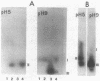
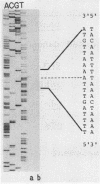
Abstract
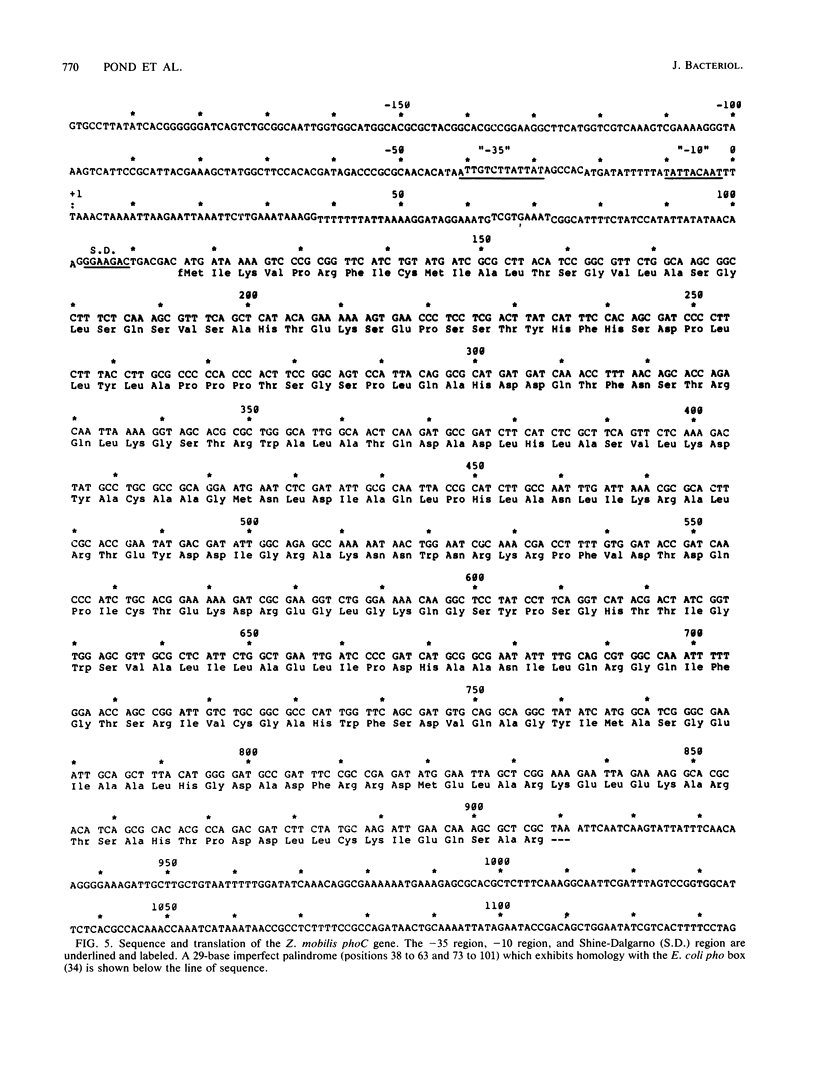
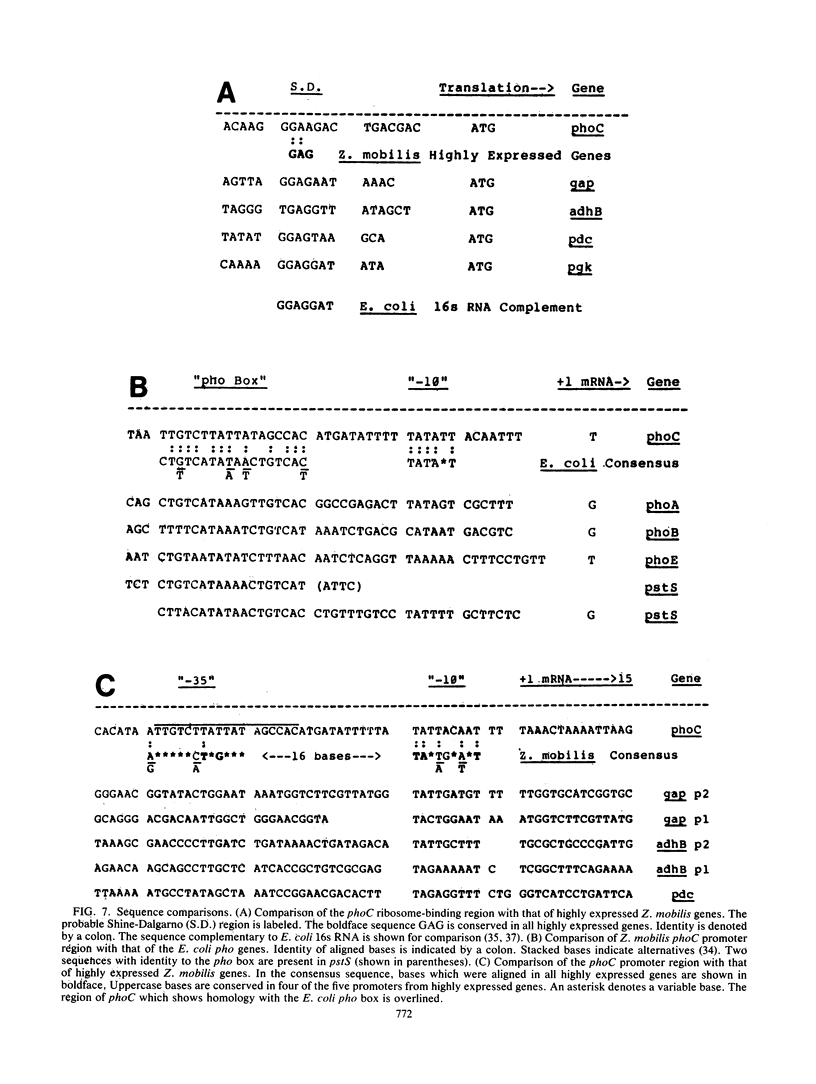
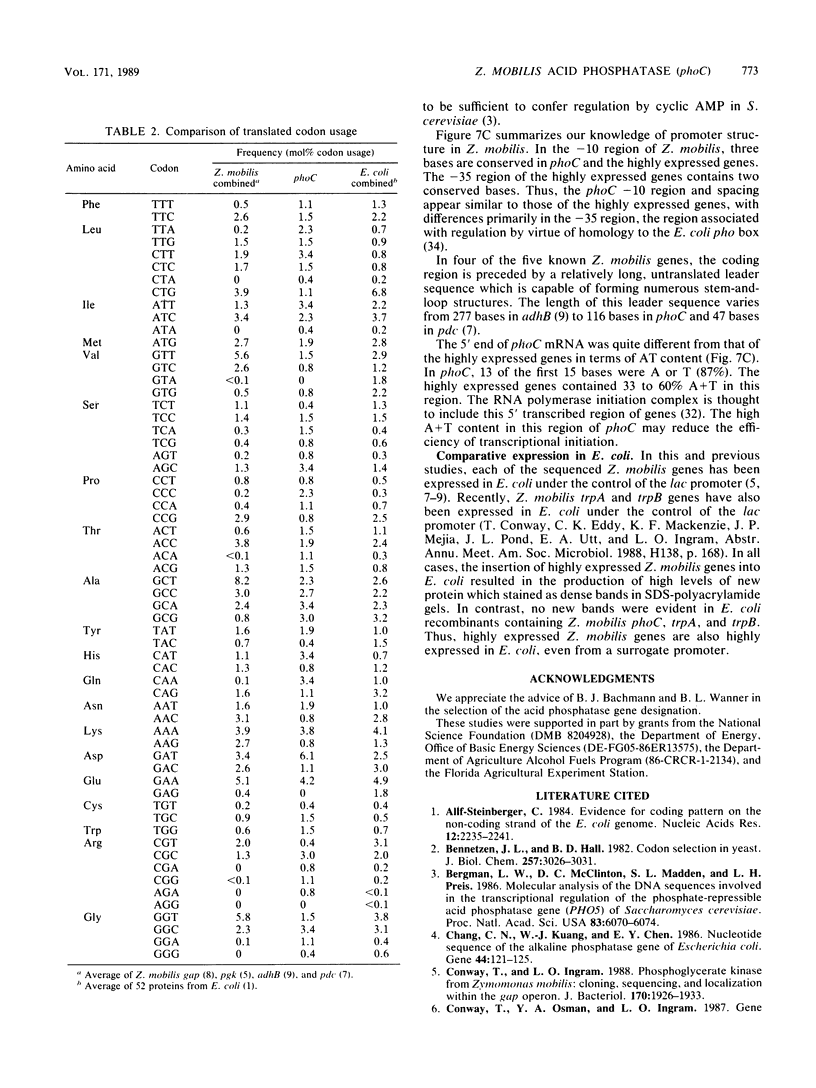
The Zymomonas mobilis gene encoding acid phosphatase, phoC, has been cloned and sequenced. The gene spans 792 base pairs and encodes an Mr 28,988 polypeptide. This protein was identified as the principal acid phosphatase activity in Z. mobilis by using zymograms and was more active with magnesium ions than with zinc ions. Its promoter region was similar to the -35 "pho box" region of the Escherichia coli pho genes as well as the regulatory sequences for Saccharomyces cerevisiae acid phosphatase (PHO5). A comparison of the gene structure of phoC with that of highly expressed Z. mobilis genes revealed that promoters for all genes were similar in degree of conservation of spacing and identity with the proposed Z. mobilis consensus sequence in the -10 region. The phoC gene contained a 5' transcribed terminus which was AT rich, a weak ribosome-binding site, and less biased codon usage than the highly expressed Z. mobilis genes.
Full text
PDF
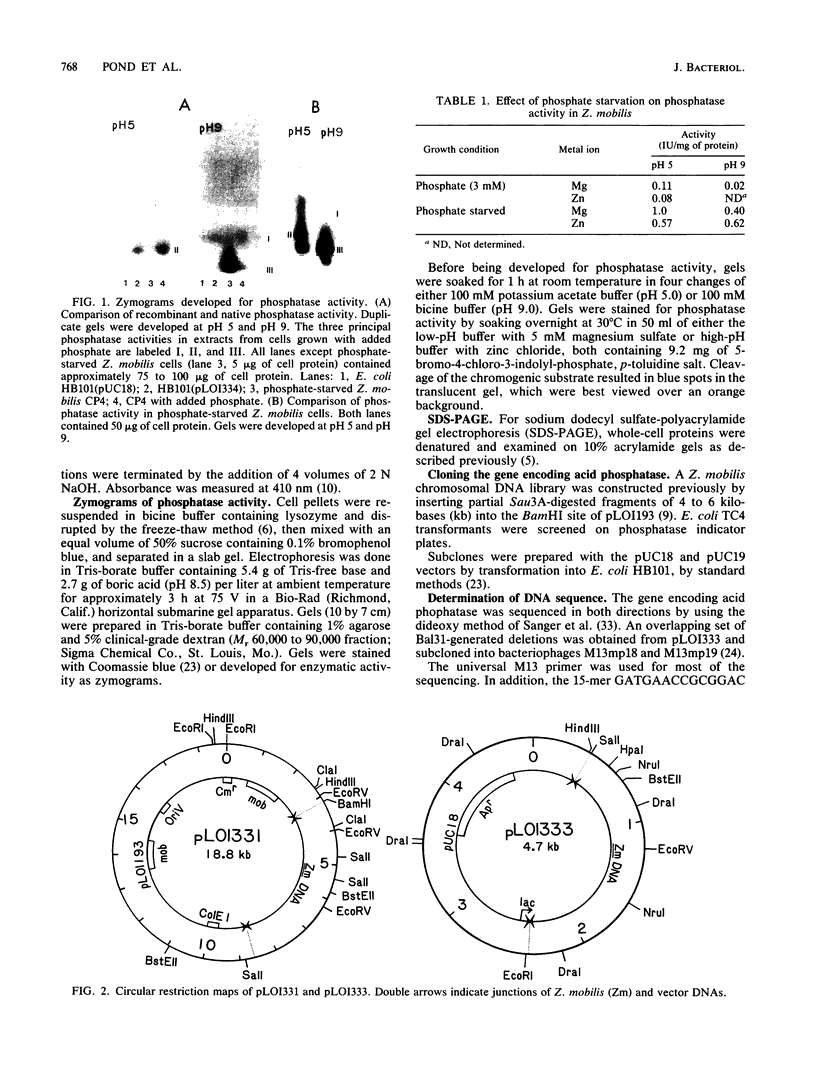
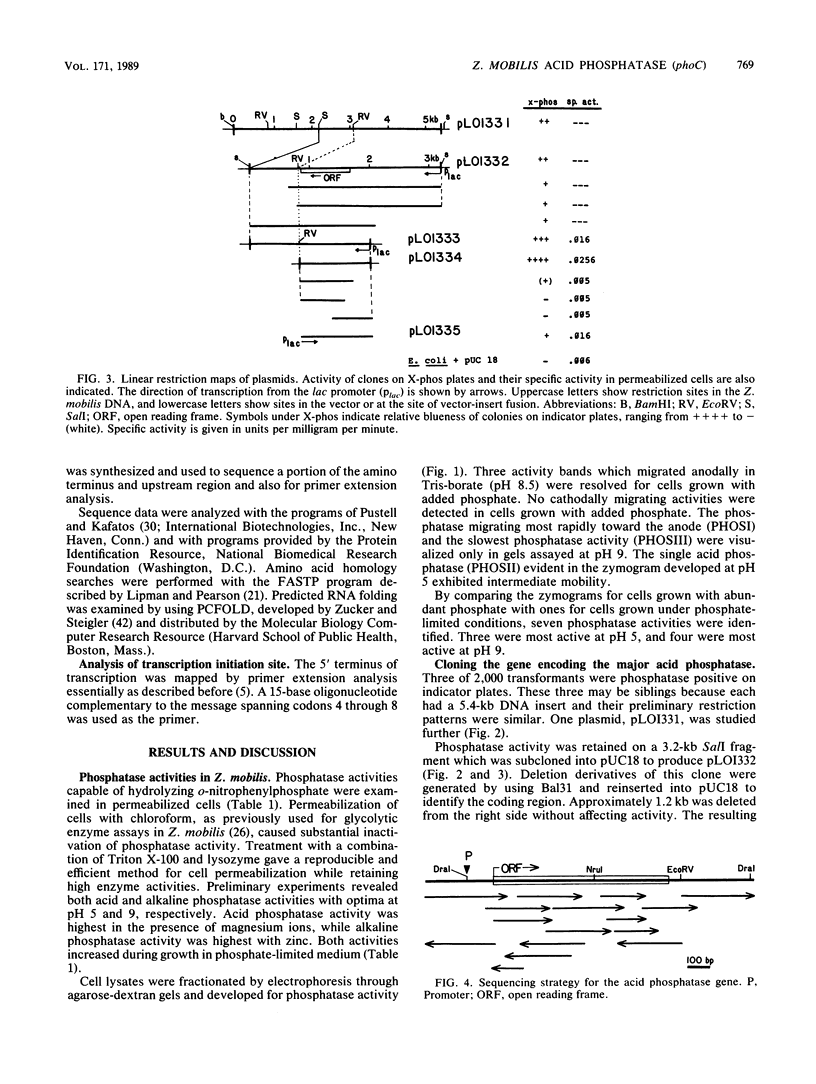


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alff-Steinberger C. Evidence for a coding pattern on the non-coding strand of the E. coli genome. Nucleic Acids Res. 1984 Mar 12;12(5):2235–2241. doi: 10.1093/nar/12.5.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bergman L. W., McClinton D. C., Madden S. L., Preis L. H. Molecular analysis of the DNA sequences involved in the transcriptional regulation of the phosphate-repressible acid phosphatase gene (PHO5) of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1986 Aug;83(16):6070–6074. doi: 10.1073/pnas.83.16.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. N., Kuang W. J., Chen E. Y. Nucleotide sequence of the alkaline phosphatase gene of Escherichia coli. Gene. 1986;44(1):121–125. doi: 10.1016/0378-1119(86)90050-8. [DOI] [PubMed] [Google Scholar]
- Conway T., Ingram L. O. Phosphoglycerate kinase gene from Zymomonas mobilis: cloning, sequencing, and localization within the gap operon. J Bacteriol. 1988 Apr;170(4):1926–1933. doi: 10.1128/jb.170.4.1926-1933.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Osman Y. A., Konnan J. I., Hoffmann E. M., Ingram L. O. Promoter and nucleotide sequences of the Zymomonas mobilis pyruvate decarboxylase. J Bacteriol. 1987 Mar;169(3):949–954. doi: 10.1128/jb.169.3.949-954.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Ingram L. O. Glyceraldehyde-3-phosphate dehydrogenase gene from Zymomonas mobilis: cloning, sequencing, and identification of promoter region. J Bacteriol. 1987 Dec;169(12):5653–5662. doi: 10.1128/jb.169.12.5653-5662.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway T., Sewell G. W., Osman Y. A., Ingram L. O. Cloning and sequencing of the alcohol dehydrogenase II gene from Zymomonas mobilis. J Bacteriol. 1987 Jun;169(6):2591–2597. doi: 10.1128/jb.169.6.2591-2597.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox G. B., Rosenberg H., Downie J. A., Silver S. Genetic analysis of mutants affected in the Pst inorganic phosphate transport system. J Bacteriol. 1981 Oct;148(1):1–9. doi: 10.1128/jb.148.1.1-9.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak H. F., Brockman R. W., Heppel L. A. Purification and properties of two acid phosphatase fractions isolated from osmotic shock fluid of Escherichia coli. Biochemistry. 1967 Jun;6(6):1743–1751. doi: 10.1021/bi00858a024. [DOI] [PubMed] [Google Scholar]
- Elliott S., Chang C. W., Schweingruber M. E., Schaller J., Rickli E. E., Carbon J. Isolation and characterization of the structural gene for secreted acid phosphatase from Schizosaccharomyces pombe. J Biol Chem. 1986 Feb 25;261(6):2936–2941. [PubMed] [Google Scholar]
- Hoekema A., Kastelein R. A., Vasser M., de Boer H. A. Codon replacement in the PGK1 gene of Saccharomyces cerevisiae: experimental approach to study the role of biased codon usage in gene expression. Mol Cell Biol. 1987 Aug;7(8):2914–2924. doi: 10.1128/mcb.7.8.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981 Sep 25;151(3):389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982 Jul 15;158(4):573–597. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- Kam W., Clauser E., Kim Y. S., Kan Y. W., Rutter W. J. Cloning, sequencing, and chromosomal localization of human term placental alkaline phosphatase cDNA. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8715–8719. doi: 10.1073/pnas.82.24.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y., Hayashi N., Toh-e A., Banno I., Oshima Y. Structural characteristics of the PHO8 gene encoding repressible alkaline phosphatase in Saccharomyces cerevisiae. Gene. 1987;58(1):137–148. doi: 10.1016/0378-1119(87)90036-9. [DOI] [PubMed] [Google Scholar]
- Kendall D. A., Bock S. C., Kaiser E. T. Idealization of the hydrophobic segment of the alkaline phosphatase signal peptide. Nature. 1986 Jun 12;321(6071):706–708. doi: 10.1038/321706a0. [DOI] [PubMed] [Google Scholar]
- Lipman D. J., Pearson W. R. Rapid and sensitive protein similarity searches. Science. 1985 Mar 22;227(4693):1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Osman Y. A., Conway T., Bonetti S. J., Ingram L. O. Glycolytic flux in Zymomonas mobilis: enzyme and metabolite levels during batch fermentation. J Bacteriol. 1987 Aug;169(8):3726–3736. doi: 10.1128/jb.169.8.3726-3736.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A., Scopes R. K., Griffiths-Smith K. Isolation and properties of the glycolytic enzymes from Zymomonas mobilis. The five enzymes from glyceraldehyde-3-phosphate dehydrogenase through to pyruvate kinase. Biochem J. 1986 Aug 15;238(1):275–281. doi: 10.1042/bj2380275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Pustell J., Kafatos F. C. A convenient and adaptable package of computer programs for DNA and protein sequence management, analysis and homology determination. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):643–655. doi: 10.1093/nar/12.1part2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Export of protein in bacteria. Microbiol Rev. 1984 Dec;48(4):290–298. doi: 10.1128/mr.48.4.290-298.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu R. S., Bollon A. P. Analysis of alpha-factor secretion signals by fusing with acid phosphatase of yeast. Gene. 1987;54(2-3):175–184. doi: 10.1016/0378-1119(87)90485-9. [DOI] [PubMed] [Google Scholar]
- Terao M., Mintz B. Cloning and characterization of a cDNA coding for mouse placental alkaline phosphatase. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7051–7055. doi: 10.1073/pnas.84.20.7051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touati E., Danchin A. The structure of the promoter and amino terminal region of the pH 2.5 acid phosphatase structural gene (appA) of E. coli: a negative control of transcription mediated by cyclic AMP. Biochimie. 1987 Mar;69(3):215–221. doi: 10.1016/0300-9084(87)90045-9. [DOI] [PubMed] [Google Scholar]
- Wanner B. L., Wilmes M. R., Young D. C. Control of bacterial alkaline phosphatase synthesis and variation in an Escherichia coli K-12 phoR mutant by adenyl cyclase, the cyclic AMP receptor protein, and the phoM operon. J Bacteriol. 1988 Mar;170(3):1092–1102. doi: 10.1128/jb.170.3.1092-1102.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]