Abstract
The mitotic cyclins promote cell division by binding and activating cyclin-dependent kinases (CDKs). Each cyclin has a unique pattern of subcellular localization that plays a vital role in regulating cell division. During mitosis, cyclin B1 is known to localize to centrosomes, microtubules, and chromatin. To determine the mechanisms of cyclin B1 localization in M phase, we imaged full-length and mutant versions of human cyclin B1-enhanced green fluorescent protein in live cells by using spinning disk confocal microscopy. In addition to centrosome, microtubule, and chromatin localization, we found that cyclin B1 also localizes to unattached kinetochores after nuclear envelope breakdown. Kinetochore recruitment of cyclin B1 required the kinetochore proteins Hec1 and Mad2, and it was stimulated by microtubule destabilization. Mutagenesis studies revealed that cyclin B1 is recruited to kinetochores through both CDK1-dependent and -independent mechanisms. In contrast, localization of cyclin B1 to chromatin and centrosomes is independent of CDK1 binding. The N-terminal domain of cyclin B1 is necessary and sufficient for chromatin association, whereas centrosome recruitment relies on sequences within the cyclin box. Our data support a role for cyclin B1 function at unattached kinetochores, and they demonstrate that separable and distinct sequence elements target cyclin B1 to kinetochores, chromatin, and centrosomes during mitosis.
INTRODUCTION
Cyclin-dependent kinases (CDKs) regulate cell cycle transitions by phosphorylating specific substrates at defined phases of the cell cycle. Together with phosphorylation and CDK inhibitory proteins, regulated synthesis and destruction of cyclin subunits ensure that CDKs are active at the proper phase of the cell cycle (Fung and Poon, 2005). In human cells, at least four different CDKs along with four cyclin classes regulate cell division (for review, see Sanchez and Dynlacht, 2005). In late G1, D-type cyclins activate CDK4/6, promoting phosphorylation of Rb, activation of E2F-dependent transcription, and DNA synthesis. In S phase, A- and E-type cyclins bind and activate CDK2, which phosphorylates substrates that induce centrosome duplication and fire origins of replication (Sherr, 2000). In mitosis, A- and B-type cyclins bind and activate CDK1 to phosphorylate proteins that mediate the dramatic changes in nuclear and cytoskeletal architecture required for chromosome segregation (Nigg et al., 1996). Even though all CDKs phosphorylate a consensus motif consisting of S/TPxK/R (Nigg, 1991), different cyclin–CDK complexes phosphorylate distinct substrates, raising the question of how substrate selection is achieved by different cyclin–CDK complexes.
Cyclins play an important role in modulating the substrate selectivity of a particular cyclin–CDK pair. B-type cyclins bestow a high intrinsic activity to CDKs resulting in the phosphorylation of any substrate containing the canonical S/TPxK/R motif (Loog and Morgan, 2005). Alternatively, A- and E-type cyclins rely on a hydrophobic patch (HP) to recruit S/TPxK/R-containing substrates to its CDK partner for phosphorylation (Schulman et al., 1998; Brown et al., 1999). This HP interacts with substrates containing an RxL/Cy motif, which is often found in substrates whose phosphorylation occurs during S phase (Schulman et al., 1998; Cross and Jacobson, 2000). HP mutations in the budding yeast S phase cyclin Clb5, but not the M phase cyclin Clb2, abolish substrate binding and phosphorylation (Cross and Jacobson, 2000; Loog and Morgan, 2005). Recent studies of human cyclin B1 suggest that residues neighboring the HP are structurally distinct from those in A- and E-type cyclins and that they may actually inhibit interactions with RxL/Cy-containing substrates (Brown et al., 2007; Petri et al., 2007). Nevertheless, the HP is highly conserved among B-type cyclins, suggesting it plays some functional role. Consistent with this notion, HP mutations in the budding yeast mitotic cyclin Clb2 disrupt its localization to the bud neck (Bailly et al., 2003), suggesting that the HP may be conserved because it plays a critical role in mediating localization of B-type cyclins.
Additional evidence suggests that the capacity of cyclin B1 to promote CDK1-dependent events at mitotic entry may rely on its subcellular localization. At mitotic entry, cyclin B1-CDK1 promotes chromosome condensation, nuclear lamina solubilization, mitotic aster assembly, and Golgi breakdown, whereas cyclin B2-CDK1 can only induce Golgi disassembly (Draviam et al., 2001). These distinct functions of cyclin B1 and cyclin B2 to induce different events of mitosis correspond well with localization of the proteins. During interphase, cyclins B1 and B2 are both concentrated in the cytoplasm; cyclin B1 shuttles in between the nucleus and the cytoplasm, whereas cyclin B2 is mainly associated with the Golgi (Jackman et al., 1995). At prophase, cyclin B1 accumulates in the nucleus (Pines and Hunter, 1994; Hagting et al., 1998) and then to condensed chromatin, spindle microtubules, centrosomes, and chromatin during prometaphase (Pines and Hunter, 1991), corresponding to localization of known CDK1 substrates, including nuclear lamins (Enoch et al., 1991; Luscher et al., 1991), microtubule-associated proteins (Ookata et al., 1995; Charrasse et al., 2000) and the chromatin-associated protein RCC1 (Arnaoutov and Dasso, 2003). In contrast, cyclin B2 remains associated with the disassembled Golgi during mitosis. Domain mapping studies have suggested that the N-terminal domain is important in dictating the distinct functions of B-type cyclins. Replacement of the cyclin B1 N-terminal domain with that of cyclin B2 is sufficient to mislocalize the chimeric protein to the Golgi (Draviam et al., 2001). This chimeric protein retains the ability to induce Golgi disassembly, but it loses the ability to induce chromosome condensation, nuclear lamina solubilization, and mitotic aster assembly (Draviam et al., 2001), demonstrating that subcellular localization of different B-type cyclins is a critical determinant of their ability to properly promote the events of mitosis. Given the potential importance of localization in modulating the biological consequences of cyclin B1-CDK1 activity, we set out to identify the sequences in cyclin B1 that mediate its localization to specific structures during mitosis.
MATERIALS AND METHODS
Cell Culture
HeLa cells, BS-C-1 monkey kidney, and 293 human embryonic kidney cells (American Type Culture Collection, Manassas, VA) were cultured in a humidified incubator at 37°C in the presence of 5% CO2 with DMEM, 10% fetal bovine serum (FBS), penicillin, and streptomycin (Invitrogen, Carlsbad, CA).
Adenovirus Construction
All adenoviruses were constructed using the AdEasy system (He et al., 1998). Enhanced green fluorescent protein (EGFP) from pEGFP-N1 (Clontech, Palo Alto, CA) was subcloned into pShuttle-CMV to create pShuttle-CMV-EGFP. Full-length human cyclin B1 was amplified by polymerase chin reaction (PCR) with pfu polymerase (Stratagene, La Jolla, CA) and subcloned into pShuttle-CMV-EGFP to make the cyclin B1-EGFP fusion construct (FL1-433). To generate the FLΔDB adenovirus, DNA encoding residues 1-41 and 51-433 of cyclin B1 was PCR amplified separately and cloned sequentially, yielding a construct that lacked the nine-amino acid Destruction box (D-box). To construct NT1-107, NT1-166, NT1-210, NT1-311, CT108-433, CT166-433, and CT211-433, DNA encoding the appropriate residues was PCR amplified, subcloned into pEGFP-N1 in frame, and then subcloned into pShuttle-CMV. The FLY170A and CT108-433, Y170A viruses were generated using the GeneTailor mutagenesis kit and primers designed to mutate the tyrosine at position 170 to an alanine (Invitrogen). DNA sequencing of both strands confirmed constructs. Adenoviral constructs were generated by recombination in BJ5183-AD1 cells (Stratagene), and infective adenoviruses were produced in 293 cells as described previously (He et al., 1998).
Time-Lapse Imaging and Microscopy
BS-C-1 cells were seeded onto no. 1.5 25-mm poly-l-lysine–coated glass coverslips or into glass-bottomed microwell dishes (MatTek, Ashland, MA). Adenovirus was added to BS-C-1 cells at the time of plating. Cells were imaged 16-18 h after infection. Mounted coverslips were imaged in a heated stage chamber at 37°C (20/20 Technology, Wilmington, NC), and microwell dishes were imaged in an enclosed incubation chamber at 37°C (Solent Scientific, Portsmouth, United Kingdom). Cells were cultured in CO2-independent L15 medium for the 20/20 system or in CO2-dependent DMEM for the Solent system. Similar results were obtained on both systems. During the course of the experiments presented in this manuscript, at least 50 FL1-433–expressing cells were imaged. At least 30 cells were imaged expressing NT1-107, NT1-166, CT108-433, and FLY170A. Images were acquired using MetaMorph imaging software (Molecular Devices, Sunnyvale, CA) on an inverted TE2000 microscope (Nikon, Melville, NY) by using a 100× 1.4 numerical aperture objective equipped with Yokogawa spinning disk confocal (PerkinElmer Life and Analytical Sciences, Boston, MA) and Orca-ER digital camera (Hamamatsu, Bridgewater, NJ). For time-lapse experiments, images were collected every 15–45 s, by using exposure times between 800 and 2000 ms, with illumination light shuttered between acquisitions. A 2 × 2 binning was used for all live-cell imaging. Figures were produced using Adobe PhotoShop 7.0 (Adobe Microsystems, Mountain View, CA).
Antibodies and Chemicals
Two of five antibodies tested reacted with endogenous cyclin B1 at the kinetochore: GNS1 and H-433 from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies that did not show kinetochore localization were H-20 and D-11 from Santa Cruz Biotechnology and Ab-3 from Calbiochem (San Diego, CA). All cyclin B1 antibodies were used a 1:1000 dilution, except the H-433 directly conjugated to fluorescein isothiocyanate (FITC) (sc-752 FITC), which was used at 1:100. Human anti-centromeric antibodies were used at a 1:250 dilution (Antibodies, Davis, CA). Hec1 antibody was used at 1:1000 (BD Biosciences Transduction Laboratories, Lexington, KY), Mad2 antibody was used at 1:100 (Abcam, Cambridge, MA), T9026 tubulin antibody was used at 1:500 (Sigma-Aldrich, St. Louis, MO), and Aurora B antibody was used at 1:500 (BD Biosciences, Franklin Lakes, NJ). Goat anti-mouse, goat anti-human, and donkey anti-rabbit secondary antibodies conjugated to Alexa-488, Alexa-568, or Alexa-647 were used at 1:1000 (Invitrogen). Monastrol was a gift from Tim Mitchison (Harvard Medical School, Boston), and it was used at a final concentration of 100 μg/ml. Nocodazole, taxol and MG132 were obtained from Sigma-Aldrich.
Immunofluorescence
Before immunostaining, 37°C media was exchanged for ice-cold media, and samples were incubated at 4°C for 10 min. After cold treatment, staining was performed as described previously (Waters et al., 1998). Cells were prepermeabilized in PHEM [60 mM piperazine-N,N′-bis(2-ethanesulfonic acid), 25 mM HEPES, 10 mM EGTA, and 4 mM MgSO4] with 0.5% Triton X-100 for 5 min, fixed in 1% formaldehyde in phosphate-buffered saline (PBS) for 15 min, and thoroughly rinsed in PBS with 0.05% Tween 20 (PBST). Cells were blocked in PBS plus 5% newborn calf serum for 1 h. All antibodies were diluted in blocking solution and staining was carried out at 37°C. Samples were washed after each step with PBST. DNA was stained using 2 μg/ml Hoechst 33342 (Invitrogen) in PBS for 2 min. Coverslips were mounted in 0.1 M n-propyl gallate (Sigma-Aldrich) in 90% glycerol in PBS. Z-series were acquired at 0.3-μm steps by using the same microscope as described for time-lapse imaging. Deconvolutions of Z-series were performed using AutoDeblur, version 8.0 (AutoQuant, Troy, NY).
Small Interfering RNA (siRNA) Transfection
HeLa cells were plated in Opti-MEM with 1% fetal bovine serum (Invitrogen) onto sterile glass coverslips at a density of 1 × 105 cells per well. Twenty-four hours after plating, cells were washed three times with Opti-MEM without FBS. GeneSilencer was used to transfect HeLa cells with modifications from the manufacturer's protocol (Genlantis, San Diego, CA). Opti-MEM, GTS diluent (GTS Gene Therapy Systems, San Diego, CA), Gene Silencer, and siRNA were mixed directly and incubated at room temperature for 15 min and then added to the cells. After 5 h of incubation, the media were replaced with DMEM plus 10% FBS. Control siRNA (Dharmacon RNA Technologies, Lafayette, CO, catalog. No. D-001210-01-20), Hec1 siRNA (Invitrogen, catalog no. 1299003; oligo HSS115931) or Mad2 siRNA (Dharmacon RNA Technologies, catalog no. KINRA-000007) were added to the cells at a final concentration of 25 nM. A mock transfection control was included in all experiments.
Immunoprecipitation
BS-C-1 cells were infected and plated at a density of 6 × 106 cells per 10-cm dish. Eighteen hours after infection, cells were washed with PBS and incubated with 1 ml of 20 mM Tris, 150 nM NaCl, 1.5 mM EDTA, 5 mM EGTA, and 0.5% NP-40 (TBST) for 30 min at 4°C. Cells were collected and centrifuged at 15,700 × g for 30 min at 4°C. Supernatant was transferred to a fresh tube, combined with 2 μl of anti-green fluorescent protein (GFP) (Ab290) (Abcam), and rotated at 4°C for 90 min. Washed and equilibrated protein A-Sepharose was added to each reaction and incubated overnight at 4°C. Reactions were washed three times with TBST and resolved by SDS-polyacrylamide gel electrophoresis. Immunoprecipitated proteins were detected by immunoblot by using cdc2 (Santa Cruz Biotechnology; sc-8395) or cyclin B1 (H-433) at 1:500 dilution or GFP antibody at 1:2000 dilution. Horseradish peroxidase-conjugated secondary antibodies were used at 1:5000 dilutions (Jackson ImmunoResearch Laboratories, West Grove, PA).
Quantitation of Kinetochore Signal
Z-series with a 0.3-μm step size were acquired of live, mitotic cells. Using MetaMorph imaging software, integrated intensity of individual kinetochores was collected from the median plane of the stack and from the three planes above and below the median plane. Fixed regions were placed over each kinetochore in the plane, in addition to a region outside the cell, the cytoplasm, and on chromatin. Chromatin associated signal was subtracted from kinetochore signal to generate normalized intensity values. Integrated intensity from 89 to 277 pairs was collected from a minimum of five different cells to calculate the normalized integrated intensity for the kinetochore signal. Confidence levels were calculated using Excel (Microsoft, Redmond, WA). Line scans measuring centrosome-associated integrated intensity were generated using MetaMorph imaging software.
RESULTS
Spinning Disk Confocal Microscopy Reveals Novel Cyclin B1 Localization to Kinetochores
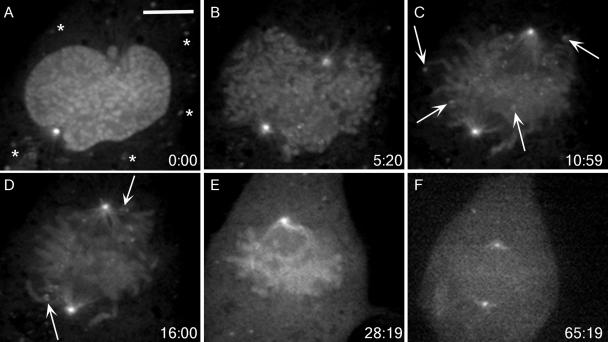
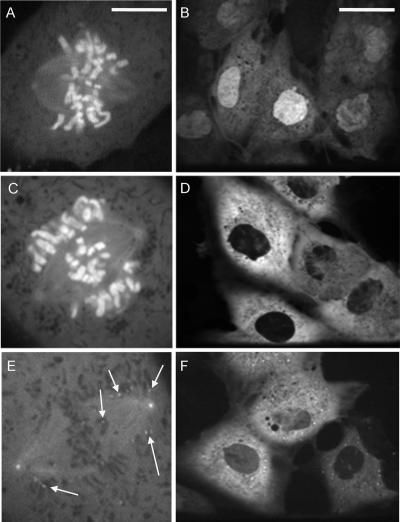
We constructed an adenovirus expressing full-length human cyclin B1 fused to EGFP (FL1-433). Using both time-lapse, wide-field fluorescence microscopy and time-lapse, spinning disk confocal microscopy, we observed FL1-433 localization to the cytoplasm and centrosomes of interphase cells, and to condensed chromatin, centrosomes, and spindle microtubules of mitotic cells, as described in a previous study (Pines and Hunter, 1991) (Figure 1). Spinning disk confocal microscopy also revealed the presence of cytoplasmic, punctate foci of FL1-433 in interphase (Figure 1A, asterisks). The size and number of these foci did not vary with viral titer, and they did not colocalize with lysosomes (data not shown). It has been reported that inactive cyclin B1–Cdc2 complexes form aggregates in starfish oocytes (Terasaki et al., 2003), and these foci may represent a similar form of sequestered cyclin B1. When using spinning disk confocal microscopy, we also observed a novel localization of FL1-433 to kinetochores during prometaphase (Figure 1, A–D, and Supplemental Video 1). This kinetochore population could first be observed at nuclear envelope breakdown (NEB) (Figure 1B). As cells progressed to metaphase, FL1-433 signal remained at kinetochores of chromosomes positioned near centrosomes, or on those clearly unattached to spindle microtubules (Figure 1, C and D, arrows). FL1-433 signal at kinetochores of chromosomes aligned at the metaphase plate was not detectable above the background of the chromatin-associated signal (Figure 1E). Local background subtraction methods (Hoffman et al., 2001) indicated that the overall decrease in FL1-433 signal intensity at kinetochores between the first indication of spindle microtubule attachment and the completion of congression was approximately fourfold (Supplemental Video 1; data not shown). After metaphase completion, we observed degradation of cyclin B1 as revealed by the global reduction of EGFP signal (Figure 1F).
Figure 1.
FL1-433 localizes to unattached kinetochores during mitosis. (A–E) BS-C-1 cells expressing FL1-433 were imaged using time-lapse, spinning disk confocal microscopy from prophase through metaphase completion. Elapsed time (minutes:seconds) from the first acquisition is shown in the lower right corner. (A) FL1-433 localizes to the nucleus and centrosomes in prophase. Asterisks indicate punctate cytoplasmic signal. (B) After NEB, FL1-433 associates with chromatin and centrosomes, and punctate kinetochore signal becomes apparent. (C) Later in prometaphase, kinetochore signal is prominent at outlying chromosomes (arrows) and along spindle microtubules. (D) As chromosomes congress to the metaphase plate, kinetochore-associated signal is diminished, but kinetochores adjacent to centrosomes continue to show strong kinetochore signal (arrows). (E) By metaphase, kinetochore signal is not observed, but chromatin and centrosome signal remain. (F) Before its degradation, FL1-433 signal is lost from chromatin. Bar, 10 μm (A). A movie of this cell is shown in Supplemental Video 1.
To rule out the possibility that adenoviral-based overexpression of FL1-433 caused artifactual kinetochore association of cyclin B1, we examined kinetochore localization in cells expressing FL1-433 at levels comparable with endogenous cyclin B1. At the viral dilution used in Figure 1, FL1-433 levels were much higher than endogenous cyclin B1 levels (Supplemental Figure S1A) and kinetochore localization was clearly visible (Supplemental Figure S1B). At a viral dilution resulting in FL1-433 expression equivalent to endogenous cyclin B1 (Supplemental Figure S1A), we were still able to observe kinetochore association of FL1-433 (Supplemental Figure S1C). These data confirm that FL1-433 localizes to kinetochores even when expressed at levels comparable with endogenous cyclin B1. However, lower dilutions resulted in a lower maximum signal intensity that necessitated exposure times incompatible with frequent image acquisition during mitosis (compare Supplemental Figure S1B with S1C).
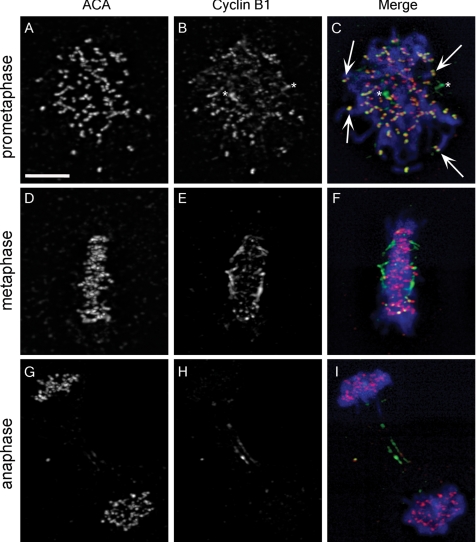
To confirm the live-cell imaging data, we determined the localization of endogenous cyclin B1 in BS-C-1 cells by indirect immunofluorescence. In these experiments, endogenous chromatin- and spindle microtubule-associated cyclin B1 was not preserved, but centrosome- and kinetochore-associated cyclin B1 was present (Figure 2). Punctate signal, which colocalized with anticentromeric antigen (ACA) signal, was observed in prometaphase cells labeled with cyclin B1 antibody confirming the localization of endogenous cyclin B1 to kinetochores (Figure 2, B and C, arrows). Preabsorption of primary antibodies with recombinant cyclin B1-CDK1 eliminated kinetochore-associated signal (Supplemental Figure S2). Consistent with live-cell imaging results, prometaphase kinetochores demonstrated robust cyclin B1 signal (Figure 2C, arrows), whereas kinetochores of fully congressed chromosomes exhibited almost no detectable signal (Figure 2, E and F). In anaphase cells, ACA signal persisted (Figure 2G), whereas cyclin B1 was undetectable (Figure 2H). Localization of endogenous cyclin B1 to kinetochores was also observed in HCT 116, MCF7, and MCF10A cell lines (data not shown), demonstrating that this novel localization is not unique to BS-C-1 cells. It is important to note that only two of five antibodies tested recognized the endogenous kinetochore population, which may explain the failure to observe cyclin B1 at kinetochores in previous fixed cell studies (Pines and Hunter, 1991; Jackman et al., 1995, 2003; Draviam et al., 2001). Both antibodies recognized epitopes in the N-terminal domain of cyclin B1, suggesting that C-terminal epitopes may become masked upon kinetochore association.
Figure 2.
Endogenous cyclin B1 localizes to kinetochores. (A, D, and G) ACA recognize kinetochores during prometaphase, metaphase, and anaphase. (B) An antibody that reacts with an N-terminal epitope in cyclin B1 has signal at kinetochores, but only during prometaphase. Arrows in C indicate colocalization of cyclin B1 and ACA. Asterisks in B and C designate centrosomes. (E and F) Cyclin B1 localization is depleted at kinetochores during metaphase, whereas ACA signal is robust (D and F). In anaphase, ACA remains at kinetochores (G and I), but cyclin B1 signal is depleted except for a reproducible population at the midzone (H and I). Merged images show DNA staining in blue, ACA staining in red, and cyclin B1 staining in green (C, F, and I). Bar, 10 μm (A).
FL1-433 Localization to Kinetochores Is Regulated by Microtubule Attachment
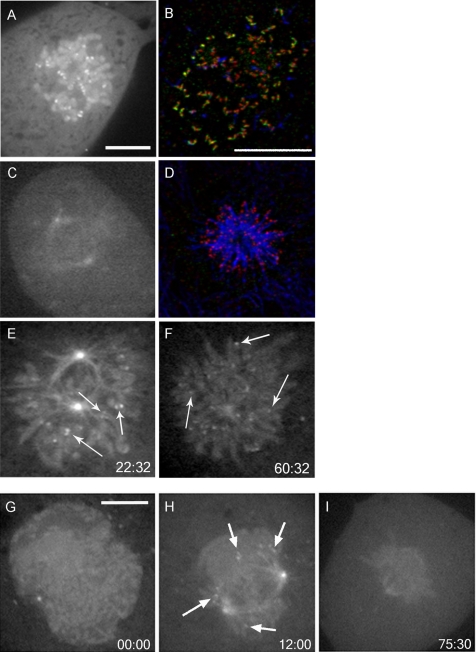
The prominent localization of cyclin B1 to prometaphase kinetochores suggested that microtubule attachment might be the principal factor regulating its localization. To address this hypothesis, we destabilized microtubules with nocodazole, thereby generating unoccupied kinetochores. Addition of nocodazole to late prometaphase or metaphase cells caused the collapse of the mitotic spindle and robust localization of FL1-433 to kinetochores at every chromatid pair (Figure 3A). Chromatin localization was also preserved. The localization of endogenous cyclin B1 to unattached kinetochores in nocodazole-treated cells was also observed in fixed cells (Figure 3B).
Figure 3.
Kinetochore localization of FL1-433 is sensitive to microtubule attachment but not tension. FL1-433–expressing BS-C-1 cells were treated with 0.5 μM nocodazole (A and B), 0.5 μM taxol (C and D), or monastrol (100 μg/ml) (E and F). (A) After nocodazole treatment, FL1-433 localizes to all kinetochores. (C) Taxol causes collapse of the spindle but no accumulation of FL1-433 at kinetochores. Cells treated with nocodazole (B) or taxol (D) were stained for cyclin B1 (green), tubulin (blue), and ACA (red), and recapitulate cyclin B1 dynamics was observed in live cells. (E and F) Cells were treated with monastrol and imaged using time-lapse, spinning disk confocal microscopy. Elapsed time (minutes:seconds) from the addition of monastrol and first acquisition is shown in the bottom right corner. (E) Early in monastrol treatment, FL1-433 localizes to centrosomes, microtubules and chromatin. FL1-433 is also present at paired kinetochores (arrows). (F) After prolonged monastrol arrest, FL1-433 is retained at centrosomes, microtubules, and chromatin. Paired FL1-433 evolves into singlets of kinetochore signal (arrows). This cell is shown in Supplemental Video 2. (G–I) Cells expressing FLΔDB were imaged using time-lapse, spinning disk confocal microscopy. Elapsed time (minutes:seconds) from the first acquisition is shown in the bottom right corner. (G) FLΔDB is enriched in the nucleus at prophase and localizes to centrosomes, spindle microtubules, chromatin, and kinetochores during mitosis (H). (I) FLΔDB is delocalized from kinetochores upon metaphase completion. Bar, 10 μm (A) for A, C, E and F; for B and D (B), and for G–I (G).
After attachment of kinetochores to spindle microtubules from opposing centrosomes, tension is generated across the chromatid pair (Nicklas et al., 1995). To determine whether FL1-433 kinetochore localization was sensitive to tension generation, we treated cells with taxol. Taxol stabilizes microtubules, thereby alleviating tension at kinetochores, but it does not ablate kinetochore–microtubule attachments (Waters et al., 1998). Metaphase-stage cells treated with taxol displayed shrinkage of the mitotic spindle but no FL1-433 relocalization to kinetochores (Figure 3C). This was also confirmed in fixed cells (Figure 3D). Together, these results show that the absence of tension is insufficient to promote cyclin B1 kinetochore relocalization, whereas lack of microtubule attachment to kinetochores causes robust cyclin B1 relocalization.
We also treated cells with monastrol, an inhibitor of the motor protein Eg5 (Mayer et al., 1999). We found that monastrol did not disrupt FL1-433 localization to kinetochores, chromatin, centrosomes, or spindle microtubules (Figure 3E), demonstrating that mitotic localization of cyclin B1 is not dependent on Eg5. Over time, kinetochore association of FL1-433 signal coalesced from two points into a single point, a pattern that remained for the duration of the experiment (Figure 3, E and F, arrows; and Supplemental Video 2). Signal coalescence is consistent with the stable attachment of microtubules to one of the kinetochores in a chromatid pair, as expected in prolonged monastrol exposure (Kapoor et al., 2000).
Cyclin B1 Localization Dynamics Are Independent of the Destruction Box
The destruction box (D-box) is a short-conserved sequence essential for recognition of cyclin B1 by the anaphase-promoting complex/cyclosome (APC/C) (Pfleger et al., 2001; Yamano et al., 2004). This D-box is required for degradation of cyclin B1 at the metaphase-to-anaphase transition. To assess the role of D-box–dependent degradation in cyclin B1 localization, we imaged a mutant in which the nine-amino acid D-box was deleted (FLΔDB). We observed that the dynamics of FLΔDB localization were identical to FL1-433: the protein was cytoplasmic during interphase and localized to chromatin, centrosomes, spindle microtubules, and kinetochores during mitosis (Figure 3, G and H). Furthermore, FLΔDB was delocalized from kinetochores at metaphase (Figure 3I), indicating that cyclin B1 dissociation does not depend on the APC/C. This protein failed to be degraded at the metaphase–anaphase transition, confirming that it is resistant to APC/C-mediated proteolysis (data not shown). Treatment with a proteasome inhibitor (MG132) did not perturb kinetochore association or dissociation of wild-type cyclin B1 (data not shown), demonstrating that the dissociation mechanism is also proteasome independent.
Cyclin B1 Localizes to the Outer Kinetochore
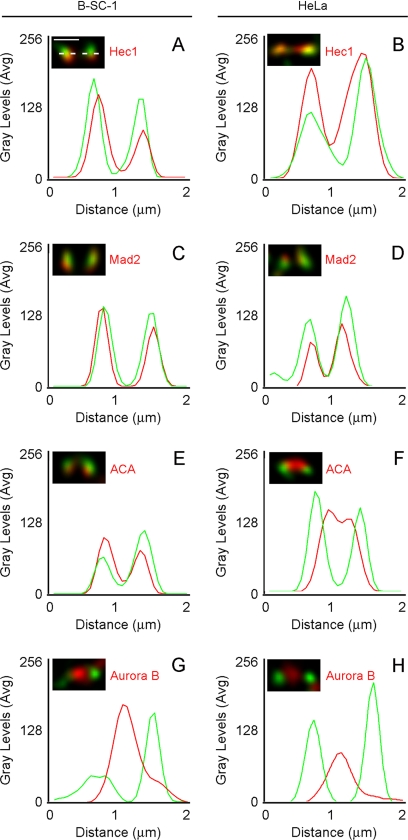
To better characterize the localization of cyclin B1-CDK1 at mitotic kinetochores, we compared its localization to that of ACA, Aurora B, Hec1, and Mad2 by immunofluorescence in BS-C-1 and HeLa cells. Relative localization of cyclin B1 was determined by line scan analysis through single kinetochore pairs in prometaphase cells. We found that localization of cyclin B1 in HeLa cells is similar to that observed in BS-C-1 cells (Figure 4). This analysis showed that cyclin B1 localized to the outer kinetochore, colocalizing with the outer kinetochore proteins Hec1 and Mad2 (Figure 4, A–D). Cyclin B1 also localized just outside of ACA signal in both cell types (Figure 4, E and F). In both cell types, cyclin B1 did not localize with Aurora B, a chromosomal passenger protein that resides in the inner centromere (Figure 4, G and H).
Figure 4.
Cyclin B1 colocalizes with Hec1 and Mad2 to the outer kinetochore. (A–H) Images of fixed B-SC-1 and HeLa cells were acquired by spinning disk confocal microscopy at 0.3-μm Z-intervals and deconvolved. (A) Kinetochore-associated signal was measured by line scan (dotted line) across the length of a kinetochore pair. Average gray levels were plotted against calibrated distance for each line scan. (A–H) Cyclin B1 signal (green) was compared with the signal from Hec1, Mad2, ACA, or Aurora B antibodies (red). The relative distributions of cyclin B1 were similar in B-SC-1 and HeLa cells. At least five kinetochore pairs from each of five separate cells were assessed to select representative pairs for this analysis. Bar, 1 μm (A).
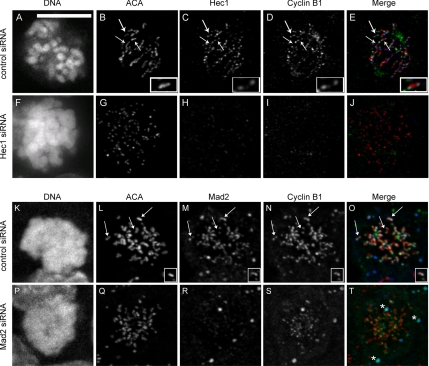
Hec1 and Mad2 Are Required for Cyclin B1 Localization to the Kinetochore
Because cyclin B1 was found to colocalize with Hec1 and Mad2 at the mitotic kinetochore, we asked whether either protein was required for cyclin B1 localization to the kinetochore. We transfected HeLa cells with siRNA targeting Hec1 or Mad2, and we examined cells by immunofluorescence 48 h later. HeLa cells depleted for Hec1 arrested in mitosis, and they displayed disorganized chromatin morphology (compare Figure 5A with F), consistent with the known role of Hec1 in microtubule attachment and chromosome congression (DeLuca et al., 2006). All cells that displayed this disorganized morphology were depleted of Hec1 as measured by immunofluorescence (Figure 5H), and they also showed depletion of cyclin B1 at the kinetochore (Figure 5I), although ACA staining was unperturbed (Figure 5G). Nocodazole treatment was insufficient to relocalize cyclin B1 to kinetochores (data not shown). Specificity of the antibody staining was confirmed by examining anaphase cells in control knockdown experiments, where we observed persistence of Hec1 kinetochore staining, but no cyclin B1 staining (Supplemental Figure 3). Furthermore, the level of Hec1 depletion correlated with cyclin B1 depletion on kinetochores in populations of cells treated with Hec1 siRNA (Supplemental Figure 4). Because Hec1 knockdown is known to displace multiple outer kinetochore proteins, including Mad2 (Martin-Lluesma et al., 2002; DeLuca et al., 2003, Lin et al., 2006), we next tested whether depletion of Mad2 was sufficient to inhibit cyclin B localization (Figure 5, K–T). Cells transfected with Mad2 siRNA were arrested in mitosis with 100 nM nocodazole and 2 μM MG132 for 1 h. Immunofluorescence of endogenous protein demonstrated that in the absence of Mad2 (Figure 5R), cyclin B1 localization at kinetochores was reduced (Figure 5S). Control experiments using primary and secondary antibodies separately demonstrated the specificity of the signal and the absence of bleed through (data not shown). These data show that cyclin B1 localization depends on Mad2 and other proteins removed upon Hec1 depletion.
Figure 5.
Cyclin B1 does not localize to kinetochores after Hec1 or Mad2 depletion. HeLa cells were transfected with control (A–E, and K–O), Hec1 (F–J), or Mad2-directed siRNA (P–T) for 48 h and then processed for immunofluorescence. In control siRNA-transfected cells, cyclin B1 (D and N) localized with Hec1 (C) or Mad2 (M) at kinetochores (B and L, arrows) in mitotic cells. A single kinetochore pair is enlarged to better show colocalization (B–E and L–O, insets). In cells depleted for Hec1 (F–J), chromatin is disorganized within the spindle (F). ACA (G) is present on kinetochores that are depleted for Hec1 (H), whereas cyclin B1 is absent from kinetochores (I). In the merged images (E and J), ACA (red), Hec1 (blue), and cyclin B1 (green) are shown. In cells depleted for Mad2 (P–T), ACA (Q) is present on kinetochores that are depleted for Mad2 (R), whereas cyclin B1 is reduced (S). In the merged images (O and T), ACA (red), Mad2 (blue), and cyclin B1 (green) are shown. Asterisks in T represent nonkinetochore signal specific to the primary antibody. Scale, 10 μm (A).
The N-Terminal Domain of Cyclin B1 Is Necessary and Sufficient for Chromatin Localization
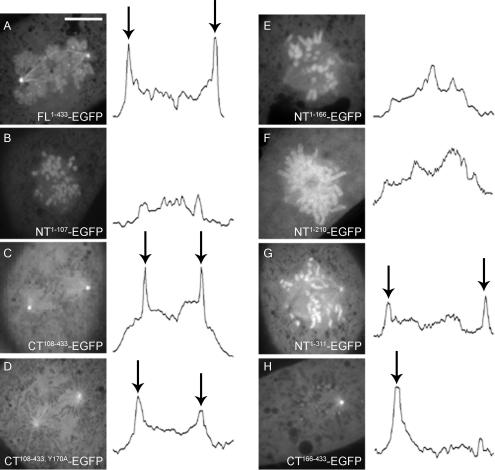
We wanted to identify sequence elements in cyclin B1 that determine localization to the kinetochore and to other mitotic structures. First, we imaged a construct consisting of the first 107 residues of cyclin B1 fused to EGFP (NT1-107). We observed that NT1-107 localized to condensed chromatin but never to kinetochores (Figure 6A), even after treatment with nocodazole (data not shown). Localization to centrosomes was greatly reduced (Figure 6A). Therefore, the N-terminal domain is sufficient to target cyclin B1 to condensed chromatin, but it is inadequate for efficient localization to centrosomes and kinetochores. Because NT1-107 is unable to bind CDK1 (Kobayashi et al., 1992 and Figure 7A), localization of NT1-107 must depend solely on cyclin B1 itself rather than indirect association mediated by CDK1. During interphase, NT1-107 localization was predominantly nuclear, presumably due to its lack of a cytoplasmic retention signal (CRS) (Figure 6B). Extension of NT1-107 to include the CRS region (NT1-166) restored its cytoplasmic localization during interphase (Figure 6D) but not its localization to centrosomes or kinetochores in mitosis (Figure 6C), demonstrating that the CRS is not a critical determinant of cyclin B1 localization during mitosis.
Figure 6.
The N-terminal domain of cyclin B1 is necessary and sufficient for localization to chromatin. (A) During mitosis, NT1-107 localizes predominantly to chromatin. Centrosome signal is severely reduced and kinetochore signal is absent. (B) NT1-107 is constitutively nuclear during interphase. NT1-166 localizes to chromatin but not to kinetochores during mitosis (C) and is cytoplasmic during interphase (D). (E) CT108-433 localizes to centrosomes, spindle microtubules, and kinetochores (arrows) during prometaphase but not to chromatin (E). (F) CT108-433 is cytoplasmic during interphase. Bar, 10 μm (A), which also applies to C and E. Bars, 10 μm (A; same scale for C and E) or 35 μm (B; same scale for D and F).
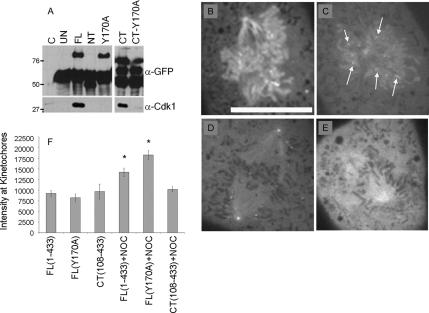
Figure 7.
Cyclin B1 localizes to kinetochores by CDK-1–dependent and – independent mechanisms. Lysates from adenovirally infected BS-C-1 cells were immunoprecipitated with an anti-GFP antibody. (A) Control BS-C-1 lysates (C, lane 1), uninfected BS-C-1 (UN), FL1-433 (FL), NT1-107 (NT), FLY170A (Y170A), CT108-433 (CT), and CT108-433, Y170A (CT-Y170A) immunoprecipitates were immunoblotted for GFP and CDK1. An immunoglobulin G-reactive band is present in each sample (α-GFP). CDK1 associates with FL1-433 and CT108-433 but not with NT1-107, FLY170A or CT108-433, Y170A (α-CDK1). By spinning disk confocal microscopy, FL1-433 (B) and FLY170A (C) localize to chromatin, centrosomes, spindle microtubules, and kinetochores. Arrows in C indicate kinetochore localization. (D) CT108-433 localizes to centrosomes, spindle microtubules, and kinetochores as shown in Figure 6F. (E) CT108-433, Y170A does not localize to kinetochores. (F) Quantitation of normalized integrated signal intensity at kinetochores of FL1-433–, FLY170A–, and CT108-433–expressing cells in untreated conditions and after treatment with 100 nM nocodazole. Error bars indicate 95% confidence levels. An asterisk represents a statistically significant difference in integrated intensity at kinetochores in the presence of 100 nM nocodazole versus untreated. Bar, 10 μm (A).
The localization pattern of NT1-107 and NT1-166 indicated that the N-terminal domain is sufficient for chromatin localization but that residues outside the N-terminal domain mediate localization to kinetochores and centrosomes. To confirm this, we generated EGFP fusions to the C-terminal domain of cyclin B1 (residues 108 through 433 or 166-433 [CT108-433, CT166-433]). Imaging experiments indicated that CT108-433 localized to the cytoplasm during interphase, as expected because it contains an intact CRS (Figure 6F). During mitosis, CT108-433 and CT166-433 localized to microtubules, centrosomes, and unattached kinetochores but never decorated chromatin (Figure 6E; data not shown). CT108-433 and CT166-433 both delocalized from kinetochores upon metaphase completion (Supplemental Video 3; data not shown). These data confirm that the N-terminal domain is necessary for chromatin localization and that sequences within residues 166-433 are necessary and sufficient for localization to kinetochores, centrosomes, and microtubules.
CDK1 Association Is Not Necessary for Cyclin B1 Localization
Because the C-terminal domain of cyclin B1 is sufficient to bind and activate CDK1 (Murray et al., 1989; Nugent et al., 1991), we asked whether CDK1 association was the principal mechanism of FL1-433 and CT108-433 localization to centrosomes, spindle microtubules, and kinetochores. We generated a single amino-acid point mutation at residue 170 (FLY170A) that was predicted to abolish CDK1 binding. Point mutations at this conserved tyrosine residue ablate the ability of cyclin B2 to bind CDK1 (Goda et al., 2001). To determine whether the mutant proteins bound CDK1, we infected BS-C-1 cells with adenovirus expressing FL1-433 or mutant versions of cyclin B1 and we used a GFP antibody to immunoprecipitate EGFP-fusion proteins. By this analysis, FL1-433 and CT108-433 bound CDK1, whereas NT1-107 and FLY170A did not (Figure 6A). Therefore, FLY170A was used to determine the requirement for CDK1 binding in localization of cyclin B1 to mitotic structures.
Time-lapse, spinning disk confocal microscopy showed that FLY170A was cytoplasmic during interphase and translocated to the nucleus before NEB (data not shown). On entry into mitosis, FLY170A localized to kinetochores, chromatin, spindle microtubules, and centrosomes (Figure 7C) in a manner that was indistinguishable from FL1-433 (Figure 7B). Loss of kinetochore signal during metaphase congression was also observed (Supplemental Video 4), indicating that CDK1 association is not required for cyclin B1 dissociation from kinetochores after microtubule attachment, or degradation of cyclin B1 at the metaphase-to-anaphase transition. Nocodazole treatment stimulated localization of FLY170A to kinetochores (data not shown). These results demonstrate that cyclin B1 can mediate its own dynamic localization during mitosis, independent of CDK1 interaction.
Although qualitative inspection showed no difference between FL1-433 and FLY170A localization, we quantitatively measured kinetochore-associated signal to validate this conclusion. Quantitation confirmed that in unperturbed cells, there was no significant difference between FL1-433, FLY170A, or CT108-433 signal at kinetochores in prometaphase (Figure 7D). When cells were arrested for at least 30 min with 100 nM nocodazole, a statistically significant increase in the kinetochore-associated signal of FL1-433 and FLY170A was observed (Figure 7D). This is consistent with literature showing that some kinetochore-associated proteins accumulate during a nocodazole arrest (Hoffman et al., 2001). However, CT108-433 kinetochore-associated signal was not increased after nocodazole-induced arrest (Figure 7D). These data suggest that although the N-terminal domain of cyclin B1 is not essential for kinetochore localization, it may play a role in stimulating association at the kinetochore under conditions of nocodazole arrest.
The normal localization of FLY170A demonstrates that cyclin B1 does not require CDK1 binding for localization to any mitotic structures. Therefore, we predicted that association of CT108-433 to kinetochores, spindle microtubules and centrosomes would also be independent of CDK1 binding. To test this idea, we generated a version of CT108-433 containing the Y170A mutation (CT108-433, Y170A). As expected, we found that this mutation abolished the interaction with CDK1 (Figure 7A). Surprisingly, CT108-433, Y170A failed to localize to kinetochores, although it remained at centrosomes and spindle microtubules (Figure 7F). Nocodazole was insufficient to localize CT108-433, Y170A to kinetochores (data not shown). These data show that CDK1 association is required for the kinetochore localization of cyclin B1 when its N-terminal domain is truncated, indicating that cyclin B1 may also be recruited to the kinetochore by a CDK1-dependent mechanism.
The Cyclin Box, but Not CDK1 Binding, Is Required for Centrosome Localization
Cyclin B1 localization to centrosomes has been proposed to be a site of cyclin B1–CDK1 activation that may regulate mitotic entry (Jackman et al., 2003). To identify sequence elements required for cyclin B1 localization to the centrosome, we measured the integrated intensity of EGFP signal across the length of the spindle and through both centrosomes by line scan in mitotic cells expressing various cyclin B1–EGFP constructs. By this method, FL1-433 and CT108-433 signal showed two peaks of centrosome-associated signal; for FL1-433, this centrosome-associated signal was roughly twice as bright as that associated with chromatin, spindle microtubules, or the cytoplasm (Figure 8A, arrows). Confirming the specificity of this analysis, NT1-107 showed no peaks of centrosome signal (Figure 8B), consistent with our qualitative analysis. FLY170A and CT108-433, Y170A localized efficiently (Figure 8, C and D, arrows), demonstrating that CDK1 interaction is not required for localization to centrosomes, even when the N-terminal domain is removed. This pattern of localization demonstrates that the mechanism of cyclin B1 localization to centrosomes is distinct from that of localization to kinetochores, where CDK1 binding is essential upon removal of the N-terminal domain (Figure 7F).
Figure 8.
A complete cyclin box is required for cyclin B1 localization to centrosomes. (A–H) Centrosome signal was measured by line scan across the length of the spindle. Representative examples of B-SC-1 mitotic cells expressing FL1-433 (A), NT1-107 (B), CT108-433 (C), CT108-433, Y170A (D), NT1-166 (E), NT1-210 (F), NT1-311 (G), and CT166-433 (H) are shown. Peaks in integrated intensity (arrows) correspond with positive centrosome signal. The presence of one centrosome in the plane of focus in H is reflected by a single peak in the corresponding line scan. Bar, 10 μm (A).
Because sequence elements within the C-terminal domain of cyclin B1, independently of those that mediate CDK1 binding, can localize the protein to centrosomes, we mapped such elements by extending the NT1-107 construct. NT1-166, which contains the CRS, and NT1-210, which contains the MRAIL residues of the HP, were both insufficient to localize cyclin B1 to centrosomes (Figure 7, E and F). However, inclusion of the entire cyclin box restored centrosome localization as revealed by NT1-311 localization (Figure 8G). Because NT1-311 cannot bind CDK1 (Figure 7; data not shown), residues 1-311 are sufficient to promote centrosome localization independently of CDK1 binding. It is possible that the addition of the remaining HP residues within the cyclin box were critical in mediating localization. Consistent with this model, we found that CT166-433 was competent to localize at centrosomes (Figure 8H, arrows), whereas a slightly shorter version, CT211-433, which lacks five essential residues of the HP, did not localize to centrosomes (Figure 9).
Figure 9.
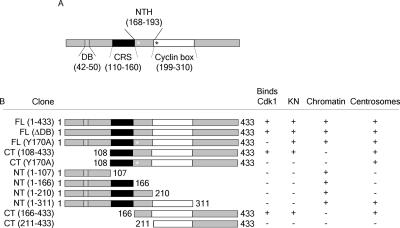
A summary of localization determinants of cyclin B1. (A) Schematic of significant sequence domains of cyclin B1. The Destruction box (DB), CRS, N-terminal helix (NTH), and cyclin box are shown. (A) Residues are indicated for each domain in parentheses. A white asterisk represents the Y170A point mutation, and a black asterisk represents the MRAIL residues in the beginning of the cyclin box. (B) The name and schematic for each cyclin B1 construct is indicated. The + and − indicate whether that particular construct binds CDK1, localizes to kinetochores (KN), chromatin or centrosomes.
DISCUSSION
Cyclin B1 Localizes to Unattached Kinetochores
To study the mechanism of cyclin B1 localization to the mitotic apparatus, we characterized the localization of wild-type and mutant versions of human cyclin B1 fused to EGFP by using time-lapse, spinning disk microscopy. We confirmed that full-length wild-type cyclin B1 (FL1-433) localizes to chromatin, spindle microtubules, and centrosomes as described in previous work (Pines and Hunter, 1991; Hagting et al., 1998; Clute and Pines, 1999). Spinning disk confocal microscopy also revealed novel localization of FL1-433 at kinetochores (Figure 1), which was confirmed by immunostaining of endogenous cyclin B1 protein (Figures 2 and 4). Kinetochore localization is sensitive to the state of microtubule attachment. Cyclin B1 is found at unattached kinetochores during prometaphase and after microtubule depolymerization with nocodazole (Figure 3). Consistent with our findings, the binding partner of cyclin B1, CDK1, has been shown to localize to kinetochores in mitosis (Rattner et al., 1990). Our data provide evidence that the CDK1 population at kinetochores is in fact active and that the abundance of the complex varies during mitotic progression.
Our mutagenesis data suggest that either cyclin B1 or CDK1 can independently recruit the cyclin B1–CDK1 complex to unattached kinetochores. Cyclin B1 can localize to kinetochores independently of CDK1 association, because a point mutation that abolishes CDK1 association (Y170A) does not perturb the normal dynamics of kinetochore association and dissociation (Figure 7). This is consistent with previous studies showing that mutations in Clb2 that ablate Cdc28 binding do not alter its normal localization to the bud neck, spindle pole body, or mitotic spindle (Bailly et al., 2003). Our data also suggest that sequences in both the N- and C-terminal domains of cyclin B1 are required for kinetochore localization in the absence of CDK1 binding, because the N-terminal domain is insufficient to localize to kinetochores (NT1-311), and the C-terminal domain fails to associate with kinetochores if it cannot bind CDK1 (CT108-433, Y170A). Together, these findings demonstrate that cyclin B1 can interact directly with components of the kinetochore.
Our data also reveal that CDK1 can target the cyclin B1–CDK1 complex to kinetochores. This mechanism is demonstrated by the localization of cyclin B1 mutants lacking the N-terminal domain. Although the CDK1-binding CT108-433 construct shows normal localization and delocalization from kinetochores (Figure 6 and Supplemental Video 3), a construct that does not bind CDK1 (CT108-433, Y170A) fails to localize to kinetochores (Figure 7). Furthermore, any construct able to bind CDK1 was also found to localize to kinetochores (Figure 9), suggesting that CDK1 binding may be sufficient for kinetochore localization. We propose that cyclin B1 association can render the entire cyclin B1–CDK1 complex sensitive to the state of microtubule attachment, perhaps through dynein-dependent transport of the complex used by spindle checkpoint proteins to leave the kinetochore (Hoffman et al., 2001; Howell et al., 2001). In this proposed model, active removal of cyclin B1 upon microtubule attachment would inactivate the kinetochore population of CDK1.
We found that cyclin B1 colocalizes with the Hec1 and Mad2 at the outer kinetochore during prometaphase (Figure 4). In addition, siRNA experiments show that either Hec1 or Mad2 depletion alters the normal kinetochore localization of cyclin B1 (Figure 5), demonstrating that both of these proteins must play a role in targeting cyclin B1 to the kinetochore. Although Hec1 depletion alters Mad1, Mad2, Zwint, Zw10, and Mps1 localization (Martin-Lluesma et al., 2002; DeLuca et al., 2003, Lin et al., 2006), Mad2 depletion has never been shown to alter the localization of other checkpoint proteins (Chung and Chen, 2002; Orr et al., 2007), and the relative dependencies of other checkpoint proteins on Mad2 kinetochore association are not sufficiently understood. Therefore, our data showing that cyclin B1 levels are reduced after Mad2 depletion is mechanistically intriguing. However, because Mad2 depletion results in a more modest effect on cyclin B1 than Hec1 depletion, we propose that additional proteins other than Mad2 and also affected by Hec1 depletion, play a role in targeting cyclin B1 to the kinetochore. Importantly, our data rule out a mechanism in which Cdc20 or APC/C-association alone recruit cyclin B1 to kinetochores (Acquaviva et al., 2004), because nondegradable versions of cyclin B1 showed normal dynamics of kinetochore localization and delocalization (Figure 3).
Our identification of cyclin B1 at unattached kinetochores corroborates previous studies proposing a positive role of CDK1 in spindle checkpoint activity in budding yeast (Kitazono and Kron, 2002; Kitazono et al., 2003) and in vertebrates (Chung and Chen, 2003; D'Angiolella et al., 2003). One potential function of kinetochore-associated cyclin B1-CDK1 could be to promote inhibition of Cdc20 by Mad2. Cdc20 contains two putative Cdk1 phosphorylation sites (Yudkovsky et al., 2000), and Cdc20 phosphorylation is required for Mad2 binding, as nonphosphorylatable versions of Cdc20 are resistant to Mad2 binding and inhibition (Chung and Chen, 2003; D'Angiolella et al., 2003). Cdc20 localizes to kinetochores from prophase through anaphase, but its association is highly dynamic, with binding times ranging from 1 to 5 s to 21 to 23 s (Kallio et al., 2002; Howell et al., 2004). We propose that localization of cyclin B1-CDK1 to unattached kinetochores could ensure the local phosphorylation of Cdc20, facilitating its binding to the active conformation of Mad2 generated at the kinetochore. In addition, other kinetochore proteins are known substrates of the cyclin B1–CDK1 complex, including INCENP (Goto et al., 2006), Bub1 (Qi et al., 2006), and APC/C subunits (Golan et al., 2002; Kraft et al., 2003). Therefore, recruitment of cyclin B1-CDK1 to unattached kinetochores could affect the activity of other components of the spindle checkpoint or regulate other processes that are essential for chromosome congression and anaphase onset.
The N-Terminal Domain Targets Cyclin B1 to Chromatin
The first 107 residues of cyclin B1 are necessary and sufficient for chromatin localization, providing a mechanism to concentrate CDK1 near chromatin-associated substrates. Because the N-terminal domain of cyclin B1 cannot bind CDK1, and the C-terminal CDK1-binding domain cannot localize to chromatin, we conclude that chromatin localization occurs independently of CDK1 binding. Possible CDK1 substrates on chromatin include condensins (Kimura et al., 1998), RCC1 and histone H1 (Arnaoutov and Dasso, 2003; Hutchins et al., 2004).
Besides targeting cyclins for APC/C-dependent degradation, other functions for the N-terminal domain of cyclin B1 have not been described. We found that removal of the conserved D-box, a motif required for interactions with Cdc20 and the APC/C (Yamano et al., 2004), did not alter chromatin localization, demonstrating that undefined sequences within the N-terminal domain are necessary for chromatin localization. The N-terminal domain is highly basic and could nonspecifically promote association with negatively charged DNA. However, cyclin A, which has access to condensed chromatin during early stages of mitosis and a similar isoelectric point, does not localize to chromatin (den Elzen and Pines, 2001). Therefore, it is likely that specific sequences within the N-terminal domain of cyclin B1, rather than charge-based mechanisms, are critical to chromatin localization.
The C-Terminal Domain Localizes Cyclin B1 to Centrosomes
Our data indicate that localization of cyclin B1 to centrosomes, like the chromatin population, is mediated directly by cyclin B1, rather than through a CDK1-dependent mechanism. Mutants of cyclin B1 that do not bind to CDK1 showed normal localization to centrosomes (Figures 8 and 9). Unexpectedly, the determinants of localization to chromatin and centrosomes are independent and separable. Whereas the N-terminal domain is necessary and sufficient for chromatin localization, the C-terminal domain of cyclin B1 is the critical mediator of centrosome association. Residues critical for centrosome localization lie within the cyclin box, because extension of the N-terminal domain to include the cyclin box (NT1-311) was sufficient to restore localization to centrosomes (Figure 8). Although the cyclin box plays an essential role in binding to CDK1, our findings indicate that its role in mediating centrosome association is independent of CDK1 binding. We propose that sequences specifically in the hydrophobic patch, which includes the MRAIL motif (positions 201–205) and a series of other conserved residues (W208, Q211, V212, L244, and V272) may be the critical determinants of cyclin B1 localization to centrosomes. The hydrophobic patch lies on a face of cyclin B1 opposite the face required for CDK1 association, explaining how it might mediate centrosome association independent of CDK1 binding. A fusion construct containing only residues 211-311 did not localize to any structure or compartment properly (data not shown), consistent with a requirement for the entire MRAIL motif and all HP residues. Consistent with our proposed HP-dependent mechanism of localization, HP mutations in Clb2 of budding yeast perturb localization to the bud neck (Bailly et al., 2003). However, because these same mutations in Clb2 do not disrupt localization to the spindle pole body, HP residues may have different roles in determining cyclin B localization in different organisms.
The Role of Cyclin B1 Localization in CDK1 Action
Our analysis shows that cyclin B1 is localized to different mitotic structures through distinct and separable mechanisms. The N-terminal domain of cyclin B1 is necessary and sufficient for chromatin localization, whereas the C-terminal domain including the HP residues mediates centrosome localization. In both cases, localization can occur independently of CDK1 binding. The requirements for kinetochore localization are more complex, because cyclin B1 localizes to kinetochores through both direct and indirect (CDK1-dependent) mechanisms.
The existence of distinct cyclin B1–CDK1 recruitment mechanisms has important implications for understanding the specificity of substrate phosphorylation during mitosis and also for regulation of cyclin B1-CDK1. For example, the affinity of cyclin B1 binding sites on chromatin, kinetochores, and centrosomes may be distinct. Therefore, cyclin B1 may be recruited to high-affinity sites during early stages of mitosis and be recruited to lower affinity sites only when cyclin B1 levels increase later in mitosis. The existence of distinct recruitment mechanisms also permits differential regulation of recruitment of cyclin B1 to each structure during mitosis. This principle is illustrated by our observation that kinetochore-associated cyclin B1 is regulated by the state of microtubule attachment, whereas levels of chromatin and centrosome-associated cyclin B1 remain steady until degradation at anaphase onset. Distinct recruitment pathways may also enable differential regulation of cyclin B1–CDK1 activity at different mitotic structures. It has been shown that the initial site of cyclin B1–CDK1 activation is at the centrosome (Jackman et al., 2003). This centrosome-localized activation relies on Plk1-dependent phosphorylation and Cdc25B-dependent dephosphorylation of cyclin B1-CDK1 (Jackman et al., 2003; Lindqvist et al., 2005). The other isoforms of Cdc25 have different patterns of localization, and they do not promote cyclin B1-CDK1 activation as efficiently in vivo (Dalal et al., 1999; Lindqvist et al., 2004, 2005; Kallstrom et al., 2005). Therefore, the existence of a specific centrosome-recruitment sequence on cyclin B1 may help concentrate the players necessary for efficient cyclin B1–CDK1 activation during mitotic entry.
Supplementary Material
ACKNOWLEDGMENTS
We thank Bert Vogelstein for the gift of the AdEasy system, Frank McKeon for the human cyclin B1 cDNA clone, and Tim Mitchison for monastrol. We also thank the Nikon Imaging Center at Harvard Medical School, where the spinning-disk confocal microscopy experiments were performed, and Dr. Jennifer Waters for advice. We thank Dr. Susan Lyman for comments on the manuscript. R.W.K. is a Damon Runyon Scholar. This work was funded by National Institutes of Health grant R01 GM-066492 (to R.W.K.), the Hellman Family Fund, and by a generous gift from Michael McKenzie in honor of his father, Harry C. McKenzie.
Abbreviations used:
- APC/C
anaphase-promoting complex/cyclosome
- ACA
anti-centromeric antigen
- CRS
cytoplasmic retention signal
- D-box
Destruction box
- HP
hydrophobic patch
- NEB
nuclear envelope breakdown.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-06-0539) on September 19, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Acquaviva C., Herzog F., Kraft C., Pines J. The anaphase promoting complex/cyclosome is recruited to centromeres by the spindle assembly checkpoint. Nat. Cell Biol. 2004;6:892–898. doi: 10.1038/ncb1167. [DOI] [PubMed] [Google Scholar]
- Arnaoutov A., Dasso M. The Ran GTPase regulates kinetochore function. Dev. Cell. 2003;5:99–111. doi: 10.1016/s1534-5807(03)00194-1. [DOI] [PubMed] [Google Scholar]
- Bailly E., Cabantous S., Sondaz D., Bernadac A., Simon M. N. Differential cellular localization among mitotic cyclins from Saccharomyces cerevisiae: a new role for the axial budding protein Bud3 in targeting Clb2 to the mother-bud neck. J. Cell Sci. 2003;116:4119–4130. doi: 10.1242/jcs.00706. [DOI] [PubMed] [Google Scholar]
- Brown N. R., Lowe E. D., Petri E., Skamnaki V., Antrobus R., Johnson L. N. Cyclin B and cyclin a confer different substrate recognition properties on CDK2. Cell Cycle. 2007;6:1350–1359. doi: 10.4161/cc.6.11.4278. [DOI] [PubMed] [Google Scholar]
- Brown N. R., Noble M. E., Endicott J. A., Johnson L. N. The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol. 1999;1:438–443. doi: 10.1038/15674. [DOI] [PubMed] [Google Scholar]
- Charrasse S., Lorca T., Doree M., Larroque C. The Xenopus XMAP215 and its human homologue TOG proteins interact with cyclin B1 to target p34cdc2 to microtubules during mitosis. Exp. Cell Res. 2000;254:249–256. doi: 10.1006/excr.1999.4740. [DOI] [PubMed] [Google Scholar]
- Chung E., Chen R. H. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol. Biol. Cell. 2002;13:1501–1511. doi: 10.1091/mbc.02-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung E., Chen R. H. Phosphorylation of Cdc20 is required for its inhibition by the spindle checkpoint. Nat. Cell Biol. 2003;5:748–753. doi: 10.1038/ncb1022. [DOI] [PubMed] [Google Scholar]
- Clute P., Pines J. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1999;1:82–87. doi: 10.1038/10049. [DOI] [PubMed] [Google Scholar]
- Cross F. R., Jacobson M. D. Conservation and function of a potential substrate-binding domain in the yeast Clb5 B-type cyclin. Mol. Cell. Biol. 2000;20:4782–4790. doi: 10.1128/mcb.20.13.4782-4790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Angiolella V., Mari C., Nocera D., Rametti L., Grieco D. The spindle checkpoint requires cyclin-dependent kinase activity. Genes Dev. 2003;17:2520–2525. doi: 10.1101/gad.267603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalal S. N., Schweitzer C. M., Gan J., DeCaprio J. A. Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol. Cell. Biol. 1999;19:4465–4479. doi: 10.1128/mcb.19.6.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J. G., Gall W. E., Ciferri C., Cimini D., Musacchio A., Salmon E. D. Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell. 2006;127:969–982. doi: 10.1016/j.cell.2006.09.047. [DOI] [PubMed] [Google Scholar]
- DeLuca J. G., Howell B. J., Canman J. C., Hickey J. M., Fang G., Salmon E. D. Nuf2 and Hec1 are required for retention of the checkpoint proteins Mad1 and Mad2 to kinetochores. Curr. Biol. 2003;13:2103–2109. doi: 10.1016/j.cub.2003.10.056. [DOI] [PubMed] [Google Scholar]
- den Elzen N., Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J. Cell Biol. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draviam V. M., Orrechia S., Lowe M., Pardi R., Pines J. The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J. Cell Biol. 2001;152:945–958. doi: 10.1083/jcb.152.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch T., Peter M., Nurse P., Nigg E. A. p34cdc2 acts as a lamin kinase in fission yeast. J. Cell Biol. 1991;112:797–807. doi: 10.1083/jcb.112.5.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T. K., Poon R. Y. A roller coaster ride with the mitotic cyclins. Semin. Cell Dev. Biol. 2005;16:335–342. doi: 10.1016/j.semcdb.2005.02.014. [DOI] [PubMed] [Google Scholar]
- Goda T., Funakoshi M., Suhara H., Nishimoto T., Kobayashi H. The N-terminal helix of Xenopus cyclins A and B contributes to binding specificity of the cyclin-CDK complex. J. Biol. Chem. 2001;276:15415–15422. doi: 10.1074/jbc.M011101200. [DOI] [PubMed] [Google Scholar]
- Golan A., Yudkovsky Y., Hershko A. The cyclin-ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1-cyclin B and Plk. J. Biol. Chem. 2002;277:15552–15557. doi: 10.1074/jbc.M111476200. [DOI] [PubMed] [Google Scholar]
- Goto H., Kiyono T., Tomono Y., Kawajiri A., Urano T., Furukawa K., Nigg E. A., Inagaki M. Complex formation of Plk1 and INCENP required for metaphase-anaphase transition. Nat. Cell Biol. 2006;8:180–187. doi: 10.1038/ncb1350. [DOI] [PubMed] [Google Scholar]
- Hagting A., Karlsson C., Clute P., Jackman M., Pines J. MPF localization is controlled by nuclear export. EMBO J. 1998;17:4127–4138. doi: 10.1093/emboj/17.14.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman D. B., Pearson C. G., Yen T. J., Howell B. J., Salmon E. D. Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol. Biol. Cell. 2001;12:1995–2009. doi: 10.1091/mbc.12.7.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., McEwen B. F., Canman J. C., Hoffman D. B., Farrar E. M., Rieder C. L., Salmon E. D. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B. J., Moree B., Farrar E. M., Stewart S., Fang G., Salmon E. D. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr. Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- Hutchins J. R., Moore W. J., Hood F. E., Wilson J. S., Andrews P. D., Swedlow J. R., Clarke P. R. Phosphorylation regulates the dynamic interaction of RCC1 with chromosomes during mitosis. Curr. Biol. 2004;14:1099–1104. doi: 10.1016/j.cub.2004.05.021. [DOI] [PubMed] [Google Scholar]
- Jackman M., Firth M., Pines J. Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J. 1995;14:1646–1654. doi: 10.1002/j.1460-2075.1995.tb07153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M., Lindon C., Nigg E. A., Pines J. Active cyclin B1-Cdk1 first appears on centrosomes in prophase. Nat. Cell Biol. 2003;5:143–148. doi: 10.1038/ncb918. [DOI] [PubMed] [Google Scholar]
- Kallio M. J., Beardmore V. A., Weinstein J., Gorbsky G. J. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J. Cell Biol. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallstrom H., Lindqvist A., Pospisil V., Lundgren A., Rosenthal C. K. Cdc25A localisation and shuttling: characterisation of sequences mediating nuclear export and import. Exp. Cell Res. 2005;303:89–100. doi: 10.1016/j.yexcr.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Kapoor T. M., Mayer T. U., Coughlin M. L., Mitchison T. J. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 2000;150:975–988. doi: 10.1083/jcb.150.5.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Hirano M., Kobayashi R., Hirano T. Phosphorylation and activation of 13S condensin by Cdc2 in vitro. Science. 1998;282:487–490. doi: 10.1126/science.282.5388.487. [DOI] [PubMed] [Google Scholar]
- Kitazono A. A., Garza D. A., Kron S. J. Mutations in the yeast cyclin-dependent kinase Cdc28 reveal a role in the spindle assembly checkpoint. Mol. Genet. Genomics. 2003;269:672–684. doi: 10.1007/s00438-003-0870-y. [DOI] [PubMed] [Google Scholar]
- Kitazono A. A., Kron S. J. An essential function of yeast cyclin-dependent kinase Cdc28 maintains chromosome stability. J. Biol. Chem. 2002;277:48627–48634. doi: 10.1074/jbc.M207247200. [DOI] [PubMed] [Google Scholar]
- Kobayashi H., Stewart H., Poon R., Adamczewski J. P., Gannon J., Hunt T. Identification of the domains in cyclin A required for binding to, and activation of, p34cdc2 and p32cdk2 protein kinase subunits. Mol. Biol. Cell. 1992;3:1279–1294. doi: 10.1091/mbc.3.11.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Herzog F., Gieffers C., Mechtler K., Hagting A., Pines J., Peters J. M. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. T., Chen Y., Wu G., Lee W. H. Hec1 sequentially recruits Zwint-1 and ZW10 to kinetochores for faithful chromosome segregation and spindle checkpoint control. Oncogene. 2006;25:6901–6914. doi: 10.1038/sj.onc.1209687. [DOI] [PubMed] [Google Scholar]
- Lindqvist A., Kallstrom H., Karlsson Rosenthal C. Characterisation of Cdc25B localisation and nuclear export during the cell cycle and in response to stress. J. Cell Sci. 2004;117:4979–4990. doi: 10.1242/jcs.01395. [DOI] [PubMed] [Google Scholar]
- Lindqvist A., Kallstrom H., Lundgren A., Barsoum E., Rosenthal C. K. Cdc25B cooperates with Cdc25A to induce mitosis but has a unique role in activating cyclin B1-Cdk1 at the centrosome. J. Cell Biol. 2005;171:35–45. doi: 10.1083/jcb.200503066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loog M., Morgan D. O. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Luscher B., Brizuela L., Beach D., Eisenman R. N. A role for the p34cdc2 kinase and phosphatases in the regulation of phosphorylation and disassembly of lamin B2 during the cell cycle. EMBO J. 1991;10:865–875. doi: 10.1002/j.1460-2075.1991.tb08019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Lluesma S., Stucke V. M., Nigg E. A. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–2270. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- Mayer T. U., Kapoor T. M., Haggarty S. J., King R. W., Schreiber S. L., Mitchison T. J. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Murray A. W., Solomon M. J., Kirschner M. W. The role of cyclin synthesis and degradation in the control of maturation promoting factor activity. Nature. 1989;339:280–286. doi: 10.1038/339280a0. [DOI] [PubMed] [Google Scholar]
- Nicklas R. B., Ward S. C., Gorbsky G. J. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J. Cell Biol. 1995;130:929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E. A. The substrates of the cdc2 kinase. Semin. Cell Biol. 1991;2:261–270. [PubMed] [Google Scholar]
- Nigg E. A., Blangy A., Lane H. A. Dynamic changes in nuclear architecture during mitosis: on the role of protein phosphorylation in spindle assembly and chromosome segregation. Exp. Cell Res. 1996;229:174–180. doi: 10.1006/excr.1996.0356. [DOI] [PubMed] [Google Scholar]
- Nugent J. H., Alfa C. E., Young T., Hyams J. S. Conserved structural motifs in cyclins identified by sequence analysis. J. Cell Sci. 1991;99:669–674. doi: 10.1242/jcs.99.3.669. [DOI] [PubMed] [Google Scholar]
- Ookata K., Hisanaga S., Bulinski J. C., Murofushi H., Aizawa H., Itoh T. J., Hotani H., Okumura E., Tachibana K., Kishimoto T. Cyclin B interaction with microtubule-associated protein 4 (MAP4) targets p34cdc2 kinase to microtubules and is a potential regulator of M-phase microtubule dynamics. J. Cell Biol. 1995;128:849–862. doi: 10.1083/jcb.128.5.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr B., Bousbaa H., Sunkel C. E. Mad2-independent spindle assembly checkpoint activation and controlled metaphase-anaphase transition in Drosophila S2 cells. Mol. Biol. Cell. 2007;18:850–863. doi: 10.1091/mbc.E06-07-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri E. T., Errico A., Escobedo L., Hunt T., Basavappa R. The crystal structure of human cyclin B. Cell Cycle. 2007;6:1342–1349. doi: 10.4161/cc.6.11.4297. [DOI] [PubMed] [Google Scholar]
- Pfleger C. M., Lee E., Kirschner M. W. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001;15:2396–2407. doi: 10.1101/gad.918201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J., Hunter T. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol. 1991;115:1–17. doi: 10.1083/jcb.115.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pines J., Hunter T. The differential localization of human cyclins A and B is due to a cytoplasmic retention signal in cyclin B. EMBO J. 1994;13:3772–3781. doi: 10.1002/j.1460-2075.1994.tb06688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi W., Tang Z., Yu H. Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol. Biol. Cell. 2006;17:3705–3716. doi: 10.1091/mbc.E06-03-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner J. B., Lew J., Wang J. H. p34cdc2 kinase is localized to distinct domains within the mitotic apparatus. Cell Motil. Cytoskeleton. 1990;17:227–235. doi: 10.1002/cm.970170309. [DOI] [PubMed] [Google Scholar]
- Sanchez I., Dynlacht B. D. New insights into cyclins, CDKs, and cell cycle control. Semin. Cell Dev. Biol. 2005;16:311–321. doi: 10.1016/j.semcdb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Schulman B. A., Lindstrom D. L., Harlow E. Substrate recruitment to cyclin-dependent kinase 2 by a multipurpose docking site on cyclin A. Proc. Natl. Acad. Sci. USA. 1998;95:10453–10458. doi: 10.1073/pnas.95.18.10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C. J. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60:3689–3695. [PubMed] [Google Scholar]
- Terasaki M., Okumura E., Hinkle B., Kishimoto T. Localization and dynamics of Cdc2-cyclin B during meiotic reinitiation in starfish oocytes. Mol. Biol. Cell. 2003;14:4685–4694. doi: 10.1091/mbc.E03-04-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters J. C., Chen R. H., Murray A. W., Salmon E. D. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 1998;141:1181–1191. doi: 10.1083/jcb.141.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano H., Gannon J., Mahbubani H., Hunt T. Cell cycle-regulated recognition of the destruction box of cyclin B by the APC/C in Xenopus egg extracts. Mol. Cell. 2004;13:137–147. doi: 10.1016/s1097-2765(03)00480-5. [DOI] [PubMed] [Google Scholar]
- Yudkovsky Y., Shteinberg M., Listovsky T., Brandeis M., Hershko A. Phosphorylation of Cdc20/fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem. Biophys. Res. Commun. 2000;271:299–304. doi: 10.1006/bbrc.2000.2622. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.