Abstract
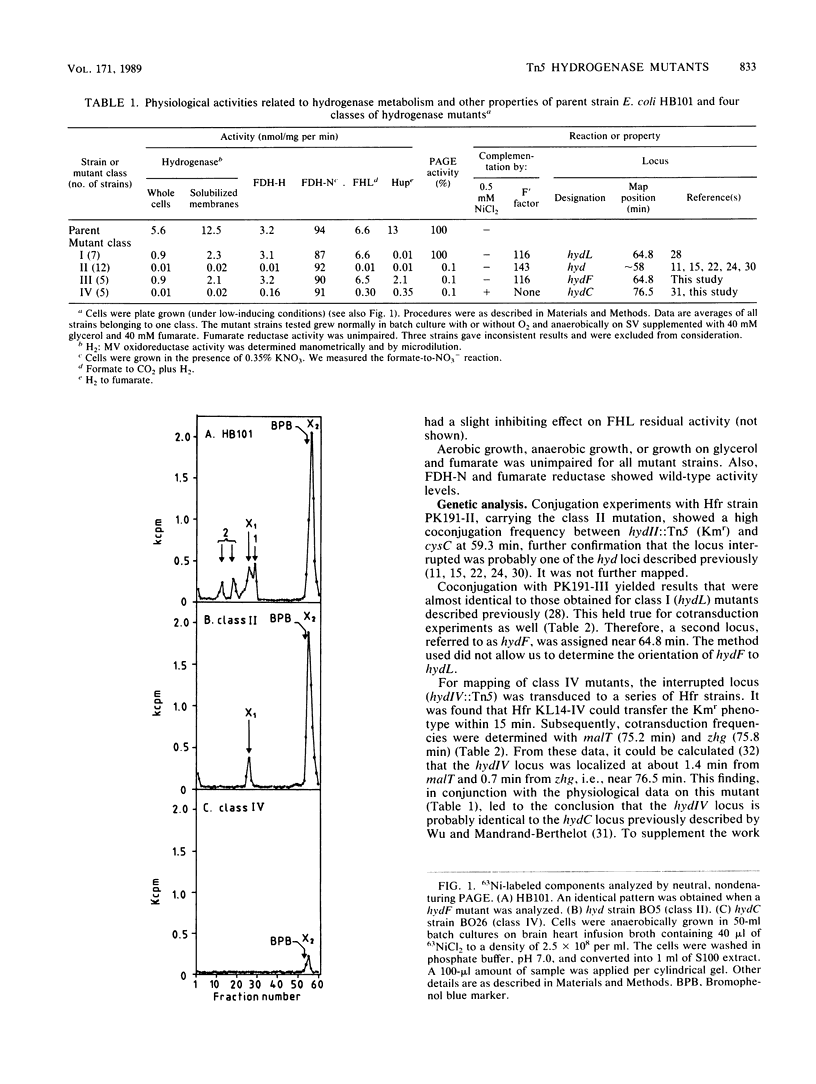
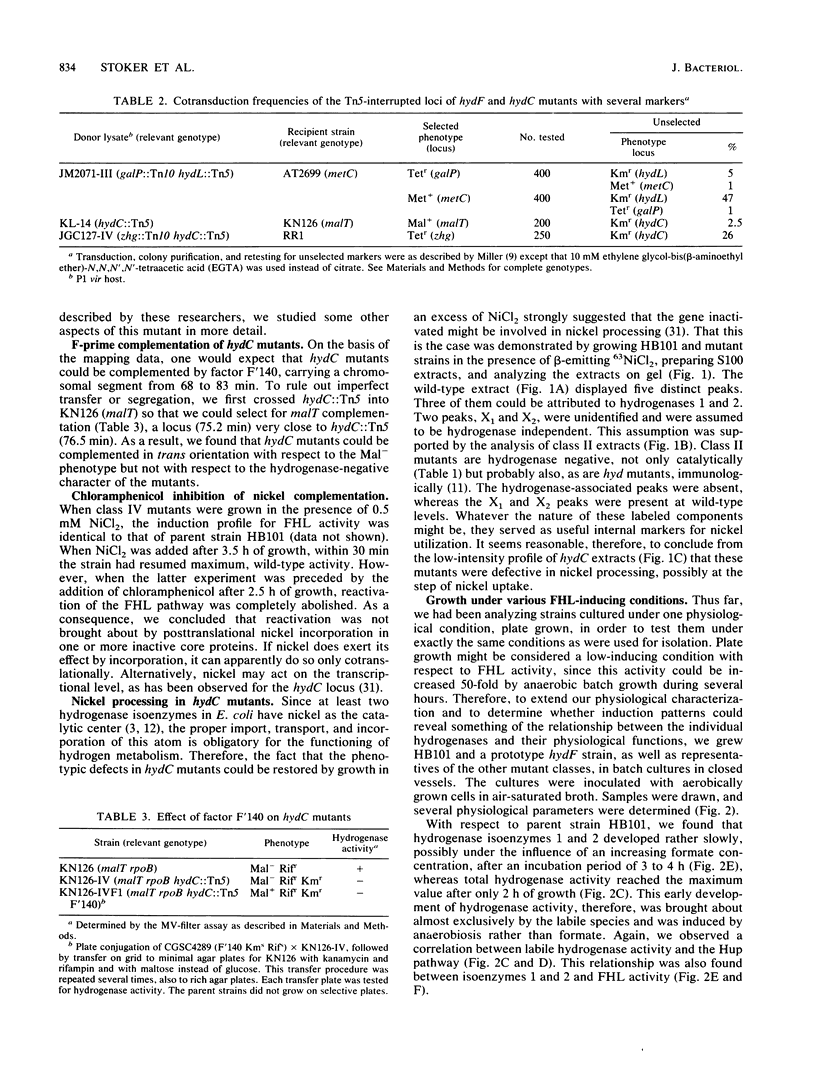
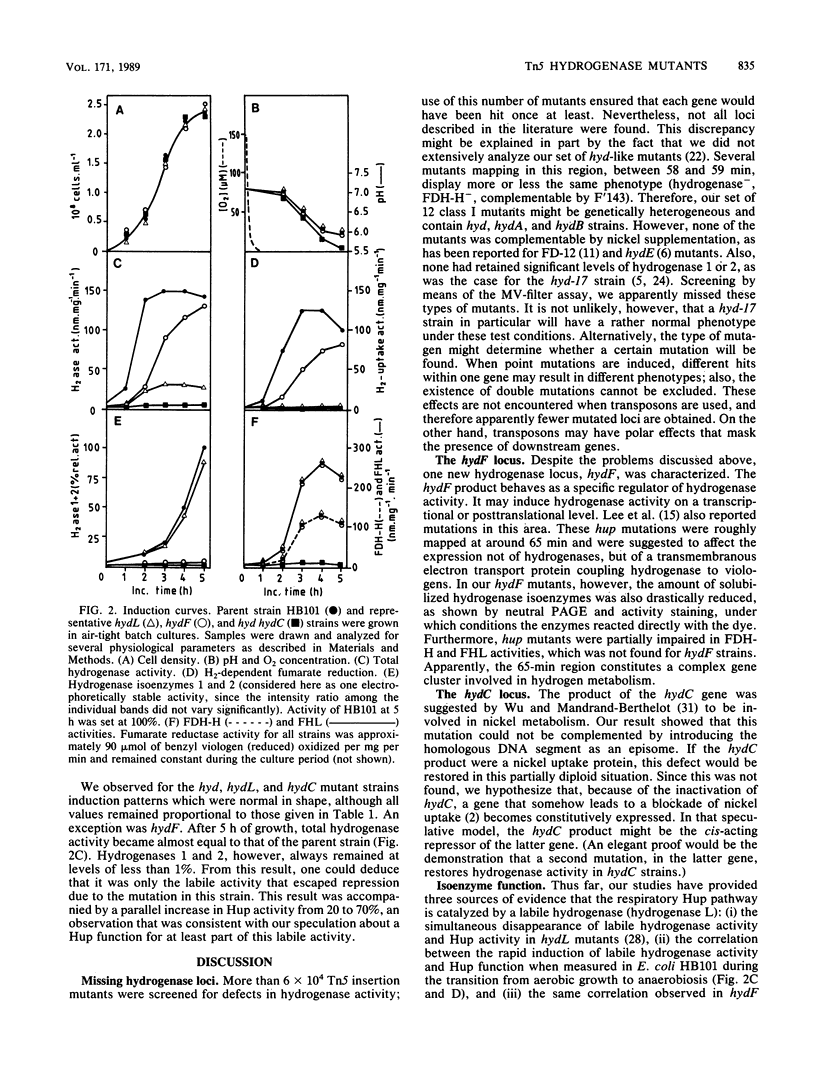
Systematic screening of 6.10(4) independent Tn5 insertion mutants of Escherichia coli yielded one new hydrogenase locus, hydF, mapping near 64.8 min, i.e., close to the hydL locus (K. Stoker, L.F. Oltmann, and A.H. Stouthamer, J. Bacteriol. 170:1220-1226, 1988). It regulated specifically the activity of the hydrogenase isoenzymes, formate dehydrogenase and lyase activities being unaffected. In hydF mutants, hydrogenase 1 and 2 activities were reduced to 1% of the parental level, whereas the electrophoretically labile part was present at about 20% of the parental level. H2 uptake was also reduced to about 20%, which suggested a relationship between these two activities. Experiments with 63Ni indicated that hydrogenase isoenzymes 1 and 2 might be present in these strains but in an inactive form. The hydF product might therefore be a posttranslational activator. At least three other mutant classes were isolated. Additional data were obtained on coisolated, nickel-restorable hydC mutants (L.F. Wu and M.-A. Mandrand-Berthelot, Biochimie 68:167-179, 1986). These strains were found to suffer a general impairment of nickel uptake. Restoration of hydrogenase activities was specific for NiCl2 and inhibited by chloramphenicol, which indicated an effect either on the transcription of hydrogenase(-associated) genes or by cotranslational incorporation in nickel-containing enzymes (e.g., in hydrogenases). The hydC mutation could not be complemented in trans, evidence that the hydC product is not a nickel transport protein but rather a cis-acting regulatory gene. Parent HB101, hydF mutants, and the other mutants were further analyzed by monitoring the induction of hydrogenase and hydrogenase-associated activities upon transition of cells from aerobic to anaerobic growth. These experiments also revealed a correlation between the early-induced H2 uptake route and labile hydrogenase activity. The formate hydrogenlyase induction patterns followed quite well the slower induction patterns of hydrogenases 1 and 2.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams M. W., Mortenson L. E., Chen J. S. Hydrogenase. Biochim Biophys Acta. 1980 Dec;594(2-3):105–176. doi: 10.1016/0304-4173(80)90007-5. [DOI] [PubMed] [Google Scholar]
- Babich H., Stotzky G. Toxicity of nickel to microbes: environmental aspects. Adv Appl Microbiol. 1983;29:195–265. doi: 10.1016/s0065-2164(08)70358-7. [DOI] [PubMed] [Google Scholar]
- Ballantine S. P., Boxer D. H. Isolation and characterisation of a soluble active fragment of hydrogenase isoenzyme 2 from the membranes of anaerobically grown Escherichia coli. Eur J Biochem. 1986 Apr 15;156(2):277–284. doi: 10.1111/j.1432-1033.1986.tb09578.x. [DOI] [PubMed] [Google Scholar]
- Ballantine S. P., Boxer D. H. Nickel-containing hydrogenase isoenzymes from anaerobically grown Escherichia coli K-12. J Bacteriol. 1985 Aug;163(2):454–459. doi: 10.1128/jb.163.2.454-459.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkmann A., Zinoni F., Sawers G., Böck A. Factors affecting transcriptional regulation of the formate-hydrogen-lyase pathway of Escherichia coli. Arch Microbiol. 1987 Jun;148(1):44–51. doi: 10.1007/BF00429646. [DOI] [PubMed] [Google Scholar]
- Chaudhuri A., Krasna A. I. Isolation of genes required for hydrogenase synthesis in Escherichia coli. J Gen Microbiol. 1987 Dec;133(12):3289–3298. doi: 10.1099/00221287-133-12-3289. [DOI] [PubMed] [Google Scholar]
- Davis N. K., Greer S., Jones-Mortimer M. C., Perham R. N. Isolation and mapping of glutathione reductase-negative mutants of Escherichia coli K12. J Gen Microbiol. 1982 Jul;128(7):1631–1634. doi: 10.1099/00221287-128-7-1631. [DOI] [PubMed] [Google Scholar]
- Glassberg J., Meyer R. R., Kornberg A. Mutant single-strand binding protein of Escherichia coli: genetic and physiological characterization. J Bacteriol. 1979 Oct;140(1):14–19. doi: 10.1128/jb.140.1.14-19.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick B. R., Wang P. Y., Schneider H., Martin W. G. Identification and partial characterization of an Escherichia coli mutant with altered hydrogenase activity. Can J Biochem. 1980 Apr;58(4):361–367. doi: 10.1139/o80-047. [DOI] [PubMed] [Google Scholar]
- Graham A., Boxer D. H., Haddock B. A., Mandrand-Berthelot A. M., Jones R. W. Immunochemical analysis of the membrane-bound hydrogenase of Escherichia coli. FEBS Lett. 1980 May 5;113(2):167–172. doi: 10.1016/0014-5793(80)80584-9. [DOI] [PubMed] [Google Scholar]
- Hausinger R. P. Nickel utilization by microorganisms. Microbiol Rev. 1987 Mar;51(1):22–42. doi: 10.1128/mr.51.1.22-42.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn P. L. Isolation of high-frequency recombining strains from Escherichia coli containing the V colicinogenic factor. J Bacteriol. 1968 Jul;96(1):205–214. doi: 10.1128/jb.96.1.205-214.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lee J. H., Patel P., Sankar P., Shanmugam K. T. Isolation and characterization of mutant strains of Escherichia coli altered in H2 metabolism. J Bacteriol. 1985 Apr;162(1):344–352. doi: 10.1128/jb.162.1.344-352.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low B. Rapid mapping of conditional and auxotrophic mutations in Escherichia coli K-12. J Bacteriol. 1973 Feb;113(2):798–812. doi: 10.1128/jb.113.2.798-812.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macy J., Kulla H., Gottschalk G. H2-dependent anaerobic growth of Escherichia coli on L-malate: succinate formation. J Bacteriol. 1976 Feb;125(2):423–428. doi: 10.1128/jb.125.2.423-428.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J. H., Ganem D., Lu P., Schmitz A. Genetic studies of the lac repressor. I. Correlation of mutational sites with specific amino acid residues: construction of a colinear gene-protein map. J Mol Biol. 1977 Jan 15;109(2):275–298. doi: 10.1016/s0022-2836(77)80034-x. [DOI] [PubMed] [Google Scholar]
- PECK H. D., Jr, GEST H. Formic dehydrogenase and the hydrogenlyase enzyme complex in coli-aerogenes bacteria. J Bacteriol. 1957 Jun;73(6):706–721. doi: 10.1128/jb.73.6.706-721.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal M. C., Casse F., Chippaux M., Lepelletier M. Genetic analysis of mutants of Escherichia coli K12 and Salmonella typhimurium LT2 deficient in hydrogenase activity. Mol Gen Genet. 1975 Nov 24;141(2):173–179. doi: 10.1007/BF00267682. [DOI] [PubMed] [Google Scholar]
- Sawers R. G., Ballantine S. P., Boxer D. H. Differential expression of hydrogenase isoenzymes in Escherichia coli K-12: evidence for a third isoenzyme. J Bacteriol. 1985 Dec;164(3):1324–1331. doi: 10.1128/jb.164.3.1324-1331.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawers R. G., Boxer D. H. Purification and properties of membrane-bound hydrogenase isoenzyme 1 from anaerobically grown Escherichia coli K12. Eur J Biochem. 1986 Apr 15;156(2):265–275. doi: 10.1111/j.1432-1033.1986.tb09577.x. [DOI] [PubMed] [Google Scholar]
- Stewart V. Nitrate respiration in relation to facultative metabolism in enterobacteria. Microbiol Rev. 1988 Jun;52(2):190–232. doi: 10.1128/mr.52.2.190-232.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker K., Oltmann L. F., Stouthamer A. H. Partial characterization of an electrophoretically labile hydrogenase activity of Escherichia coli K-12. J Bacteriol. 1988 Mar;170(3):1220–1226. doi: 10.1128/jb.170.3.1220-1226.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waugh R., Boxer D. H. Pleiotropic hydrogenase mutants of Escherichia coli K12: growth in the presence of nickel can restore hydrogenase activity. Biochimie. 1986 Jan;68(1):157–166. doi: 10.1016/s0300-9084(86)81080-x. [DOI] [PubMed] [Google Scholar]
- Wu L. F., Mandrand-Berthelot M. A. Genetic and physiological characterization of new Escherichia coli mutants impaired in hydrogenase activity. Biochimie. 1986 Jan;68(1):167–179. doi: 10.1016/s0300-9084(86)81081-1. [DOI] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Alphen L., Lugtenberg B., van Boxtel R., Hack A. M., Verhoef C., Havekes L. meoA is the structural gene for outer membrane protein c of Escherichia coli K12. Mol Gen Genet. 1979 Jan 31;169(2):147–155. doi: 10.1007/BF00271665. [DOI] [PubMed] [Google Scholar]


