker F., Wickner R. B., Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc. Natl. Acad. Sci. USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shorter J., Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- Shorter J., Lindquist S. Destruction or potentiation of different prions catalyzed by similar Hsp104 remodeling activities. Mol. Cell. 2006;23:425–438. doi: 10.1016/j.molcel.2006.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondheimer N., Lindquist S. Rnq1, an epigenetic modifier of protein function in yeast. Mol. Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- Sondheimer N., Lopez N., Craig E. A., Lindquist S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001;20:2435–2442. doi: 10.1093/emboj/20.10.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparrer H. E., Santoso A., Szoka F. C., Jr, Weissman J. S. Evidence for the prion hypothesis: induction of the yeast [PSI+] factor by in vitro- converted Sup35 protein. Science. 2000;289:595–599. doi: 10.1126/science.289.5479.595. [DOI] [PubMed] [Google Scholar]
- Taneja V., Maddelein M. L., Talarek N., Saupe S. J., Liebman S. W. A non-Q/N-rich prion domain of a foreign prion, [Het-s], can propagate as a prion in yeast. Mol. Cell. 2007;27:67–77. doi: 10.1016/j.molcel.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True H. L. The battle of the fold: chaperones take on prions. Trends Genet. 2006;22:110–117. doi: 10.1016/j.tig.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Tuite M. F., Koloteva-Levin N. Propagating prions in fungi and mammals. Mol. Cell. 2004;14:541–552. doi: 10.1016/j.molcel.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Weibezahn J., et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Wickner R. B. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae [see comments] Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- Wickner R. B., Edskes H. K., Ross E. D., Pierce M. M., Baxa U., Brachmann A., Shewmaker F. Prion genetics: new rules for a new kind of gene. Annu. Rev. Genet. 2004;38:681–707. doi: 10.1146/annurev.genet.38.072902.092200. [DOI] [PubMed] [Google Scholar]
- Zhou P., Derkatch I. L., Liebman S. W. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI+] and [PIN+] Mol. Microbiol. 2001;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.