Abstract
Cytokinesis in metazoan cells requires a set of antiparallel microtubules that become bundled upon anaphase onset to form a structure known as the central spindle. Bundling of these microtubules requires a protein complex, centralspindlin, that consists of the CYK-4/MgcRacGAP Rho-family GTPase-activating protein and the ZEN-4/MKLP1 kinesin-6 motor protein. Centralspindlin, but not its individual subunits, is sufficient to bundle microtubules in vitro. Here, we present a biochemical and genetic dissection of centralspindlin. We show that each of the two subunits of centralspindlin dimerize via a parallel coiled coil. The two homodimers assemble into a high-affinity heterotetrameric complex by virtue of two low-affinity interactions. Conditional mutations in the regions that mediate complex assembly can be readily suppressed by numerous second site mutations in the interacting regions. This unexpected plasticity explains the lack of primary sequence conservation of the regions critical for this essential protein–protein interaction.
INTRODUCTION
Cell division in animal cells is dependent upon a structure known as the central spindle. As cells initiate anaphase, this structure assembles between the segregating chromosomes. The central spindle consists of antiparallel microtubules (MTs) that are bundled at their plus ends, through the action of microtubule-associated proteins (MAPs) and kinesin family motors (for review, see Glotzer, 2005). Successful completion of cytokinesis in animal cells requires the bundled microtubules of the central spindle (Wheatley and Wang, 1996). In addition, the central spindle plays an important role in regulating cleavage plane positioning (Dechant and Glotzer, 2003; Bringmann and Hyman, 2005; Werner et al., 2007).
Central spindle assembly is mediated by several distinct protein complexes that act cooperatively to bundle antiparallel microtubules during anaphase. These complexes include centralspindlin, which contains a kinesin-6 motor protein, ZEN-4/MKLP1, and a Rho-family GTPase-activating protein (GAP) protein, CYK-4/MgcRacGAP (Mishima et al., 2002). Localization of centralspindlin requires the multisubunit Aurora B kinase complex, which may play a direct role in microtubule bundling (Kaitna et al., 2000; Severson et al., 2000; Pereira and Schiebel, 2003). Finally, the PRC1 MAP is also implicated in microtubule bundling during anaphase (Mollinari et al., 2002, 2005; Kurasawa et al., 2004; Verbrugghe and White, 2004; Verni et al., 2004) in part through the ability of this factor to bind to the kinesin-4 motor protein Kif4 (Kurasawa et al., 2004; Mollinari et al., 2005). In vitro microtubule bundling activity has been demonstrated for both centralspindlin and PRC1 (Mishima et al., 2002; Mollinari et al., 2002); however, these two complexes are each required for the assembly of robust, organized microtubule bundles, with these components at restricted, well-defined positions within the central spindle. These complexes not only act to organize microtubule bundles but also serve to recruit additional factors that regulate initiation of cytokinesis and a variety of components that facilitate completion of cytokinesis, or abscission (Gromley et al., 2005).
Centralspindlin is of particular interest among these complexes because the CYK-4 subunit contains a GAP domain that can induce GTP hydrolysis by RhoA, Rac, and Cdc42, at varying efficiencies (Toure et al., 1998; Jantsch-Plunger et al., 2000). In addition, CYK-4 also directly binds the guanine nucleotide exchange factor (GEF) for RhoA, ECT2 (Yuce et al., 2005; Zhao and Fang, 2005; Kamijo et al., 2006; Nishimura and Yonemura, 2006), but ECT2 is not a stoichiometric component of this complex. Cells depleted of the kinesin component of centralspindlin, ZEN-4/MKLP-1, often initiate cleavage furrow formation, but they invariably fail to complete cytokinesis. However, human cells depleted of the RhoGAP subunit of centralspindlin, CYK-4, do not even initiate cytokinesis (Yuce et al., 2005; Zhao and Fang, 2005; Kamijo et al., 2006; Nishimura and Yonemura, 2006). Because centralspindlin controls furrow initiation by promoting RhoA activation and regulates completion of cytokinesis, it is important to define its biochemical function and the mechanism by which it becomes so discretely localized on the central spindle.
The components of centralspindlin have been purified to near homogeneity under stringent conditions. The two subunits are present in stoichiometric amounts, with the native molecular weight of centralspindlin being ∼300 kDa (Mishima et al., 2002). Under stringent purification conditions, no additional factors copurified in significant amounts, suggesting that the complex is a tetramer. Because each component can individually oligomerize, the centralspindlin tetramer is likely a heterotetramer. The complex has been reconstituted from recombinant sources and the regions required for the interaction between CYK-4 and ZEN-4 have been roughly mapped. Microtubule bundling by centralspindlin requires the presence of both subunits and their ability to interact (Mishima et al., 2002). The nature of the interaction domain is particularly intriguing, because it is not well conserved at the primary sequence level, even though the interaction between these proteins has been documented in a variety of metazoan species (Mishima et al., 2002; Somers and Saint, 2003).
Although the broad outlines of how the centralspindlin complex assembles and bundles microtubules have been defined, critical questions remain. In particular, structural and functional studies would benefit from a description of the minimal interacting domains of CYK-4 and ZEN-4. In addition, the question of whether their interaction requires dimeric forms of CYK-4 and ZEN-4 remains open. To address these issues, we have conducted a detailed structure–function analysis of centralspindlin. We demonstrate that both CYK-4 and ZEN-4 form parallel coiled coils, and we show that their ability to independently dimerize contributes substantially to stabilization of centralspindlin. We show that CYK-4 and ZEN-4 form a high-affinity complex and that both of the temperature-sensitive mutations in these genes destabilize this protein complex. Through the use of genome-wide genetic screens, we have identified additional critical features of the interaction interface.
MATERIALS AND METHODS
Nematode Strains and Alleles
The following strains or alleles were used in this study: N2 (Bristol), zen-4(or153ts), EH135 (unc-44(e362) bli-6(sc16) IV), SU62 (unc-44(e362) lag-1 (q385)/zen-4(w35) IV), and MG367 (zen-4(w35) bli-6(sc16)/unc-44(el260) lag-1(q385)). Some strains in this study were obtained from Caenorhabditis Genetics Center, which is funded by the National Institutes of Health National Center for Research Resources. For rescue assays, simple transgenic arrays were prepared in MG367 as described previously (Mello and Fire, 1995) by using pRF4 at 80 ng μl−1 and MP66 (zen-4), VP01 (zen-4 D520N), and VP02 (zen-4 D735N) at 10 ng μl−1.
Isolation of zen-4 (or153ts) Suppressors
Suppressors of zen-4(or153ts) mutations were obtained by mutagenesis of zen-4(or153ts) animals with 30 μM ethane methyl sulfonate (EMS). After recovery, F1 embryos were isolated by bleaching and allowed to self-fertilize at permissive temperature (16°C). When F2 animals reached adulthood, the population was shifted to 25°C, and fertile animals were selected, a single isolate was kept from each pool of F1 embryos. Approximately 500,000 F1 genomes were screened, and 14 suppressor mutations were isolated, all of which were viable at 25°C.
Protein Preparation
ZEN-4 fragments were cloned into pCBD-TEV and expressed in Escherichia coli as N-terminal chitin binding domain (CBD) fusion proteins. CYK-4 fragments were cloned into pGEX-4-TEV and expressed in E. coli as N-terminal glutathione transferase (GST) fusion proteins. Lysis buffer (buffer A) contained 10 mM HEPES, pH 7.7, 1 mM EGTA, 1 mM MgCl2, 0.1% (wt/vol) Triton X-100, 250 mM NaCl, 10 μg ml−1 leupeptin/pepstatin, 1 mM dithiothreitol (DTT), and 1 mM phenylmethylsulfonylfluoride (PMSF). Extracts were clarified by centrifugation at 19,000 rpm, and the recombinant proteins were affinity purified with chitin or glutathione Sepharose beads, and eluted with tobacco etch virus (TEV) protease.
For bicistronic expression of ZEN-4 and CYK-4, the fragments were cloned into pGEX-6, as N-terminal CBD fusions of ZEN-4 and N-terminal GST fusions of CYK-4 (Sessa et al., 2005). After E. coli expression, GST-CYK-4 was purified on glutathione Sepharose beads and eluted with reduced glutathione, followed by incubation of the eluates with chitin beads. The recombinant proteins were eluted from chitin beads with TEV protease and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Coomassie Blue staining.
In Vitro Translation and Binding Assay
CYK-4 and ZEN-4 were expressed by in vitro transcription and translation system with reticulocyte lysate by using the TNT-coupled reticulocyte lysate system (Promega, Madison, WI) in a 20-μl reaction. Protein fragments of CYK-4 or ZEN-4 tagged with CBD at the N terminus were expressed from the T7 promoter of pCBD-TEV. After translation, the binding reaction was set up in 100 μl of buffer B and incubated at 20°C for 30 min. ZEN-4 protein was immunoprecipitated with specific antibodies, by addition of 0.2 μg antibody to the reactions and incubated on ice for 1 h, followed by incubation with 5 μl of protein A-Sepharose beads at 4°C for 1 h. The beads were subsequently washed with buffer B. Proteins bound to the beads were analyzed by SDS-PAGE followed by autoradiography with a phosphorimager. Buffer B consists of 20 mM HEPES, pH 7.7, 10 mM EDTA, 2 mM MgCl2, 0.5% (wt/vol) Triton X-100, 150 mM NaCl2, 10 μg ml−1 leupeptin/pepstatin, 1 mM DTT, and 1 mM PMSF.
Microtubule Bundling Assay
The centralspindlin complex was reconstituted by mixing equimolar (∼1 μM) amounts of recombinant CYK-4 and ZEN-4 and incubating for 20 min at room temperature. For microtubule bundling assay, samples were mixed with Taxol-stabilized microtubules (final 4 μM) in buffer C and incubated for 20 min at room temperature (Mishima et al., 2002). The reaction was fixed with glutaraldehyde, and the microtubules were sedimented on a coverslip as described previously (Desai and Walczak, 2001). Microtubules were fixed in cold methanol and stained with anti-tubulin monoclonal antibody (DM1α), and images were acquired with a 63×/1.4 numerical aperture objective on a Zeiss Axioplan II and a CoolSnapFX charge-coupled device camera that was controlled with MetaMorph (Molecular Devices, Sunnyvale, CA). The quantification of results was performed by analyzing 10 random fields and integrating pixel intensities above a fixed threshold for all images that excluded single microtubules. Buffer C consists of 125 mM KCl, 80 mM PIPES, 1 mM EGTA, 1 mM MgCl2, 1 mM DTT, and 2 mM adenyl-5′-yl imidodiphosphate, pH 6.8.
Surface Plasmon Resonance
The affinity and the rates of interaction between ZEN-4 and CYK-4 were measured with a BIAcore 3000 biosensor (BIAcore, Uppsala, Sweden). CYK-4, with an N-terminal GST tag, was immobilized to the sensor chip (CM5) via an anti-GST antibody that had been amine coupled with the dextran matrix in 10 mM sodium acetate, pH 5.0. The control surface was prepared similarly except that CYK-4 GST was not immobilized on the chip. Kinetic experiments were performed at 16 and 25°C, by using flow rates of 5–20 μl min−1. ZEN-4 was added into the flow in buffer A (but with 25 μM ATP) at various concentrations (3–300 nM) during the binding phase. The amount of protein bound to the sensor chip was monitored by the change in refractive index (given in arbitrary units). For the measurement of kinetic parameters, the amount of immobilized GST-CYK-4 was minimized, to avoid saturation and avidity effects. After each binding experiment, the sensor chip was regenerated by sequential washing with 10 mM glycine, pH 2.0, followed by two washes with running buffer. Triplicate injections of each concentration of ZEN-4 were performed. For data analysis, BIAevaluate software (BIAcore) was used, and the sensorgram association and dissociation curves were locally or globally fit to a two-site model. Running buffer consists of buffer A without protease inhibitors or DTT.
Isothermal Titration Calorimetry
Binding of monomeric ZEN-4 to dimeric CYK-4 was measured using a MicroCal VP isothermal titration calorimeter. Protein samples used for the titration were prepared by dialyzing overnight against 2 l of working buffer A without Triton X-100 and protease inhibitors. Measurements were performed at 25°C by using the working buffer A minus Triton X-100 and protease inhibitors as a reference. A sample cell containing dimeric CYK-4 (10 μM) was titrated by repeated injection of monomeric ZEN-4 (100 μM). The collected data were analyzed with the Origin software (OriginLab, Northampton, MA) by using a simple single-site model.
Binding Studies
Binding of different CYK-4 truncation products was measured by a standard pull-down assay. CYK-4 fragments were purified as maltose binding protein (MBP) fusions, and they were retained on the amylose beads. Recombinant ZEN-4(1-585)GFP was added at different concentrations to the beads in a 100-μl reaction, and it was incubated at room temperature for 2 h in buffer B. In the case of CYK-4(1-120) and CYK-4(1-36)GCN-4, the fluorescence remaining in the supernatant after incubation was measured with a Safire2 (Tecan, Reading, United Kingdom) microplate reader. The bound fraction was calculated by subtracting the unbound fraction from the total. In the case of CYK-4(1-36), the bound fraction was analyzed by SDS-PAGE and Coomassie Blue staining, followed by quantification of band intensities with the Odyssey (Li-Cor, Lincoln, NE) infrared imaging system. In both cases, a graph of bound fractions versus increasing concentrations of ligand was generated, and the KD was estimated using nonlinear regression in which the data were fit to an equation describing a rectangular hyperbola (y = Bmax · x/(KD + x)). In addition, the data were plotted on a Scatchard plot, and the KD value was estimated from the slope (−1/KD). Similar results were obtained by the two methods. Protein concentrations were estimated by the Bradford assay (Bio-Rad, Hercules, CA) using bovine serum albumin as a standard. Binding of different ZEN-4 products was also measured by a standard pull-down assay. CYK-4 fragments were purified as GST fusions and retained on the glutathione-Sepharose beads. Recombinant wild-type ZEN-4(1-585), ZEN-4(1-585)D520N, ZEN-4(1-539), and ZEN-4(1-539)GCN4 were added to the beads in a 100-μl reaction and incubated for 2 h at 25°C in buffer A without protease inhibitors or DTT. The beads were subsequently washed two times, and the bound fraction was analyzed by SDS-PAGE and Coomassie Blue staining.
Gel Filtration Chromatography
The diffusion coefficients of different recombinant ZEN-4 fragments were analyzed by size exclusion chromatography using a Superdex 200 column (30 ml) on an ÁKTA FPLC system (GE Healthcare, Chalfont St. Giles, United Kingdom) in buffer A (250 mM KCl and 25 μM ATP, minus Triton X-100, leupeptin/pepstatin, and PMSF). The fractions were analyzed by SDS-PAGE and Coomassie Blue staining.
Cross-Linking
Recombinant ZEN-4 and CYK-4 fragments containing cysteine residues at N or C termini were incubated in a buffer A (minus ATP, Triton X-100, leupeptin/pepstatin, and PMSF), containing 5 mM oxidized and 4 mM reduced glutathione. The reaction was quenched with 100 mM iodoacetamide and analyzed by SDS-PAGE and Coomassie Blue staining.
RESULTS
Characterization of zen-4(or153ts)
To gain further insight into the nature of centralspindlin, we took a genetic approach starting with the previously identified temperature-sensitive loss-of-function allele zen-4(or153ts). zen-4(or153ts) animals exhibit penetrant embryonic lethality at the restrictive temperature (25°C), but they are completely viable and have a high brood size at the permissive temperature (16°C) (Severson et al., 2000). Because this is a conditional, loss-of-function allele, we reasoned that suppressors of the high temperature lethality could lead to the identification of novel factors that act in conjunction with centralspindlin or identify key residues within the complex that stabilize its activity, either directly or indirectly.
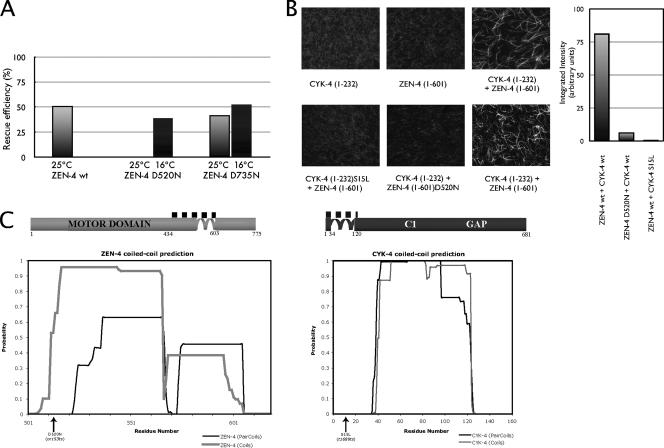
Because the zen-4(or153ts) allele contains two mutations, each of which encodes for a Asp (D)-to-Asn (N) substitution at positions 520 and 735 (Severson et al., 2000), we first wanted to ascertain whether one of the two mutations is primarily responsible for the phenotype. We generated multiple independent transgenic lines expressing either wild-type ZEN-4, ZEN-4 D520N, or ZEN-4 D735N, in a background containing the null zen-4(w35) allele, which causes embryonic lethality associated with cytokinesis defects; zen-4(w35) was linked to the visible recessive marker bli-6. We assayed for the ability of the transgene to rescue the null allele, which could be conveniently scored by counting blistered animals. We found that both ZEN-4 and ZEN-4 D735N could rescue zen-4(w35) at 25°C, whereas ZEN-4 D520N could not (Figure 1A). Therefore, the D520N point mutation in zen-4(or153ts) is responsible for causing the mutant phenotype. Additionally, ZEN-4 D520N could rescue the null allele at 16°C, the permissive temperature for zen-4(or153ts), indicating that this substitution is solely responsible for the conditional nature of this allele.
Figure 1.
ZEN-4 D520N impairs microtubule bundling in vitro and is responsible for the in vivo phenotype of zen-4(or153ts). (A) zen-4(or153ts) D520N fails to rescue zen-4(w35) at 25°C. Simple transgenic arrays marked by rol-6(su1006) were prepared and the efficiency of zen-4 D520N and D735N versus wt ZEN-4 to rescue zen-4(w35) was assayed. The rescue efficiency was scored by counting the blistered [bli-6(sc16)] animals, both at the restrictive (25°C; gray) and the permissive temperature (16°C; black) as described previously (Mishima et al., 2004). At 25°C ZEN-4 D520N could not rescue the null, whereas both wt and ZEN-4 D735N efficiently rescued the null. (B) The D520N point mutation in zen-4(or153ts) inhibits MT bundling. MT bundling assays were performed with constituents of centralspindlin alone, and with reconstituted centralspindlin complex, comparing wt and mutant complexes. The quantification of results was performed by the analysis of 10 random fields, followed by integrating pixel intensities above a fixed threshold. (C) Schematic representation of ZEN-4 (light gray) and CYK-4 (dark gray) proteins. The CYK-4/ZEN-4 interaction domains are indicated with a dotted line above the illustrations. The plots represent the predictions of the coiled coils of ZEN-4 and CYK-4, by COILS (Lupas et al., 1991) and PAIRCOILS (Berger et al., 1995).
ZEN-4 D520N Is Defective in MT Bundling
Before performing the screen, we wanted to identify the biochemical nature of the defect caused by the D520N substitution mutation. The first question we asked was whether ZEN-4 D520N is defective in microtubule bundling, because the primary function of centralspindlin is to bundle microtubules of the central spindle. We performed a series of in vitro MT bundling assays, comparing ZEN-4(1-603) and ZEN-4(1-603)D520N in complexes with CYK-4(1-232). As shown previously, a complex containing ZEN-4(1-603) and CYK-4(1-232) is sufficient to bundle microtubules (Mishima et al., 2002). In contrast, a complex containing ZEN-4 D520N and CYK-4(1-232) cannot bundle microtubules (Figure 1B). This assay requires complex formation between CYK-4 and ZEN-4, because CYK-4 S15L, the product of the temperature-sensitive allele cyk-4(t1689ts), which is defective in ZEN-4 binding, is similarly defective in the in vitro MT bundling assay (Mishima et al., 2002). These data indicate that the phenotype of zen-4(or153ts) is likely to be directly linked to its inability to bundle microtubules.
ZEN-4 D520N Is a Dimer
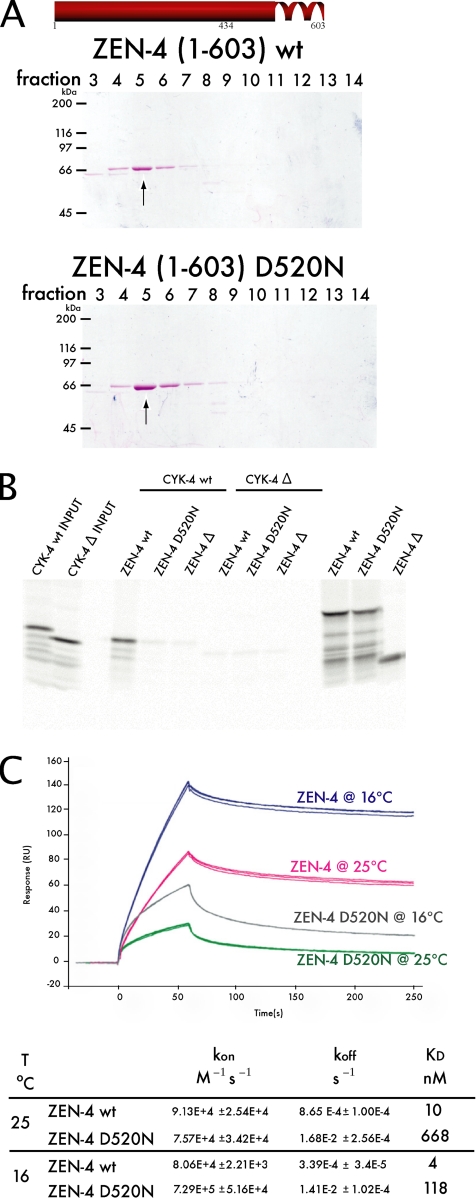
In previous studies, binding of CYK-4 and ZEN-4 was not separable from ZEN-4 dimerization (Mishima et al., 2002). Because D520N maps within the previously predicted coiled coil domain (as predicted by COILS; Lupas et al., 1991), we tested whether ZEN-4 D520N is a dimer or a monomer (Figure 1C). Therefore, we compared the behavior of ZEN-4(1-603), a dimer, ZEN-4(1-603)D520N, and ZEN-4(1-434), which is a monomer, on size exclusion chromatography. We found that ZEN-4(1-603)D520N elutes in the same fraction as ZEN-4(1-603), indicating that D520N does not prevent dimerization (Figure 2A). Thus, the primary defect in zen-4(or153ts) embryos is not due to the failure of ZEN-4 to form dimers.
Figure 2.
The primary defect of zen-4(or153ts) is due to the failure of ZEN-4 D520N to form a stable complex with CYK-4. (A) The ability of ZEN-4 D520N to form a dimer was tested by size exclusion chromatography. ZEN-4(1-603)D520N eluted in the same fraction as wt ZEN-4(1-603), thus ZEN-4 D520N is a dimer. (B) CYK-4(1-120) and ΔCYK-4(34-120) fragments were expressed as 35S-labeled proteins by in vitro translation. Also shown (far right lanes) are parallel reactions of ZEN-4 translation reactions with [35S]methionine, as controls for the efficiency of translation. Unlabeled ZEN-4(434-775) wt and D520N fragments, and the negative control ΔZEN-4(475-775), were added and incubated for 1 h. ZEN-4 was subsequently immunoprecipitated with an anti-ZEN-4 antibody. Whereas wt ZEN-4 can bind wt CYK-4, ZEN-4 D520N cannot. Therefore, ZEN-4 D520N is defective in CYK-4 binding, yet is a dimer (A). (C) The affinity and the rates of interaction between ZEN-4 and CYK-4 were measured through SPR. Sensorgrams of triplicate runs representing 100 nM ZEN-4 injected are shown. The affinity of ZEN-4 D520N for CYK-4 is reduced by 60-fold from wt ZEN-4/CYK-4 affinity at 25°C. However, at 16°C the mutant's affinity for CYK-4 improves.
ZEN-4 D520N Is Defective in CYK-4 Binding
Thus far, we have established that ZEN-4 D520N is defective in MT bundling, yet it is still able to self-associate into dimers. Given this, we turned our attention to the ability of the mutant ZEN-4 to bind CYK-4, because D520N maps near the previously determined ZEN-4/CYK-4 binding interface. We compared the CYK-4 binding activity of ZEN-4(434-775) and ZEN-4(434-775)D520N. Whereas ZEN-4(434-775) interacts strongly with CYK-4, ZEN-4(434-775)D520N does not (Figure 2B). To gain further insight into how structure affects the function of centralspindlin, we measured the binding affinities of wild-type and mutant centralspindlin complex, by using surface plasmon resonance (SPR) technology. CYK-4(1-120) was immobilized on the chip matrix, and ZEN-4(1-603) flowed free in solution. We measured binding kinetics at 25°C [the nonpermissive temperature for zen-4(or153ts)] and at 16°C (the permissive temperature). The affinity of wild-type (wt) ZEN-4 for CYK-4 was 10 nM at 25°C, compared with ZEN-4 D520N that had an affinity of 668 nM for CYK-4 (Figure 2C). At the permissive temperature, ZEN-4 D520N bound CYK-4 with an affinity of 118 nM, revealing that the interaction of this ZEN-4 variant and CYK-4 is temperature-sensitive in vitro. This result strongly indicates that the destabilization of the complex formation between ZEN-4 D520N and CYK-4 is the primary defect in zen-4(or153ts) animals.
Isolation of zen-4(or153ts) Suppressors
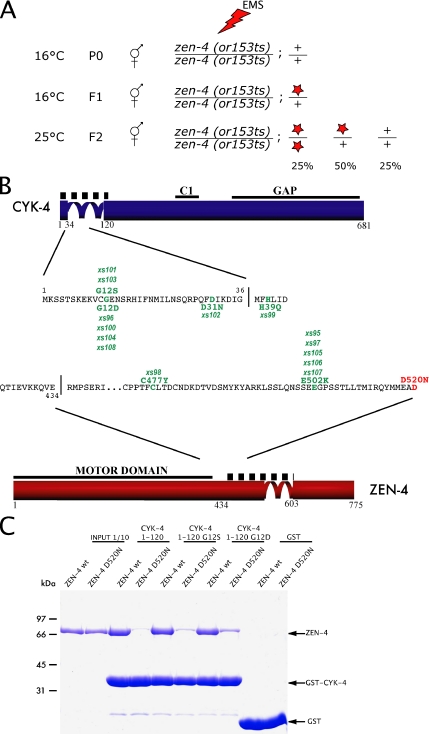
To identify mutations in the C. elegans genome that are capable of restoring viability to zen-4(or153ts) animals, we mutagenized large numbers of zen-4(or153ts) animals and propagated them for two generations at the permissive temperature (16°C). When the F2 animals reached adulthood, the plates were shifted to 25°C to impose a selection for fertile animals carrying putative suppressor mutations (Figure 3A). Fourteen independent suppressors were identified. Before performing crosses to map the putative suppressors, we sequenced the ZEN-4 and CYK-4 loci from PCR products amplified from the suppressor strains to determine whether any of the suppressors contain intragenic mutations in zen-4 or extragenic mutations in cyk-4, as a related screen using cyk-4(t1689) resulted in many such mutations (Mishima et al., 2002). Surprisingly, each of the suppressor mutants contained a single missense mutation in either the CYK-4 interaction region of ZEN-4 or the ZEN-4 interaction region of CYK-4 (Figure 3B). The use of EMS as a mutagen likely prevented the isolation of simple revertants of the ZEN-4 D520N point mutation, because it induces G/C-A/T transitions (Anderson, 1995). Several criteria suggest that these mutations were responsible for the genetic suppression rather than bystander mutations from the mutagenesis. First, each strain contained a single substitution in either cyk-4 or zen-4; second, all mutations were substitution mutations, no silent mutations or mutations in introns were identified; and third, all of the mutations fell near the region previously implicated in complex formation. As evidence that the screen was extensive, three of six individual mutations were independently isolated multiple times. The data from this screen provide further proof that the interaction between ZEN-4 and CYK-4 is fundamental for the in vivo function of centralspindlin. Interestingly, two mutations, D31N in CYK-4 and E502K within ZEN-4, were found as suppressors of both cyk-4(t1689ts) and zen-4(or153ts). Because we have determined that the primary defect in both of these alleles is a defect in complex formation, we expect that these mutations may stabilize the complex. To directly test this conjecture, we assessed whether some of the extragenic suppressor mutations in cyk-4 that rescue zen-4(or153ts) restore binding to ZEN-4 D520N. Indeed, the binding of ZEN-4 D520N to CYK-4 G12S and CYK-4 G12D was significantly improved compared with wild-type CYK-4 (Figure 3C). Interestingly, the binding of these variants of CYK-4 to wild-type ZEN-4 was unimpaired (Figure 3C).
Figure 3.
Isolation of zen-4(or153ts) suppressors. (A) Schematic view of the zen-4(or153ts) suppressor screen. Homozygous zen-4(or153ts) animals were mutagenized at 16°C. After recovery, the F1 embryos were isolated by bleaching and allowed to self-fertilize at permissive temperature (16°C). When F2 animals reached adulthood, the population was shifted to 25°C and fertile animals were selected. Star depicts a suppressor mutation. (B) Approximately 500,000 F1 genomes were screened and 14 suppressor mutations (point mutations indicated in green) were isolated, all of which were viable at 25°C. Some mutations were isolated multiple times. The D520N mutation of zen-4(or153ts) is indicated in red. (C) CYK-4 fragments purified as GST fusions, and retained on the glutathione Sepharose beads, were used in a standard pull-down assay. Recombinant wild type or D520N ZEN-4(1-585) were added to the beads in a 100-μl reaction, and incubated for 2 h at 25°C. The beads were subsequently washed and the bound fraction was analyzed by SDS-PAGE and Coomassie Blue staining. CYK-4 fragments containing the suppressor substitutions (G12S and G12D) bound to both ZEN-4 D520N and wild-type ZEN-4, whereas wild-type CYK-4 could only bind to wild-type ZEN-4. The negative control, GST, did not bind either form of ZEN-4.
ZEN-4 Dimerization Is Separable from CYK-4 Binding
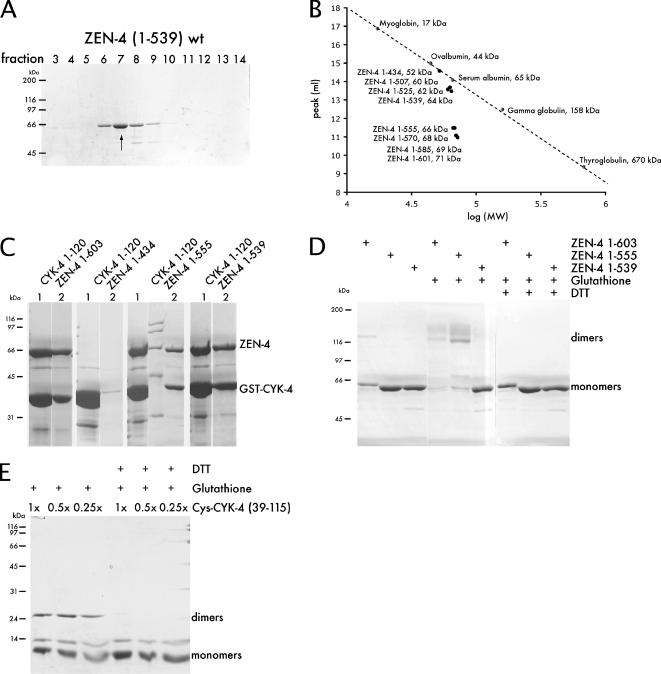
The results from this screen prompted a reevaluation of the coiled coil boundaries within ZEN-4. In previous studies we used the COILS algorithm to predict that the ZEN-4 coiled coil began at residue 508 (Lupas et al., 1991). The finding that D520N impairs complex formation yet retains the ability to dimerize suggests that parts of the coiled coil may be directly involved in CYK-4 binding, or possibly the prediction of the coiled coil domain could have been incorrect. To test this latter possibility, we constructed a series of deletion alleles of ZEN-4 and examined their oligomeric state by performing gel filtration chromatography. We found that ZEN-4(1-539), lacking the predicted coiled coil region, eluted significantly later from a gel filtration column compared with ZEN-4(1-555), which behaved similarly to ZEN-4(1-603), which is a dimer (Figures 2A and 4A). ZEN-4(1-539) and smaller truncations eluted from the column at the position predicted for a monomeric globular protein (Figure 4B). Interestingly, the PAIRCOILS algorithm predicts that the coiled coil starts at residue 537, rather than at residue 507 as predicted by COILS (Lupas et al., 1991; Berger et al., 1995) (Figure 1C). These data suggest that the coiled coil is likely to start at approximately position 540, and this places the critical residue, D520, within the noncoiled coil CYK-4 interaction domain.
Figure 4.
CYK-4 and ZEN-4 form parallel coiled coil dimers, yet dimerization is not strictly required for their interaction. (A) Purified fragments of ZEN-4(1-603) and (1-539) were subjected to size exclusion chromatography (refer to Figure 2A, because these samples were analyzed in the same experiment). ZEN-4(1-539) eluted in a much earlier fraction than ZEN-4(1-603), thus it behaves as a monomer. (B) Summary of size exclusion chromatography with a panel of ZEN-4 deletion derivatives. The peak of the elution fractions of ZEN-4 derivatives are plotted versus log (MW) and compared with standards. (C) CBD ZEN-4 fragments (1-539), (1-555), and (1-603) and GST CYK-4(1-120) were coexpressed from a bicistronic vector. GST-CYK-4 was purified on glutathione Sepharose beads and eluted with reduced glutathione, followed by incubation of the eluates with chitin beads and elution with TEV protease. Both monomeric ZEN-4(1-539) and dimeric ZEN-4(1-555 or 1-603) copurified with CYK-4. (D) Recombinant ZEN-4 fragments containing a cysteine residue at position 547 were incubated under oxidizing conditions. The reaction was quenched and the ability of ZEN-4 to form disulfide bridges was analyzed. ZEN-4(603) as well as ZEN-4(1-555) efficiently formed dimers, whereas ZEN-4(1-539) did not. Thus, ZEN-4 forms a parallel coiled coil dimer. (E) CYK-4 preparations (36-119 with an N-terminal cysteine shown here), were incubated under oxidizing conditions. The reaction was quenched and the ability of CYK-4 to form disulfide bridges was analyzed. CYK-4 also forms a parallel coiled coil dimer.
We next sought to test whether the monomeric form of ZEN-4, ZEN-4(1-539), is competent to bind to CYK-4. We therefore coexpressed GST-CYK-4 in a bicistronic operon with CBD-ZEN-4 in E. coli, and performed sequential affinity chromatography steps. Monomeric ZEN-4(1-539) copurified with stoichiometric amounts of CYK-4 as was observed with dimeric ZEN-4(1-555) or ZEN-4(1-601) (Figure 4C). Although monomeric ZEN-4 binds to CYK-4 with lower affinity compared with dimeric ZEN-4 (see below), these data establish that ZEN-4 dimerization is not strictly essential for CYK-4 binding.
CYK-4 and ZEN-4 Are Parallel Coiled Coil Dimers
The arrangement of the individual subunits within the native centralspindlin complex is a feature that may govern mechanistic attributes of central spindle assembly, such as the mechanism of bundling antiparallel microtubules. We therefore wanted to determine whether the coiled coils in ZEN-4 and CYK-4 are in a parallel or an antiparallel configuration. The fact that ZEN-4(1-555) is a dimer indicates that the coiled coil in ZEN-4 is in a parallel configuration, because this truncation roughly bisects the coiled coil region and were it in an antiparallel configuration, and in register, this truncation would eliminate the overlap. To independently address the question, we used disulfide bridge formation under oxidizing conditions followed by nonreducing gel electrophoresis to assay for dimer formation and thereby infer the arrangement of the coiled coils. ZEN-4 contains a cysteine at position 547 that is predicted to be in the “a” register (i.e., internal) of the coiled coil. If the helices in the ZEN-4 dimer are antiparallel, there is only a low probability that cysteine 547 of one monomer would be sufficiently close to cysteine 547 of the adjacent monomer to allow disulfide bridge formation under oxidizing conditions (Falke et al., 1988), whereas if they are in a parallel configuration these cysteine residues would be predicted to pack against each other. Under oxidizing conditions, a substantial fraction of ZEN-4(1-603) and ZEN-4(1-555) form covalent dimers, yet ZEN-4(1-539) does not, suggesting that the coiled coil is in a parallel configuration (Figure 4D). This was confirmed by engineering a derivative, ZEN-4(1-556)C547VGGC, that contains a Gly-Gly-Cys extension at the C terminus and assaying for dimer formation (Oakley and Kim, 1998). This ZEN-4 derivative also forms covalent dimers under oxidizing conditions, whereas, ZEN-4(1-539)GGC does not form covalent dimers under oxidizing conditions (Supplemental Figure S1). We conclude that the coiled coil of ZEN-4 is in a parallel configuration.
We used a similar methodology to assess the orientation of the CYK-4 coiled coil. The region of CYK-4 predicted to form a coiled coil comprises residues 36-120. We engineered a series of expression constructs of the CYK-4 coiled coil region, and we added a cysteine in the predicted “b”-position at the C terminus or in the “c”-orientation at the N terminus. CYK-4 derivatives containing either a C-terminal cysteine, an N-terminal cysteine, or a cysteine at both the N and C termini were assayed for disulfide bridge formation under oxidizing conditions. All the CYK-4 derivatives, C39-115, 39-115C, and C39-115C, formed covalent dimers under oxidizing conditions (Figure 4E; data not shown). In addition, the efficiency of dimer formation was independent of protein concentration over at least a fourfold range (Figure 4E), providing further evidence for specificity. These data establish that CYK-4 dimerizes via a parallel coiled coil. Thus, both CYK-4 and ZEN-4 form parallel coiled coil dimers.
Organization and Function of the Coiled Coil
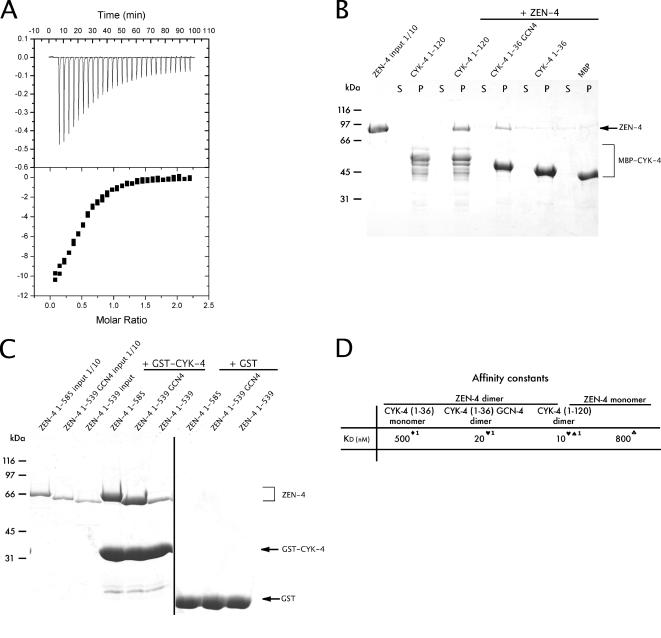
To further probe the relationship between the structure and function of centralspindlin, we wanted to assess whether and how the dimerization of each component contributes to complex formation. Because our genetic and biochemical data establish that a high-affinity centralspindlin complex is required in vivo, we wanted to determine how coiled coil-mediated dimerization of individual subunits contributes to the overall stability of the centralspindlin complex. To measure the affinity constants of monomeric ZEN-4(1-539) for dimeric CYK-4(1-120), we used isothermal titration calorimetry (ITC). This method was chosen instead of SPR, because it measures binding of proteins free in solution and would therefore eliminate avidity affects that can be induced by immobilization of ligands on the chip matrix used for SPR experiments. We measured an 800 nM dissociation constant for monomeric ZEN-4(1-539) for dimeric CYK-4(1-120) (Figure 5A), or ∼80-fold weaker than the affinity of dimeric ZEN-4(1-603) for dimeric CYK-4(1-120). Thus, dimerization of ZEN-4 greatly impacts the overall stability of centralspindlin.
Figure 5.
Coiled coils in CYK-4 and ZEN-4 play a stabilizing role in centralspindlin complex formation. (A) Binding of monomeric ZEN-4(1-539) to dimeric CYK-4(1-120) was measured using ITC. Measurements were performed at 25°C using the working buffer as a reference. The protein concentration was 10 μM, and the ligand concentration (ZEN-4) was 100 μM. Using a simple one-site model, the affinity of monomeric ZEN-4 for dimeric CYK-4 was calculated at 800 nM. (B) CYK-4 fragments purified as MBP fusions and retained on the amylose beads were used in a standard pull-down assay. Recombinant ZEN-4(1-585) GFP was added to the beads in a 100-μl reaction, and it was incubated at room temperature for 2 h. The beads were subsequently washed, and the bound fraction was analyzed by SDS-PAGE and Coomassie Blue staining. CYK-4 containing a coiled coil, able to form dimers, could bind ZEN-4, whereas CYK-4 lacking a coiled coil could not, nor could the negative control, MBP. S, supernatant; P, pellet. (C) CYK-4 fragments purified as GST fusions and retained on the glutathione Sepharose beads were used in a standard pull-down assay. Recombinant ZEN-4(1-585), ZEN-4(1-539) GCN4, and ZEN-4(1-539) were added to the beads in a 100-μl reaction, and they were incubated at room temperature for 2 h. The beads were subsequently washed, and the bound fraction was analyzed by SDS-PAGE and Coomassie Blue staining. ZEN-4 containing a coiled coil, either native or engineered, could bind CYK-4, whereas ZEN-4 lacking a coiled coil could not. The negative control, GST, did not bind any of the ZEN-4 fragments. (D) Comparison of binding affinities between monomeric and dimeric ZEN-4/CYK-4 constructs. Symbols indicate the method by which the binding constant was determined: ♣, ITC; ♦, pull-down assay; ♥, pull-down assay with fluorescence intensity measured; ♠, SPR; and 1, approximate KD.
We next investigated the extent to which dimerization of CYK-4 affects centralspindlin stability. We constructed a MBP fusion of CYK-4(1-36), which lacks the coiled coil, CYK-4(1-120), and CYK-4(1-36)GCN4, which contains an exogenous parallel coiled coil (O'Shea et al., 1991). The latter fusion protein was generated to assess whether the coiled coil contributes solely through dimerization. In semiquantitative binding experiments with 1 μM ZEN-4(1-585)GFP and 0.5 μM CYK-4 derivatives, we found that CYK-4(1-120) bound well to ZEN-4 as did CYK-4(1-36)GCN4, but the construct lacking a coiled coil did not bind significantly more ZEN-4 than the negative control, MBP (Figure 5B). To obtain quantitative measures of binding affinities, we performed pull-down assays after incubation with ZEN-4(1-585)GFP and measured the fluorescence remaining in the supernatant. As seen previously using SPR, these experiments show that CYK-4(1-120) and ZEN-4(1-585) form a complex with a dissociation constant of ∼10 nM (Figure 5D). Interestingly, the affinity of the CYK-4(1-36)GCN4 induced dimer was only twofold lower that wild-type CYK-4 dimer. The binding affinity of monomeric CYK-4(1-36) for ZEN-4(1-585)GFP was outside the range of this assay, therefore titration binding assays with CYK-4 monomer constructs were performed. CYK-4(1-36) at 1 μM was incubated with increasing concentrations of ZEN-4(1-585)GFP, ranging from 0 to 3 μM, followed by a pull-down assay. Monomeric CYK-4(1-36) bound to ZEN-4(1-585) with a dissociation constant of ∼500 nM, or 50-fold higher than of the CYK-4 dimer for the ZEN-4 dimer. Thus, the CYK-4 coiled coil domain contributes significantly to complex formation. However, because the CYK-4 coiled coil can be effectively substituted by an exogenous dimerization domain, it is not likely to directly participate in binding; rather, dimerization may primarily serve to increase the avidity of the complex. To extend these findings, we used a similar methodology to assess the effect of the ZEN-4 dimerization on centralspindlin stability. We constructed CBD fusions of ZEN-4(1-585), ZEN-4(1-539), which lacks the coiled coil, and ZEN-4(1-539)GCN4, which contains an exogenous parallel coiled coil. As in the aforementioned scenario, ZEN-4(1-539)GCN4 showed significantly improved binding to CYK-4 compared with monomeric ZEN-4(1-539) (Figure 5C).
DISCUSSION
The centralspindlin complex plays myriad roles in cytokinesis, including bundling antiparallel microtubules, regulating furrow formation, and promoting abscission (Powers et al., 1998; Raich et al., 1998; Jantsch-Plunger et al., 2000; Gromley et al., 2005; Yuce et al., 2005; Werner et al., 2007). We have used a combination of biochemistry and genetics to more precisely define the interactions between CYK-4 and ZEN-4 and to establish the molecular organization of this complex that lies at the nexus of cytokinesis. Our results indicate that low-affinity interactions serve as the interface between centralspindlin subunits. However, each subunit of centralspindlin contributes two such interfaces that are linked through a parallel coiled coil. This results in the cooperative assembly of a stable, high-affinity heterotetramer. These observations refine our understanding of this critical complex and provide insight into how it bundles microtubules.
Because centralspindlin localizes to overlapping antiparallel microtubules, and it is required for their bundling (Jantsch-Plunger et al., 2000), it is of particular interest to establish how this complex specifically recognizes microtubules arranged in this configuration. One possibility is that the complex is organized in an antiparallel configuration, as is thought to be the case for the tetrameric form of the Eg5 kinesin complex that plays an analogous role, stabilizing the bipolar mitotic spindle before anaphase. Therefore, we used multiple approaches to establish the orientation of the coiled coils in CYK-4 and ZEN-4. First, we used a deletion approach. If a protein is arranged in a parallel coiled coil, it could retain its ability to dimerize when the coiled coil region is truncated by greater than half, whereas an antiparallel coiled coil would not dimerize if the coiled coil region were bisected. We found that both CYK-4 and ZEN-4 could dimerize even when the regions predicted to form the coiled coil were reduced to less than half their original lengths. To unambiguously demonstrate that CYK-4 and ZEN-4 each form parallel coiled coils, we assessed cross-linking between either a native cysteine close to one end of the coiled, between an engineered cysteine residue at the extreme ends of the predicted coiled coils, or both, and we demonstrated that these cysteine residues could efficiently form a disulfide bridge (Oakley and Kim, 1998). Formation of a disulfide bridge requires that the two sulfhydryl groups be within 0.7 nm, which is unlikely to occur unless the proteins formed a parallel coiled coil in register (Falke et al., 1988). Furthermore, in the case of CYK-4 we found that the efficiency of dimer formation was independent of protein concentration, suggesting that the molecules exist as dimers in their native state, as expected for a coiled coil dimer. Finally, the parallel nature of the CYK-4 coiled coil is substantiated by the fact that CYK-4(1-120) and CYK-4(1-36)GCN4, which contains a well characterized parallel coiled coil (O'Shea et al., 1991), bind to ZEN-4 with near equal affinity. Similarly, the improved CYK-4 binding of ZEN-4(1-539)GCN4 relative to ZEN-4(1-539), further confirms the parallel nature of the ZEN-4 coiled coil.
In previous work, we defined ZEN-4 (434-603) as the minimal region capable of binding to CYK-4 (Mishima et al., 2002). This region contained a coiled-coil region shown to mediate homodimerization. In this current work, using more sensitive assays, we have refined the CYK-4 binding region of ZEN-4 to (434-539). Significantly, this separates two biochemical functions of ZEN-4, which previously seemed linked: dimerization and CYK-4 binding. We have shown, however, that monomeric ZEN-4 binds to CYK-4 dimer ∼80× less well compared with dimeric ZEN-4. Likewise, we previously demonstrated that the minimal ZEN-4 binding region of CYK-4 includes residues 1-120. Using stringent assays, we were previously unable to detect an interaction between the N-terminal 35 amino acids of CYK-4 and ZEN-4. However, here we have used less stringent assays, and we show that CYK-4 (1-36) can in fact bind ZEN-4, but with significantly lower affinity (∼0.5 μM). Furthermore, the high-affinity binding of CYK-4(1-36)GCN4 to ZEN-4 demonstrates that the difference in affinity between these two CYK-4 derivatives can largely be ascribed to the loss of dimerization as opposed to loss of specific binding interfaces.
In an effort to further understand the molecular interactions involved in central spindle assembly, we performed a genome-wide screen for mutations that ameliorate the temperature sensitivity of the zen-4(or153ts) allele. Although this allele contained two substitution mutations, we determined that the defect was due to just one of the two substitutions, D520N. Previous predictions suggested that residue 520 lies within the coiled coil region, and it seemed surprising that a conservative change would disrupt coiled coil formation. We therefore directly defined the regions of the protein that are sufficient to form a stable coiled coil. The data presented here suggest that D520 likely lies outside of the coiled coil region. Indeed, our experimental results fit well with the prediction of the PAIRCOILS algorithm (Berger et al., 1995).
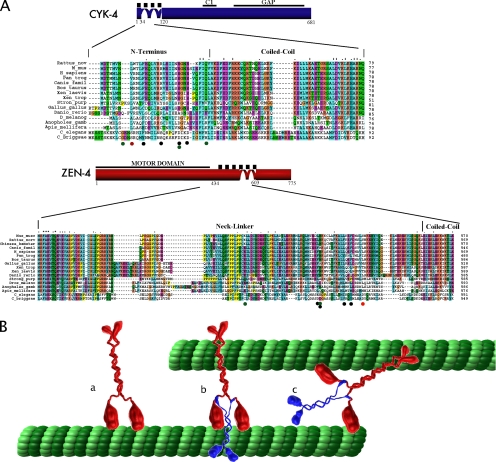
The only mutations we found that could suppress the zen-4(or153ts) allele were located within the ZEN-4/CYK-4 interface. This is similar to our previous characterization of suppressors of cyk-4(t1689ts). The combined data from these two screens reveal 12 substitutions that restore the interaction between CYK-4 and ZEN-4 when it is perturbed by either of these substitution alleles. Interestingly, some substitution mutations were found to suppress both the cyk-4 temperature-sensitive mutation and the zen-4 temperature-sensitive mutation, suggesting that some of these suppressor mutations may not accommodate the altered amino acid, but rather strengthen the interaction overall. Our genetic and biochemical rescue of ZEN-4 D520N by mutations in CYK-4 strengthen this argument and further substantiate that the interaction between CYK-4 and ZEN-4 is critical for the function of centralspindlin in cytokinesis. Strikingly, of the two substitutions within CYK-4, which rescued complex formation, the G12D allele more effectively restored binding to ZEN-4 D520N than did the G12S substitution. The former substitution was identified four times in the screen, whereas the allele that more weakly restores binding was only isolated twice. Additionally, the D31N allele, which was only isolated once, did not detectably restore binding under the conditions tested (data not shown). Additionally, the suppressor mutations all fall within the biochemically defined minimal interacting regions in CYK-4 and ZEN-4 corresponding to 1-36 in CYK-4 and 434-539 ZEN-4. It is notable that two independent screens for temperature-sensitive, maternal-effect lethal mutations both identified mutations that disrupt the CYK-4/ZEN-4 binding interface and not other sites within either gene. Furthermore, multiple substitutions can suppress each of these defects, suggesting remarkable plasticity in the CYK-4/ZEN-4 binding interface. These findings are particularly intriguing in light of the facts that these regions are not strongly conserved during evolution, yet there is direct evidence that the orthologous proteins interact in C. elegans, Drosophila, and human cells (Mishima et al., 2002; Somers and Saint, 2003). Presumably, these regions interact in all metazoan species, despite the poor conservation at the level of primary sequence (Figure 6A). It will be of significant interest to obtain high-resolution structural information for this interaction interface and the refinement of the minimal interaction domain reported here should facilitate these future studies.
Figure 6.
Centralspindlin: functional insights from structural aspects of the complex. (A) Sequence comparison of the CYK-4/ZEN-4 interaction domains among vertebrates and invertebrates. The original temperature-sensitive mutations are indicated by red dots; suppressors of zen-4(or153ts) are indicated by green dots; and suppressors of cyk-4(t1689ts) are indicated by black dots (Mishima et al., 2002). The accession numbers for the aligned sequences are as follows: ZEN-4/MKLP1[C_elegans (NP_741411.1); C_briggsae (emb CAE61601.1); H_sapiens (AAH51826.1); Danio_rerio (AAF00594.1); Strong_purp (AAG18582.1); Dros_melano (CAA12181.1); Mus_musc (NP_077207); Canis_famil (XP_535528.2); Chinese_hamster (CAA58558.1); Xen_trop (NP_001011104.1); Gallus_gallus (NP_001025725.1); Anopheles_gamb (XP_308170.2); Apis_mellifera (XP_624886.1); Xen_laevis (AAH84928.1); Tetraodon_nigriv (CAF99883.1); Rattus_norv (XP_236313.3); Bos_taurus (XP_593824.2)]; CYK-4/MgcRacGAP [C_Briggsae (CAE71360.1); D_melanog (CAB96203.1); C_elegans (NP_499845.1); M_mus (NP_036155.1); H_sapiens (NP_037409.2); Rattus_nov (XP_235650.2); Canis_famil (XP_543675.2); Bos_taurus (XP_592496.2); Xen_laevis (AAH70771.1); Xen_trop (NP_001001236.1); Tetraodon_nigrov (CAG03964.1); Gallus_gallus (XP_424490.1); Stron_purp (XP_783360.1); Danio_rerio (AAH95799.1); Pan_trog (XP_509531.1); Apis_mellifera (XP_393627.2); Anopholes_gamb (XP_319660.2)]. (B) Schematic representation of ZEN-4 (a) and the centralspindlin complex (b). Model of possible mode of action of the complex, and the structural effects that CYK-4 may have on ZEN-4 (c).
The data presented here indicates that a rather weak (∼0.5 μM KD) interaction connects a monomer of the RhoGAP CYK-4 to the monomer of the kinesin motor protein ZEN-4. However, because both CYK-4 and ZEN-4 individually assemble into parallel coiled coil dimers, the heterotetrameric form of centralspindlin is a highly stable (10 nM KD) complex. In view of the results presented here, and assuming that centralspindlin forms a symmetrical complex, we propose the working model depicted in Figure 6B and Supplemental Movie 1. In vivo, bundling of microtubules in the central spindle requires that CYK-4 and ZEN-4 bind each other with high affinity. At this juncture, we do not yet know whether CYK-4 directly participates in microtubule bundling or whether it allosterically modulates the structure and biochemical properties of ZEN-4. Future biophysical analysis of centralspindlin will be necessary to answer this important, open question. The work presented here provides a rich context for future structural studies of centralspindlin and its components.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Kaitna for help in the initial stages of this study, Mohammed Yousef and Elena Solomaha for assistance at the Biophysics Core Facility, and Georg Kaltenbrunner for artistic contribution to the figures. This project was supported by National Institutes of Health grant R0I GM-074743. M.M. was supported by Cancer Research UK program grant C19769/A6356.
Abbreviations used:
- GAP
GTPase-activating protein
- ITC
isothermal titration calorimetry
- MAP
microtubule-associated protein
- MT
microtubule
- SPR
surface plasmon resonance
- wt
wild type.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-05-0468) on October 17, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Anderson P. Mutagenesis. Methods Cell Biol. 1995;48:31–58. [PubMed] [Google Scholar]
- Berger B., Wilson D. B., Wolf E., Tonchev T., Milla M., Kim P. S. Predicting coiled coils by use of pairwise residue correlations. Proc. Natl. Acad. Sci. USA. 1995;92:8259–8263. doi: 10.1073/pnas.92.18.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H., Hyman A. A. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436:731–734. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- Dechant R., Glotzer M. Centrosome separation and central spindle assembly act in redundant pathways that regulate microtubule density and trigger cleavage furrow formation. Dev. Cell. 2003;4:333–344. doi: 10.1016/s1534-5807(03)00057-1. [DOI] [PubMed] [Google Scholar]
- Desai A., Walczak C. E. Assays for microtubule-destabilizing kinesins. Methods Mol. Biol. 2001;164:109–121. doi: 10.1385/1-59259-069-1:109. [DOI] [PubMed] [Google Scholar]
- Falke J. J., Dernburg A. F., Sternberg D. A., Zalkin N., Milligan D. L., Koshland D.E.J. Structure of a bacterial sensory receptor. A site-directed sulfhydryl study. J. Biol. Chem. 1988;263:14850–14858. [PubMed] [Google Scholar]
- Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- Gromley A., Yeaman C., Rosa J., Redick S., Chen C. T., Mirabelle S., Guha M., Sillibourne J., Doxsey S. J. Centriolin anchoring of exocyst and SNARE complexes at the midbody is required for secretory-vesicle-mediated abscission. Cell. 2005;123:75–87. doi: 10.1016/j.cell.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger V., Gönczy P., Romano A., Schnabel H., Hamill D., Schnabel R., Hyman A. A., Glotzer M. CYK-4, A Rho family GTPase activating protein (GAP) required for central spindle formation and cytokinesis. J. Cell Biol. 2000;149:1391–1404. doi: 10.1083/jcb.149.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitna S., Mendoza M., Jantsch-Plunger V., Glotzer M. Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr. Biol. 2000;10:1172–1181. doi: 10.1016/s0960-9822(00)00721-1. [DOI] [PubMed] [Google Scholar]
- Kamijo K., Ohara N., Abe M., Uchimura T., Hosoya H., Lee J. S., Miki T. Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol. Biol. Cell. 2006;17:43–55. doi: 10.1091/mbc.E05-06-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurasawa Y., Earnshaw W. C., Mochizuki Y., Dohmae N., Todokoro K. Essential roles of KIF4 and its binding partner PRC1 in organized central spindle midzone formation. EMBO J. 2004;23:3237–3248. doi: 10.1038/sj.emboj.7600347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A., Van Dyke M., Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mello C., Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Mishima M., Kaitna S., Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev. Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- Mishima M., Pavicic V., Gruneberg U., Nigg E. A., Glotzer M. Cell cycle regulation of central spindle assembly. Nature. 2004;430:908–913. doi: 10.1038/nature02767. [DOI] [PubMed] [Google Scholar]
- Mollinari C., Kleman J. P., Jiang W., Schoehn G., Hunter T., Margolis R. L. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J. Cell Biol. 2002;157:1175–1186. doi: 10.1083/jcb.200111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C., Kleman J. P., Saoudi Y., Jablonski S. A., Perard J., Yen T. J., Margolis R. L. Ablation of PRC1 by small interfering RNA demonstrates that cytokinetic abscission requires a central spindle bundle in mammalian cells, whereas completion of furrowing does not. Mol. Biol. Cell. 2005;16:1043–1055. doi: 10.1091/mbc.E04-04-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura Y., Yonemura S. Centralspindlin regulates ECT2 and RhoA accumulation at the equatorial cortex during cytokinesis. J. Cell Sci. 2006;119:104–114. doi: 10.1242/jcs.02737. [DOI] [PubMed] [Google Scholar]
- O'Shea E. K., Klemm J. D., Kim P. S., Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- Oakley M. G., Kim P. S. A buried polar interaction can direct the relative orientation of helices in a coiled coil. Biochemistry. 1998;37:12603–12610. doi: 10.1021/bi981269m. [DOI] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science. 2003;302:2120–2124. doi: 10.1126/science.1091936. [DOI] [PubMed] [Google Scholar]
- Powers J., Bossinger O., Rose D., Strome S., Saxton W. A nematode kinesin required for cleavage furrow advancement. Curr. Biol. 1998;8:1133–1136. doi: 10.1016/s0960-9822(98)70470-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich W. B., Moran A. N., Rothman J. H., Hardin J. Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol. Biol. Cell. 1998;9:2037–2049. doi: 10.1091/mbc.9.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L. B., Schneider T. R., Stukenberg P. T., Musacchio A. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- Severson A. F., Hamill D. R., Carter J. C., Schumacher J., Bowerman B. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr. Biol. 2000;10:1162–1171. doi: 10.1016/s0960-9822(00)00715-6. [DOI] [PubMed] [Google Scholar]
- Somers W. G., Saint R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev. Cell. 2003;4:29–39. doi: 10.1016/s1534-5807(02)00402-1. [DOI] [PubMed] [Google Scholar]
- Toure A., Dorseuil O., Morin L., Timmons P., Jegou B., Reibel L., Gacon G. MgcRacGAP, a new human GTPase-activating protein for Rac and Cdc42 similar to Drosophila rotundRacGAP gene product, is expressed in male germ cells. J. Biol. Chem. 1998;273:6019–6023. doi: 10.1074/jbc.273.11.6019. [DOI] [PubMed] [Google Scholar]
- Verbrugghe K. J., White J. G. SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr. Biol. 2004;14:1755–1760. doi: 10.1016/j.cub.2004.09.055. [DOI] [PubMed] [Google Scholar]
- Verni F., Somma M. P., Gunsalus K. C., Bonaccorsi S., Belloni G., Goldberg M. L., Gatti M. Feo, the Drosophila homolog of PRC1, is required for central-spindle formation and cytokinesis. Curr. Biol. 2004;14:1569–1575. doi: 10.1016/j.cub.2004.08.054. [DOI] [PubMed] [Google Scholar]
- Werner M., Munro E., Glotzer M. Astral signals spatially bias cortical Myosin recruitment to break symmetry and promote cytokinesis. Curr. Biol. 2007;17:1286–1297. doi: 10.1016/j.cub.2007.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley S. P., Wang Y. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J. Cell Biol. 1996;135:981–989. doi: 10.1083/jcb.135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuce O., Piekny A., Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J. Cell Biol. 2005;170:571–582. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W. M., Fang G. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc. Natl. Acad. Sci. USA. 2005;102:13158–13163. doi: 10.1073/pnas.0504145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.