Abstract
R1162 is efficiently comobilized during conjugative transfer of the self-transmissible plasmid R751. Bacteriophage M13 derivatives that contain two directly repeated copies of oriT, the site on R1162 DNA required in cis for mobilization, were constructed. Phage DNA molecules underwent recombination during infection of Escherichia coli, with the product retaining a single functional copy of oriT. Recombination was strand specific and depended on R1162 gene products involved in mobilization, but did not require the self-transmissible plasmid vector. Two genes were identified, one essential for recombination and the other affecting the frequency of recombination. Recombination of bacteriophage DNA could form the basis of a simple model for some of the events occurring during conjugation without the complexity of a true mating system.
Full text
PDF
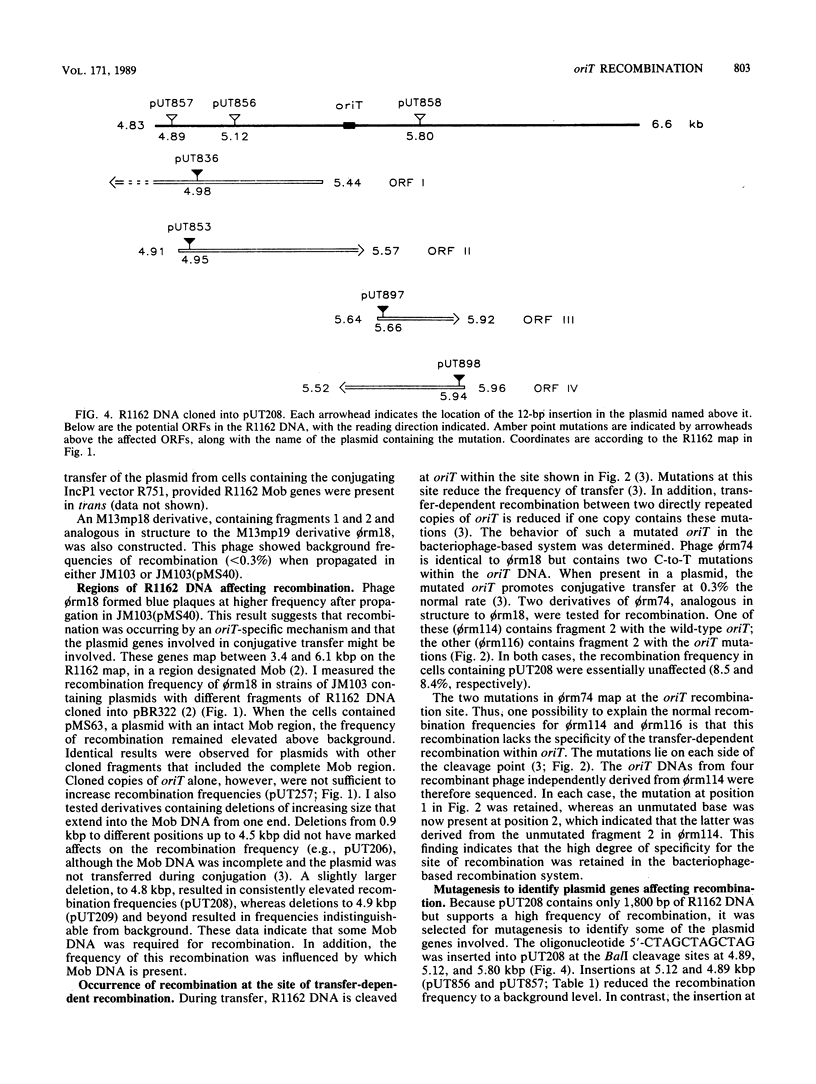
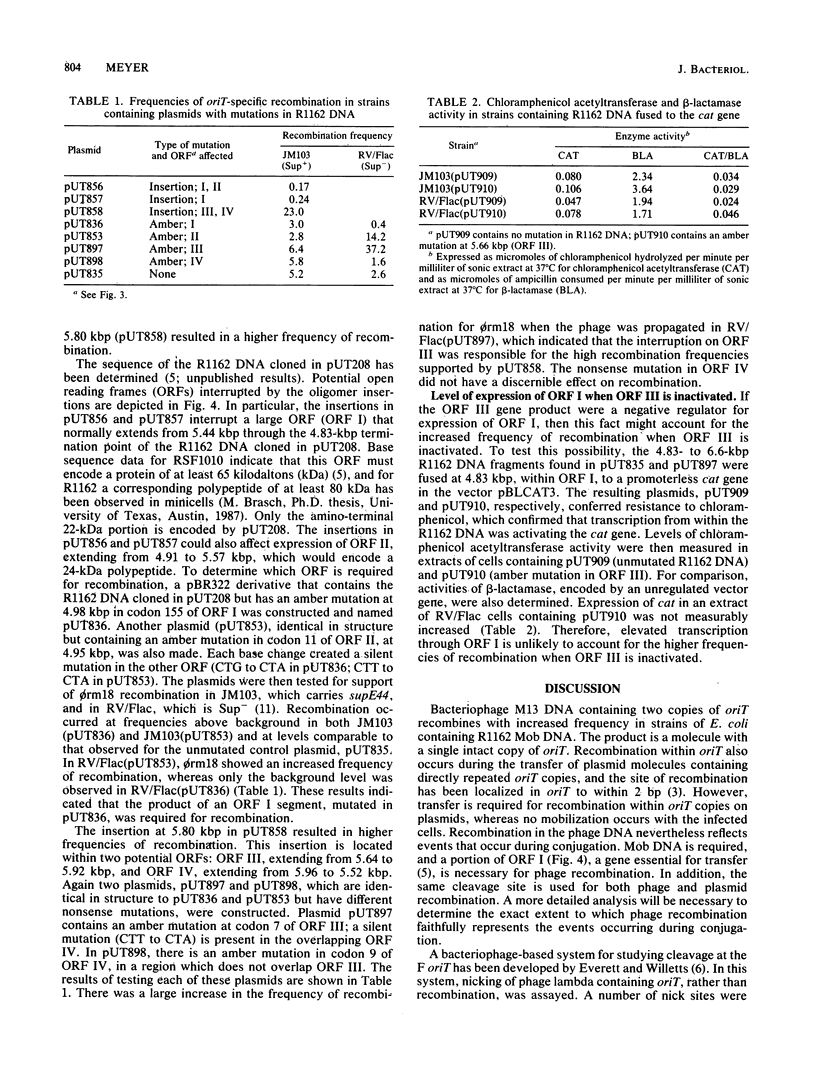
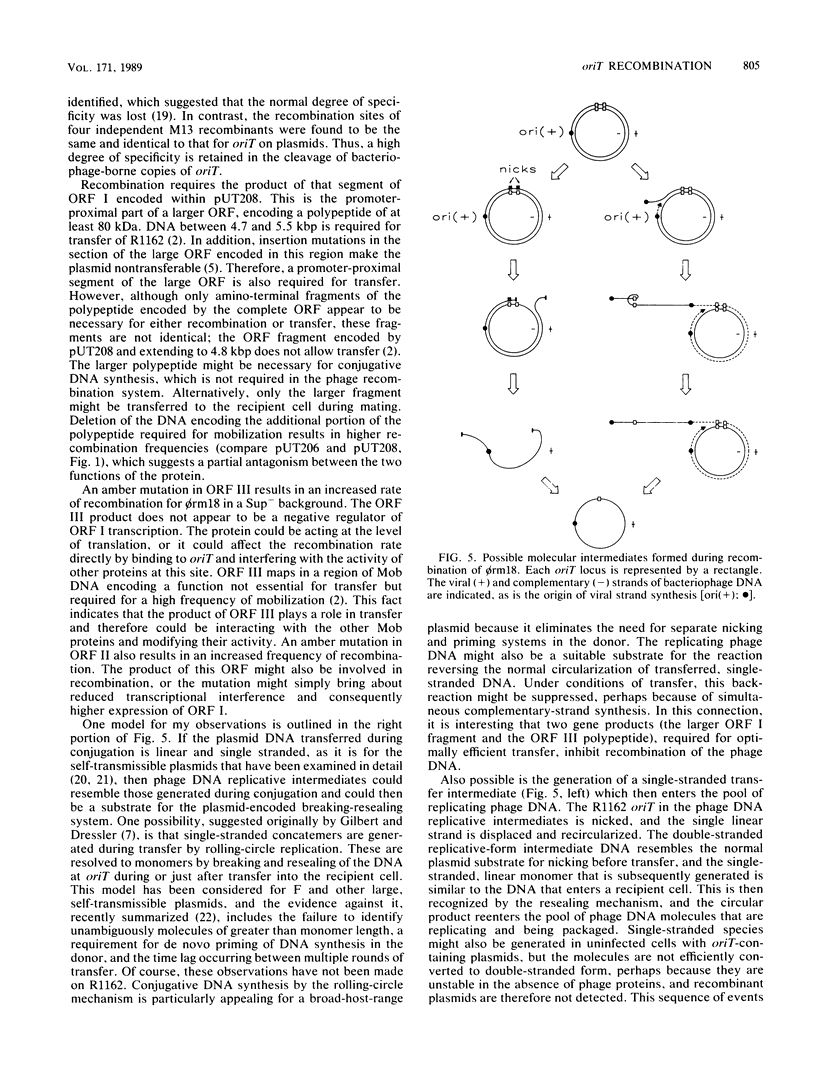

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barth P. T., Grinter N. J. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974 Nov;120(2):618–630. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasch M. A., Meyer R. J. A 38 base-pair segment of DNA is required in cis for conjugative mobilization of broad host-range plasmid R1162. J Mol Biol. 1987 Dec 5;198(3):361–369. doi: 10.1016/0022-2836(87)90286-5. [DOI] [PubMed] [Google Scholar]
- Brasch M. A., Meyer R. J. Genetic organization of plasmid R1162 DNA involved in conjugative mobilization. J Bacteriol. 1986 Aug;167(2):703–710. doi: 10.1128/jb.167.2.703-710.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Derbyshire K. M., Hatfull G., Willetts N. Mobilization of the non-conjugative plasmid RSF1010: a genetic and DNA sequence analysis of the mobilization region. Mol Gen Genet. 1987 Jan;206(1):161–168. doi: 10.1007/BF00326552. [DOI] [PubMed] [Google Scholar]
- Everett R., Willetts N. Characterisation of an in vivo system for nicking at the origin of conjugal DNA transfer of the sex factor F. J Mol Biol. 1980 Jan 15;136(2):129–150. doi: 10.1016/0022-2836(80)90309-5. [DOI] [PubMed] [Google Scholar]
- Gilbert W., Dressler D. DNA replication: the rolling circle model. Cold Spring Harb Symp Quant Biol. 1968;33:473–484. doi: 10.1101/sqb.1968.033.01.055. [DOI] [PubMed] [Google Scholar]
- Goebel W., Helinski D. R. Nicking activity of an endonuclease. I. Transfer ribonucleic acid complex of Escherichia coli. Biochemistry. 1970 Nov 24;9(24):4793–4801. doi: 10.1021/bi00826a025. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckow B., Schütz G. CAT constructions with multiple unique restriction sites for the functional analysis of eukaryotic promoters and regulatory elements. Nucleic Acids Res. 1987 Jul 10;15(13):5490–5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy M. H., Fiandt M., Szybalski W. Electron microscopy of polar insertions in the lac operon of Escherichia coli. Mol Gen Genet. 1972;119(3):207–222. doi: 10.1007/BF00333859. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Meyer R., Hinds M., Brasch M. Properties of R1162, a broad-host-range, high-copy-number plasmid. J Bacteriol. 1982 May;150(2):552–562. doi: 10.1128/jb.150.2.552-562.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOVICK R. P. Micro-iodometric assay for penicillinase. Biochem J. 1962 May;83:236–240. doi: 10.1042/bj0830236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Thompson R., Taylor L., Kelly K., Everett R., Willetts N. The F plasmid origin of transfer: DNA sequence of wild-type and mutant origins and location of origin-specific nicks. EMBO J. 1984 May;3(5):1175–1180. doi: 10.1002/j.1460-2075.1984.tb01947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vapnek D., Rupp W. D. Asymmetric segregation of the complementary sex-factor DNA strands during conjugation in Escherichia coli. J Mol Biol. 1970 Nov 14;53(3):287–303. doi: 10.1016/0022-2836(70)90066-5. [DOI] [PubMed] [Google Scholar]
- Vapnek D., Rupp W. D. Identification of individual sex-factor DNA strands and their replication during conjugation in thermosensitive DNA mutants of Escherichia coli. J Mol Biol. 1971 Sep 28;60(3):413–424. doi: 10.1016/0022-2836(71)90178-1. [DOI] [PubMed] [Google Scholar]
- Willetts N., Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiol Rev. 1984 Mar;48(1):24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]