Abstract
Cytosolic 5′nucleotidase II (cN-II) catalyses both the hydrolysis of a number of nucleoside monophosphates (e.g., IMP + H2O→inosine + Pi), and the phosphate transfer from a nucleoside monophosphate donor to the 5′position of a nucleoside acceptor (e.g., IMP + guanosine →inosine + GMP). The enzyme protein functions through the formation of a covalent phosphoenzyme intermediate, followed by the phosphate transfer either to water (phosphatase activity) or to a nucleoside (phosphotransferase activity). It has been proposed that cN-II regulates the intracellular concentration of IMP and GMP and the production of uric acid. The enzyme might also have a potential therapeutic importance, since it can phosphorylate some anti-tumoral and antiviral nucleoside analogues that are not substrates of known kinases. In this review we summarise our recent studies on the structure, regulation and function of cN-II. Via a site-directed mutagenesis approach, we have identified the amino acids involved in the catalytic mechanism and proposed a structural model of the active site. A series of in vitro studies suggests that cN-II might contribute to the regulation of 5-phosphoribosyl-1-pyrophosphate (PRPP) level, through the so-called oxypurine cycle, and in the production of intracellular adenosine, formed by ATP degradation.
Key words: IMP, GMP, oxypurine cycle, phosphotransferase, purine analogues, purine salvage, site direct mutagenesis, 5′nucleotidase
Introduction
Cytosolic 5′-nucleotidase II (cN-II) belongs to a family of proteins catalysing the hydrolysis of intracellular nucleotides. Members of this family differ widely in cellular location, substrate specificity, regulation and amino acid sequence [1]. cN-II has been described as a bifunctional enzyme, since it catalyses both the hydrolysis of IMP and GMP and the transfer of phosphate group from a mononucleotide donor to a nucleoside acceptor such as inosine, guanosine, deoxyinosine and several nucleoside analogues in use as antiviral or antineoplastic purine prodrugs [2, 3]. Furthermore, it seems to be responsible of the resistance to several purine derivative drugs. Therefore, its activity has been indicated as predictive of patient survival in acute myeloid leukaemia [4, 5]. The bovine recombinant enzyme presents a 99% amino acid identity with human cN-II [6]. In previous work we have shown that the enzyme catalyses the hydrolase/phosphotransferase reaction through the formation of a covalent enzyme-phosphate intermediate and have described the complex regulation of the enzyme activity, depending on intracellular energy charge [7, 8]. Here, we report our more recent results obtained on the structure, function, and role of this intriguing enzyme. By a site-directed mutagenesis approach we have identified the amino acid residues involved in the catalytic mechanism and proposed a structural model of the active site [9]. In vitro results suggest that cN-II might be involved also in the regulation of PRPP (5-phosphoribosyl-1-pyrophosphate) concentration [10] and in the production of intracellular adenosine, formed by ATP degradation [11].
Enzyme distribution, purification and kinetics
The existence of cytosolic 5′-nucleotidase was first reported in the liver of some vertebrates such as chicken, rat, pig and frog. Subsequently, its existence in chicken heart, rat heart and kidney, pig lung and several human tissues, such as malignant lymphocytes, placenta, erythrocytes, lymphoblasts and colon carcinoma, has been assessed by its isolation from these tissues [12]. In mammals the activities, revealed by immunotitration to eliminate the interference of non-specific phosphatases and other nucleotidases, are high in testis and spleen and low in skeletal muscle and erythrocytes; in human tissues, specific activity is higher in fibroblasts and granulocytes than in erythrocytes. Furthermore, T and B lymphoblastoid cells display a higher activity than do peripheral lymphocytes [13]. The cN-II gene is located on chromosome 10 [4], and its mRNA is ubiquitously expressed in human tissues. The specific activity of cytosolic 5′-nucleotidase is significantly increased in human colon carcinoma extracts compared to the activity measured in the apparently normal peritumoral tissue from the same patients; this confirms the previous observation that the enzyme is more expressed in cells with active DNA synthesis and/or high turnover rate of nucleic acids and their precursors [14]. cN-II has been purified from many eukaryotic sources. The various enzymes exhibit similar kinetic characteristics, substrate specificity and regulation, despite the large differences reported in the molecular mass [13]. The apparent molecular mass of the enzyme estimated by gel filtration ranges from 165 kDa for the Artemia enzyme [15] to 265 kDa for the pig lung enzyme purified in the presence of protease inhibitors [16], whereas the subunit molecular masses ranges from 51 kDA to 62 kDa, indicating a tetrametric structure for 5-N. The enzyme is specific for IMP (Km 0.1 mM), GMP and their corresponding deoxy-derivatives. It is activated by ATP, ADP, 2,3-bis-phosphoglycerate, decavanadate and, to a smaller extent, by other nucleoside diphosphates and triphosphates (Table 1). The enzyme activity depends on adenylate energy charge and phosphate concentration. The inhibition by inorganic phosphate (Pi) is counteracted by ATP at physiological concentrations and, to a smaller extent, by ADP [3, 6, 7]. In 1982 Worku and Newby [17] proposed that the hydrolysis of the monophosphate proceeds via the formation of an enzyme-phosphate intermediate, as for many phosphate-transferring enzymes; this hypothesis was based on kinetic evidence. The formation of a phosphorylated intermediate was later demonstrated directly by trapping the 32P-labelled phosphoenzyme, followed by electrophoresis and autoradiography [7]. The best phosphate acceptor in the phosphotransferase reaction is inosine, with a Km of approximately 1 mM. The Km values of calf thymus cN-II for both inosine and IMP were unaffected by the presence of activators, while Vmax was increased approximately 10 times. The inhibitor Pi causes a Km increase of about 4 times for both substrates.
Table 1.
Structural and functional characteristics of cN-II (SDS sodium dodecyl sulphate, BPG bis-phosphoglycerate).
| Distribution | Ubiquitous |
| MW subunit | |
| (SDS electrophoresis) | 512–62 kDa |
| (predicted from mRNA) | 66 kDa |
| Quaternary structure | Tetramer |
| Substrates | |
| IMP | Km 0.1 mM |
| Inosine | Km 1 mM |
| Activators: ATP, ADP, BPG | At millimolar concentrations |
| Inhibitor: Pi | At millimolar concentrations |
Enzyme structure
The expression of the cloned bovine cDNA led to a recombinant protein with functional properties identical to the protein purified from calf thymus. The coding nucleotide sequences reveals a very high degree of similarity, 94%, to the coding region of the human cN-II [9]. The recombinant bovine enzyme expressed and purified in E. coli, being identical to human protein for functional and regulatory properties and being an abundant source of cN-II, has been selected as a source for our structural, regulatory and functional studies. cN-II is the first member of the cytosolic 5′-nucleotidases whose reaction mechanism has been elucidated [6, 7]. During catalysis, this enzyme was demonstrated to become phosphorylated on the first aspartate of its DMDYT sequence. A similar motif DXDX (T/V) (motif I) is present in all members of the HAD (l-2-haloacid dehalogenase) superfamily, where the nucleophilic attack of this aspartate is essential for the catalytic machinery [9, 18, 19]. P-type ATPase/phosphotransferase members of the HAD superfamily share a similar structural fold and a common reaction mechanism, which requires the formation of a covalent enzyme-phosphate intermediate. Furthermore, crystallographic and site-directed mutagenesis studies on these proteins demonstrated that a series of other common amino acids always occurs in their active site, thus determining the occurrence of two additional sequence motifs common to all members of the HAD family. The first one (motif II) is characterised by a threonine/serine residue included in a hydrophobic region; the second one (motif III) presents a conserved lysine and a pair of aspartic acid residues (Table 2). Recently, the resolution of the crystal structure of mdN, a dimeric mitochondrial nucleotidase specific for deoxynucleotides, has been reported, demonstrating this enzyme as the first example of a 5′- nucleotidase belonging to the HAD superfamily [20]. On this basis, a large number of proteins differing in catalytic activity on various substrates, polypeptide length (from 200 to 1,400 amino acids), domain arrangement, oligomerisation and conformational change following ligand binding, have been related to the HAD/P-type ATPases/phosphotransferases superfold [21]. However, no structural data on cN-II are available at the moment.
Table 2.
Amino acid sequence of the cN-II three motifs involved in the active site. Amino acid residues that have been mutated are indicated in bold type.
| Motif | Amino acid sequence |
|---|---|
| Motif I | 51 DMDYTL |
| Motif II | 244 VFLATNS |
| Motif III | 291K.....347 YIGDHIFGDIL |
To verify the predicted role for the conserved residues present in the three motifs of cN-II, 13 mutated cN-II products were constructed and expressed. Extracts were prepared 16 h after the addition of isopropylthiogalactoside, and recombinant wild type and cN-II mutants were purified. In all cases sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis showed a single component, migrating with an apparent mass of 60 kDa. Purified proteins were used in kinetics measurements.
Both conservative and non-conservative substitution of D52 and D54 (motif I) completely abolished enzyme activity and formation of cN-II-phosphate intermediate [6]. Replacement at other positions of this motif strongly affected cN-II function. In fact, substitution at position 56 (mutant T56R) resulted in a protein devoid of phosphatase and phosphotransferase activities. In contrast, mutagenesis of M53 had a less severe effect; however, while nonconservative substitutions (M53N) caused only a strong decrease in catalytic efficiency, conservative substitution (M53I) determined effects both on enzyme activity and affinity toward IMP. This latter mutant exhibited the lowest phosphatase vs phosphotransferase activity ratio observed so far; it also showed a sigmoidal dependence on Mg2+ concentration. These results indicate the essential role of these residues for proper D52 and D54 orientation and effective cN-II catalysis, confirming the function of motif I as deduced by previous chemical labelling and site-directed mutagenesis experiments.
The conserved amino acid present in motif II common to all HAD superfamily members is always a serine or threonine residue; this amino acid is important for a proper orientation of the substrate within the active site through specific hydrogen bonding. The sequence alignment of HAD superfamily members indicated the T249 as an essential residue for cN-II activity. However, another amino acid (S251) with similar properties occurs closely in motif II. In order to identify the residue present in motif II unambiguously, we prepared conservative and non-conservative mutants of both amino acids. Mutant T249V showed strongly reduced enzyme activity and an alteration of Km values for both substrates. In contrast, a conservative substitution (mutant T249S) yielded kinetic behaviour more similar to wild type enzyme. On the other hand, the effect on enzyme activity exerted by nonconservative mutation of residue 251 (mutant S251A) was by far less pronounced than that produced by the conservative S251T mutation. These results demonstrate the essential role of the T249 hydroxyl group for cN-II catalysis, confirming the nature of motif II deduced by multiple sequence alignments.
Motif III in the HAD superfamily is characterised by the presence of a conserved lysine residue and two negatively charged residues that are involved in the stabilisation of the negatively charged reaction intermediate and metal ion coordination, respectively. As expected on the basis of the proposed alignment, mutation of K292 (mutant K292R and K292M) strongly affected cN-II activity. Similarly, mutation of two aspartate residues (D351 and D356) resulted in very poor phosphatase and phosphotransferase activities and significantly reduced affinity towards Mg2+. As expected, conservative mutations (mutant D351E and D356E) had a less pronounced effect than non-conservative mutations. Furthermore, D356N showed a tenfold increase of the Km value for inosine, suggesting a possible role for this amino acid in the interaction with the second substrate. All these data confirmed the nature of the residues present in motif III as deduced by sequence alignment.
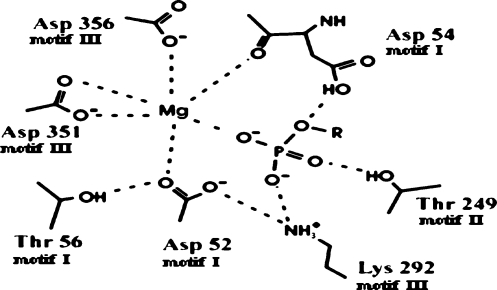
A general comparison of all kinetically characterised mutants showed that the phosphatase vs phosphotransferase activity ratio was significantly altered in two cases: mutant S251T and mutant M53I. In the first case this phenomenon was not justified simply on the basis of the slight alterations of Km value measured for both substrates. In the latter case the observed value for this parameter was in line with the tenfold increase of Km for IMP. A model of the cN-II active site structure, designed on the basis of our results, is presented in Figure 1.
Figure 1.
Schematic representation of the proposed active site of cN-II. Modified from Allegrini et al., 2004 [9]
Physiological aspects of cN-II
The apparent inhibition of cN-II by its product inosine [22] led to the discovery of the exchange of the nucleoside moiety between inosine and IMP and to the proposal that cN-II catalyses the transfer of the phosphate group of IMP to inosine by forming a phosphorylated enzyme as intermediate [17]. Studies on the enzyme-substrate specificity revealed that cN-II may, indeed, utilise IMP as phosphate donor for a number of nucleosides and nucleoside analogues, such as 2′-3′-dideoxyinosine and 8-azaguanosine [3, 23]. The respective importance of hydrolase and phosphotransferase activities of cN-II in cellular metabolism is unclear. The reader is referred to the recent review of Bretronnet et al. [24] for a discussion on this important functional aspect of cN-II. At least in vitro, cN-II mediated xanthosine (Xao) phosphorylation (IMP + Xao → XMP + Ino) appears to be the only possible pathway for salvaging Xao [25]. This reaction can bypass the NAD+-mediated reduction of IMP into XMP, catalysed by IMP dehydrogenase (IMPDH), an enzyme targeted for human antiviral, immunosuppressive, and antineoplastic chemotherapy [26, 27]. cN-II may also enable dIMP formation from IMP as a donor and deoxyinosine as an acceptor of phosphate, followed by conversion of dIMP to dAMP or dGMP. This pathway bypasses the ribonucleotide reductase activity [14].
cN-II is involved in the maintenance of the intracellular PRPP pool through the oxypurine cycle
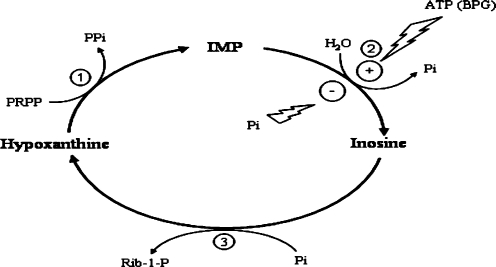
The regulation of PRPP synthesis by PRPP synthetase and its utilisation by the phosphoribosyl transferases are the major factors maintaining the intracellular level of PRPP, a high-energy sugar phosphate, with a high potential of 5-phosphoribosyl transfer [28]. An additional factor is the “oxypurine cycle,” also named the “purine nucleoside cycle” [29] The oxypurine cycle is a cyclic pathway, composed of hypoxanthine-guanine phosphoribosyltransferase (HPRT) nucleoside phosphorylase and the cN-II (Figure 2). Because its net reaction is the hydrolysis of PRPP, the cycle has been proposed to play a role in the metabolic regulation of intracellular PRPP levels [30]. Rat brain cytosol is particularly suitable for investigating the kinetics of the cycle, because neither IMP nor inosine acts as a substrate of ATP-mediated kinases [10, 31], and xanthine oxidase is virtually absent in rat brain [32]. As evidenced in Figure 2, the velocity of the “oxypurine cycle,” and, therefore, the rate of PRPP disappearance and ribose-1-phosphate (Rib 1-P) formation, is modulated by ATP, acting as an activator of cN-II, and by Pi, acting as an inhibitor. (2,3-bis-phosphoglycerate (2,3-BPG) may substitute ATP as an activator.
Figure 2.
The oxypurine cycle. The enzymes participating in the cycle are: 1 HPRT; 2 cN-II; 3 purine nucleoside phosphorylase (PNP). The net reaction is the hydrolysis of 5-PRPP to Rib 1-P and inorganic pyrophosphate (PPi). The allosteric activation by ATP and 2-3-BPG, and the inhibition by Pi, modulate the cycle rate. Modified from Leach et al., 1998 [33]
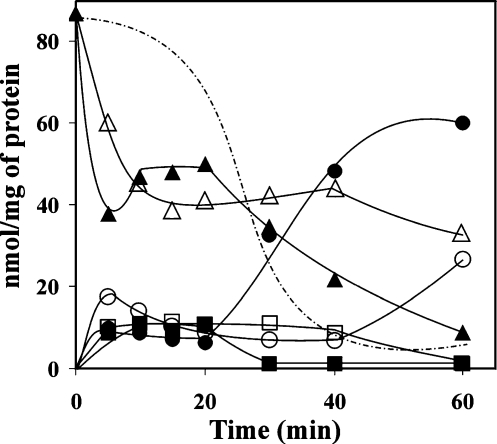
Studies on the kinetics of the in vitro reconstituted cycle and in rat brain cytosolic preparations [10] have shown that the three components, IMP, inosine and hypoxanthine, attain a steady-state level. Strikingly, at the end of this steady state period, whose duration depends on the initial PRPP concentration, hypoxanthine abruptly accumulates, at the expense of IMP degradation (Figure 3). We explain these results as follows. While PRPP is being degraded to inorganic pyrophosphate (PPi) and Rib 1-P, the intermediates are continuously recycled, but, as soon as PRPP disappears, HPRT becomes inactive. Then the cycle “collapses”, causing IMP degradation by cN-II, and hypoxanthine accumulation by purine nucleoside phosphorylase. As a consequence, the duration of the state-state period may be considered as a measure of the rate of the PRPP consumption. The velocity of the cycle, as measured by the rate of PRPP disappearance, responds to protein concentration as well as to the allosteric activators of cN-II and to Pi, the allosteric inhibitor. We conclude that modulation of cN-II might, in turn, regulate the oxypurine nucleoside cycle. It might be speculated that, at normal low [Pi]/[ATP] ratios, as found in well-oxygenated cells, cN-II is fully active, and the velocity of the cycle is maximal. As a consequence the PRPP pool is maintained at a low level, thus avoiding unnecessary salvage or de novo nucleotide synthesis. During ischaemia, the [Pi]/[ATP] ratio rises drastically [33, 34], the situation is reversed, and enough PRPP is made available for purine nucleotide salvage and/or synthesis during reperfusion. This process, which in our opinion represents the real purine salvage, is favoured by the increase in purine base levels occurring in ischaemia [34] and by the increase in PRPP that accompanies massive ATP degradation, as it occurs in rat brain during anoxia-like conditions [35].
Figure 3.
The oxypurine cycle as catalysed by rat brain extract. The reaction mixture contained 100 µM [14C]-inosine, 1 mM PRPP (closed symbols) or 2 mM PRPP (open symbols), 5 mM KH2PO4, 8.3 mM MgCl2, 4.5 mM BPG, 5 mM Tris-HCl buffer, pH 7.4, and a brain cytosolic preparation, 1.15 mg of protein/ml. Circles hypoxanthine, triangles inosine, squares IMP. The dotted line represents the time course of PRPP disappearance at 2 mM initial concentration. During PRPP consumption the three components of the cycle attain a steady-state level whose duration depends on the initial PRPP concentration, suggesting that, during PRPP degradation to PPi and Rib-1-P, the intermediates are continuously recycled. Modified from Barsotti et al., 2003 [10]
A widely accepted tenet is that, under normal conditions, the PRPP pool is maintained at a low level, to avoid excessive and unbalanced nucleotide synthesis. We notice PRPP, the “fuel” of the cycle, is an obligate precursor of purine salvage, while Rib 1-P, one of the cycle products (Figure 2) may be used for uracil salvage and 5-fluorouracil (5-FU) activation, a process involving pyrimidine base (or base analogue) ribosylation, catalysed by uridine phosphorylase, followed by multiple phosphorylation steps [36–39]. The observation that uracil is readily converted into uridine in the presence of PRPP, in the absence of any detectable uracil phosphoribosyltransferase activity and provided that hypoxanthine is present in the reaction mixture, led to the proposal that Rib 1-P needed to ribosylate uracil or 5-FU might well arise from PRPP breakdown occurring in the oxypurine cycle.
Modulation of adenylate deaminase and cN-II affects adenosine production in rat brain
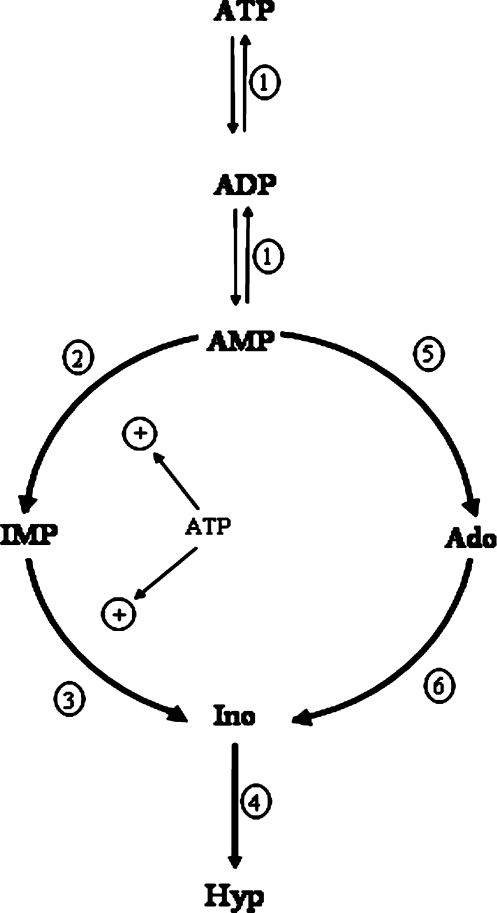
Adenosine is a neuroactive nucleoside and a homoeostatic cellular modulator. Its main source is ATP breakdown [40]. While extracellular ATP breakdown follows a unique pathway, composed of a set of membrane-bound ectonucleotidases, intracellular ATP breakdown follows two distinct routes [11] (Figure 3). In the first route, ATP ↔ ADP ↔ AMP interconversion, catalysed by adenylate kinase, is followed by AMP deamination and cN-II-mediated IMP dephosphorylation (the “IMP pathway”). In the second route, AMP is dephosphorylated by the AMP preferring cytosolic 5′-nucleotidase cN-1A (cytosolic AMP preferring 5′-nucleotidase, form A) to adenosine, and adenosine is deaminated to inosine (the “adenosine pathway”). Recent studies [11] have established that the nature of the ATP catabolic route (i.e., if it follows the IMP or the adenosine pathway) is concentration dependent. Thus, at 3.6 mM (a normoxic concentration), ATP breakdown follows the IMP pathway, with little, if any, adenosine production. Moreover, IMP, but not AMP or adenosine, accumulates in cytosolic preparations of rat brain. This can be explained by the deactivation of cN-II, prompted by the disappearance of ATP, an allosteric activator of the enzyme. (Figure 4). This condition also increases the Km value for the substrate IMP from micromolar to millimolar value [8]. Moreover, the production of the small amount of adenosine after prolonged incubation was clearly preceded by inosine and hypoxanthine formation, thus excluding a precursor-product relationship between adenosine and inosine plus hypoxanthine. Any inosine and hypoxanthine would, therefore, originate from IMP, rather than adenosine. We have proposed that, as far as intracellular ATP is maintained at normoxic concentration, its catabolism follows the IMP pathway, and only a minimal amount of adenosine, if any, is generated in rat brain from ATP. As a consequence, the ratios of the inosine plus hypoxanthine levels to adenosine cannot be considered to indicate the rate of metabolic degradation from adenosine to inosine and hypoxanthine in normoxic conditions. This view is in accordance with the strong activation exerted by physiological ATP concentration on both adenylate deaminase and cN-II [41–43]. It is also supported by the low Km value of adenosine kinase for adenosine, as compared to that of adenosine deaminase. Accordingly, adenosine at 1 µM, a reasonable physiological normoxic concentration, is anabolised mostly to AMP, ADP and ATP, rather than catabolised to inosine. Only at higher concentrations is adenosine catabolised into inosine and hypoxanthine [11].
Figure 4.
Pathways of intracellular ATP catabolism in rat brain. The enzymes participating in these pathways are: 1 adenylate kinase, 2 adenylate deaminase, 3 cytosolic IMP-GMP specific 5′-nucleotidase (cN-II), 4 purine nucleoside phosphorylase, 5 cytosolic AMP specific 5′nucleotidase, 6 adenosine deaminase. Enzymes 2 and 3 are activated by physiological ATP concentrations. Modified from Barsotti and Ipata, 2004 [11]
At an initial concentration of 0.5 mM, AMP immediately and transiently accumulates, followed by an accumulation of adenosine, inosine and hypoxanthine. This suggests a precursor-product product relationship between AMP and adenosine, inosine and hypoxanthine and explains the apparently paradoxical observation that more adenosine is produced at 0.5 mM than at 3.6 mM initial ATP concentration [11]. Hydrolysis by S-adenosylmethionine hydrolase, another possible source of adenosine, does not significantly contribute to adenosine production in rat brain [44, 45].
Abbreviations
- cN-1A
cytosolic AMP preferring 5′nucleotidase, form A
- cN-II
cytosolic 5′nucleotidase II
- HAD
L-2-haloacid dehalogenase
- HPRT
hypoxanthine-guanine phosphoribosyltransferase
- IMPDH
IMP dehydrogenase
- Ino
inosine
- PNP
purine nucleoside phosphorylase
- PRPP
5-phosphoribosyl-1-pyrophosphate
- Rib 1-P
ribose-1-phosphate
- Xao
xanthosine
- 2,3-BPG
2,3-bis-phosphoglycerate
- 5-FU
5-fluorouracil
References
- 1.Bianchi V, Spychala J (2003) Mammalian 5′nucleotidases. J Biol Chem 278:46195′6198 [DOI] [PubMed]
- 2.Johnson MA, Fridland A (1989) Phosphorylation of 2′3′dideoxyinosine by cytosolic 5′nucleotidase of human lymphoid cells. Mol Pharmacol 36:291′95 [PubMed]
- 3.Banditelli S, Baiocchi C, Pesi R et al (1996) The phosphotransferase activity of cytosolic 5′nucleotidase, a purine analog phosphorylating enzyme. Int J Biochem Cell Biol 28:711′20 [DOI] [PubMed]
- 4.Galmarini CM, Jordheim L, Dumontet C (2003) Role of IMP-selective 5′nucleotidase (cN-II) in hematological malignancies. Leuk Lymphoma 44:1105′111 [DOI] [PubMed]
- 5.Galmarini CM, Thomas X, Graham K et al (2003) Deoxycytidine kinase and cN-II nucleotidase expression in blast cells predict survival in acute myeloid leukaemia patients treated with cytarabine. Br J Haematol 122:53′0 [DOI] [PubMed]
- 6.Allegrini S, Scaloni A, Ferrara L et al (2001) Bovine cytosolic 5′nucleotidase acts through the formation of an aspartate 52-phosphoenzyme intermediate. J Biol Chem 276:33526′3532 [DOI] [PubMed]
- 7.Baiocchi C, Pesi R, Camici M et al (1996) Mechanism of the reaction catalysed by cytosolic 5′nucleotidase/phosphotransferase: formation of a phosphorylated intermediate. Biochem J 317:797′01 [DOI] [PMC free article] [PubMed]
- 8.Pesi R, Turriani M, Allegrini S, Scolozzi C et al (1994) The bifunctional cytosolic 5′nucleotidase: regulation of the phosphotransferase and nucleotidase activities. Arch Biochem Biophys 312:75′0 [DOI] [PubMed]
- 9.Allegrini S, Scaloni, Careddu MG et al (2004) Mechanistic studies on bovine cytosolic 5′nucleotidase II, an enzyme belonging to the HAD superfamily. Eur J Biochem 271:4881′891 [DOI] [PubMed]
- 10.Barsotti C, Pesi R, Felice F et al (2003) The purine nucleoside cycle in cell-free extracts of rat brain: evidence for the occurrence of an inosine and a guanosine cycle with distinct metabolic roles. Cell Mol Life Sci 60:786′93 [DOI] [PMC free article] [PubMed]
- 11.Barsotti C, Ipata PL (2004) Metabolic regulation of ATP breakdown and of adenosine production in rat brain extracts. Int J Biochem Cell Biol 36:2214′225 [DOI] [PubMed]
- 12.Itoh R (1993) IMP/GMP 5′nucleotidase. Comp Biochem Physiol 105B:13′9 [DOI] [PubMed]
- 13.Itoh R, Yamada K (1991) Determination of cytoplasmic 5′nucleotidase which preferentially hydrolyses 6-hydroxypurine nucleotides in pig, rat and human tissues by immunotitration. Int J Biochem 23:461′65 [DOI] [PubMed]
- 14.Tozzi MG, Camici M, Pesi R et al (1991) Nucleoside phosphotransferase activity of human colon carcinoma cytosolic 5′nucleotidase. Arch Biochem Biophys 291:212′17 [DOI] [PubMed]
- 15.Pinto RM, Canales J, Faraldo A et al (1987) 5′Nucleotidase from Artemia embryos. Purification and properties. Comp Biochem Physiol 86B:49′3 [DOI] [PubMed]
- 16.Itoh R, Yamada K (1990) Pig lung 5′nucleotidase: effect of diadenosine 5′5′″P1, P4-tetraphosphate and its related compounds. Int J Biochem 22:231′38 [DOI] [PubMed]
- 17.Worku Y, Newby AC (1982) Nucleoside exchange catalysed by the cytoplasmic 5′nucleotidase. Biochem J 205:503′10 [DOI] [PMC free article] [PubMed]
- 18.Collet JF, Stroobant V, Pirard M et al (1998) A new class of phosphotransferase phosphorylated on an aspartate residue in an amino-terminal DXDX (T/V) motif. J Biol Chem 273:14107′4112 [DOI] [PubMed]
- 19.Aravind L, Galperin MY, Koonin EV (1998) The catalytic domain of the P-type ATPase has the haloacid dehalogenase fold. Trends Biol Sci 23:127′29 [DOI] [PubMed]
- 20.Rinaldo-Matthis A, Rampazzo C, Reichard P et al (2002) Crystal structure of a human mitochondrial deoxyribonucleotidase. Nat Struct Biol 9:779′87 [DOI] [PubMed]
- 21.Wang W, Cho HS, Kim R et al (2002) Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic ‘snapshots′of intermediate states. J Mol Biol 319:421′31 [DOI] [PubMed]
- 22.Itoh R, Mitsui A, Tsushima K (1967) 5′Nucleotidase of chicken liver. Biochim Biophys Acta 146:151′59 [DOI] [PubMed]
- 23.Turriani M, Pesi R, Nardone A et al (1994) Cytosolic 5′nucleotidase/nucleoside phosphotransferase: a nucleoside analog activating enzyme? J Biochem Toxicol 9:51′7 [DOI] [PubMed]
- 24.Bretronnet AS, Jordheim LP, Dumontet C et al (2005) Regulation and activity of cytosolic 5′nucleotidase II. A bifunctional allosteric enzyme of the haloacid dehalogenase superfamily involved in cellular metabolism. FEBS Lett 579:3363′368 [DOI] [PubMed]
- 25.Barsotti C, Pesi R, Giannecchini M et al (2005) Evidence for the involvement of cytosolic 5′nucleotidase (cN-II) in the synthesis of guanine nucleotides from xanthosine. J Biol Chem 280:1365′369 [DOI] [PubMed]
- 26.Pankiewicz KW, Patterson SE, Blank PL et al (2004) Cofactor mimics as selective inhibitors of NAD-dependent inosine monophosphate dehydrogenase (IMPDH)—the major therapeutic target. Curr Med Chem 11:887′00 [DOI] [PubMed]
- 27.Zimmermann AG, Gu JJ, Laliberte J et al (1998) Inosine-5′monophosphate dehydrogenase: regulation of expression and role in cellular proliferation and T lymphocyte activation. Prog Nucleic Acid Res Mol Biol 61:181′09 [DOI] [PubMed]
- 28.Becker MA (2001) Phosphoribosylpyrophosphate synthetase and the regulation of phosphoribosylpyrophosphate production in human cells. Prog Nucleic Acid Res Mol Biol 69:115′47 [DOI] [PubMed]
- 29.Barankiewicz J, Gelfand EW, Issekutz A et al (1982) Evidence for the active purine nucleoside cycle in human mononuclear cells and cultured fibroblasts. J Biol Chem 257:11597′1600 [PubMed]
- 30.Berman PA, Human L (1990) Regulation of 5-phosphoribosyl 1-pyrophosphate and of hypoxanthine uptake and release in human erythrocytes by oxypurine cycling. J Biol Chem 265:6562′568 [PubMed]
- 31.Mascia L, Cotrufo T, Cappiello M et al (1999) Ribose 1-phosphate and inosine activate uracil salvage in rat brain. Biochim Biophys Acta 1472:93′8 [DOI] [PubMed]
- 32.Markley HG, Faillace LA, Mezey E (1973) Xanthine oxidase activity in rat brain. Biochim Biophys Acta 309:23′1 [DOI] [PubMed]
- 33.Leach RM, Sheehan DW, Chaco VP et al (1998) Effects of hypoxia, energy state and pH in resting pulmonary and femoral arterial smooth muscles. Am J Physiol 275:L1051–L1060 [DOI] [PubMed]
- 34.Phillis JW, O’Regan MH, Estevez AY et al (1996) Cerebral energy metabolism during severe ischemia of varying duration and following reperfusion. J Neurochem 67:1525′531 [DOI] [PubMed]
- 35.Barsotti C, Tozzi M, Ipata PL (2002) Purine and pyrimidine salvage in whole rat brain. Utilization of ATP-derived ribose-1-phosphate and 5-phosphoribosyl-1-pyrophosphate generated in experiments with dialyzed cell-free extracts. J Biol Chem 277:9865′869 [DOI] [PubMed]
- 36.Mascia L, Cappiello M, Cherri S, Ipata PL (2000) In vitro recycling of α-D-ribose 1- phosphate for the salvage of purine bases. Biochim Biophys Acta 1474:70′4 [DOI] [PubMed]
- 37.Barsotti C, Ipata PL (2002) Pathways for α-D-ribose utilization for nucleobase salvage and 5-fluorouracil activation in rat brain. Biochem Pharmacol 63:117′22 [DOI] [PubMed]
- 38.Mascia L, Ipata PL (2001) Activation pathways of 5-fluorouracil in rat organs and in PC12 cells. Biochem Pharmacol 62:213′21 [DOI] [PubMed]
- 39.Cao D, Russell R, Zhang D et al (2002) Uridine phosphorylase (−/−) murine embryonic stem cells clarify the key role of this enzyme in the regulation of the pyrimidine salvage pathway and in the activation of fluoropyrimidines. Cancer Res 62:2313′317 [PubMed]
- 40.Latini C, Pedata F (2001) Adenosine in the central nervous system: release mechanisms and extracellular concentrations. J Neurochem 79:463′84 [DOI] [PubMed]
- 41.Chapman AG, Atkinson DE (1973) Stabilization of adenylate energy charge by the adenylate deaminase reaction. J Biol Chem 248:8309′312 [PubMed]
- 42.Setlow B, Lowenstein JM (1967) Adenylate deaminase: II. Purification and some regulatory properties of the enzyme from calf brain. J Biol Chem 242:607′15 [PubMed]
- 43.Van den Berghe G, Van Pottelsberghe CD, Hers HG (1977) A kinetic study of the soluble 5′nucleotidase of rat liver. Biochem J 162:611′16 [DOI] [PMC free article] [PubMed]
- 44.Latini S, Corsi C, Pedata F et al (1995) The source of adenosine outflow during ischemia and electrical stimulation. Neurochem Int 79:239′44 [DOI] [PubMed]
- 45.Pak M, Haas HL, Decking UKM (1994) Inhibition of adenosine kinase increases endogenous adenosine and depresses neuronal activity in hippocampal slices. Neuropharmacology 33:1049′053 [DOI] [PubMed]