Abstract
The Puf family of RNA-binding proteins directs cell fates by regulating gene expression at the level of translation and RNA stability. Here, we report that the C. elegans pumilio homolog, puf-9, controls the differentiation of epidermal stem cells at the larval-to-adult transition. Genetic analysis reveals that loss-of-function mutations in puf-9 enhance the lethality and heterochronic phenotypes caused by mutations in the let-7 microRNA (miRNA), while suppressing the heterochronic phenotypes of lin-41, a let-7 target and homolog of Drosophila Brat. puf-9 interacts with another known temporal regulator hbl-1, the C. elegans ortholog of hunchback. We present evidence demonstrating that puf-9 is required for the 3’UTR-mediated regulation of hbl-1, in both the hypodermis and the ventral nerve cord. Finally, we show that this regulation is dependent on a region of the hbl-1 3’UTR that contains putative Puf family binding sites as well as binding sites for the let-7 miRNA family, suggesting that puf-9 and let-7 may mediate hypodermal seam cell differentiation by regulating common targets.
Keywords: puf-9, hbl-1, let-7, C. elegans, Pumilio, hunchback, development, microRNA
Introduction
As early as the first embryonic cell divisions, spatial and temporal control mechanisms establish asymmetric gene expression patterns necessary for proper cell fate decisions during development. In multi-cellular organisms cell fates are often determined by the precise control of gene expression at a post-transcriptional level. In many cases cis-regulatory elements in the 3’ untranslated region (3’UTR) of an mRNA direct these control mechanisms by the binding of specific regulatory factors.
The Puf family of of RNA-binding proteins is named for its founding members, Drosophila Pumilio and C. elegans fem-3 Binding Factor (FBF) (Zhang et al., 1997), and is comprised of a complex group of proteins that are found in diverse organisms, including yeast, flies, nematodes, frogs, and humans (Edwards et al., 2001; Wang et al., 2001; Zamore et al., 1997). Despite their widespread abundance, the molecular mechanism by which Puf proteins regulate their target RNAs is still largely unknown. Previously characterized Puf proteins have been found to act by binding to specific regulatory elements in the 3’UTR’s of their target RNAs, leading to either translational repression or degradation of the target mRNA (Wickens et al., 2002). Puf proteins share an evolutionarily conserved C-terminal motif, termed the Pumilio-homology domain (Pum-HD) (Zamore et al., 1997). This domain consists of eight imperfect repeats and two flanking conserved regions that form an arc of supercoiled α-helices and retains the RNA binding activity of the protein (Zamore et al., 1997). Drosophila Pumilio (Pum) is one of the most well characterized members of the Puf family and has been assigned multiple developmental roles, including the establishment of axis formation and germline stem cell maintenance (Wickens et al., 2002). For example, Pum negatively regulates the maternal gap gene hunchback in the embryonic syncytium (Murata and Wharton, 1995; Tautz, 1988) to establish a gradient of Hunchback protein that is essential for the formation of anterior structures of the fly (Wu et al., 2001). Pum participates in the post-transcriptional repression of hunchback mRNA by binding to a defined 36nt motif in the hunchback 3’ UTR called the Nanos Response Elements (NRE) (Murata and Wharton, 1995; Wharton and Struhl, 1991) and acts in a multi-protein complex that includes Nanos (Nos) and Brain Tumor (Brat) (Sonoda and Wharton, 2001).
The C. elegans genome encodes nine pumilio and three nanos homologs (Zhang et al., 1997). Like Drosophila, the characterized C. elegans pumilio genes play definitive roles in regulating stem cell maintenance in the germline. For instance, puf-8 is required to maintain germline cells in a meiotically arrested state during spermatogenesis (Subramaniam and Seydoux, 2003). In PUF-8 depleted animals, primary spermatocytes dedifferentiate and proliferate, leading to germline tumors (Subramaniam and Seydoux, 2003). FBF-1 and FBF-2, two other C. elegans Puf proteins collectively referred to as FBF, target the fem-3 3’UTR and play a largely redundant role to promote mitotic divisions and define the contributing region of stem cells in the adult germline (Crittenden et al., 2002; Lamont et al., 2004). Deletion of the fbf-1 and fbf-2 gene products leads to the failure of germline stem cell maintenance, due to premature entry into meiosis (Crittenden et al., 2002).
The C. elegans developmental timing, or heterochronic, pathway also displays many examples of negative gene regulation at the post-transcriptional level. Heterochronic genes coordinately establish cell fate profiles unique to a particular stage and act as timing switches to control cellular identities in many different tissues (Ambros and Horvitz, 1984; Banerjee and Slack, 2002). In heterochronic mutants, the hypodermal seam cells, which are a set of specified cells that run down the length of the animal on each side, will inappropriately adopt cellular fates in either a retarded or precocious manner relative to wild type developmental stages. During wild type development the seam cells divide in a stem-cell-like manner, such that they are continuously renewed during each larval stage. Like stem cells, seam cells are characterized by asymmetric cell divisions where one daughter cell differentiates and one retains its stem cell identity. For example, following each division, the anterior seam cell daughter differentiates and fuses with the large hypodermal syncytium (hyp7), while the posterior seam cell daughter retains its identity and continues to divide at the subsequent larval stages. This self-renewing division cycle continues until the fourth larval stage when the seam cells terminally differentiate and permanently exit the cell cycle, thus defining the larval-to-adult (L/A) switch.
The L/A switch is controlled by a cascade of heterochronic gene interactions and heterochronic mutants often display defects in the cuticle of the animal at the L/A switch. The first larval (L1) and adult stages of C. elegans post-embryonic development are characterized by an epidermal structure called lateral alae, which are cuticular ridges that run down each side of the animal. Alae are secreted by the underlying seam cells in response to a variety of differentiation factors, including the lin-29 transcription factor (Ambros, 1989; Papp et al., 1991) and the let-7 miRNA (Reinhart et al., 2000), which act to promote adult fates in the hypodermis (Ambros, 1989; Pasquinelli et al., 2000).
Loss-of-function (lf) mutations in let-7 and lin-29 result in retarded developmental phenotypes where the seam cells continue to divide at the adult stage, thereby failing to differentiate and secrete alae (Ambros, 1989; Reinhart et al., 2000). Strong let-7(lf) mutants also display an adult lethality due to vulval bursting (Reinhart et al., 2000). RNA interference (RNAi) of two known let-7 target genes, hbl-1 and lin-41, can partially suppress the lethality and alae defects associated with let-7 mutant animals (Abrahante et al., 2003; Lin et al., 2003; Pasquinelli et al., 2000; Slack et al., 2000), thus supporting the idea that let-7 acts in an opposing manner to hbl-1 and lin-41 to regulate the L/A switch. let-7 inhibits its targets, hbl-1 and lin-41 in a post-transcriptional manner by binding to sequences in their 3’UTRs (Vella et al., 2004a) and has been shown in vivo to bind directly to the lin-41 3’UTR at specific regulatory sequences called let-7 complementary sites (LCS) (Vella et al., 2004a; Vella et al., 2004b).
In hbl-1 or lin-41 loss-of-function mutants the seam cells precociously exit the self-renewing cell cycle, differentiate and secrete alae during the fourth larval stage (Abrahante et al., 2003; Lin et al., 2003; Pasquinelli et al., 2000; Slack et al., 2000). Thus, hbl-1 and lin-41 normally prevent cells from attaining adult fates prematurely. HBL-1 is the ortholog of the Drosophila transcription factor, Hunchback (Hb) (Fay et al., 1999), while LIN-41 is closely related to Drosophila Brain Tumor (Brat) (Slack and Ruvkun, 1998)
In a wild type animal, hbl-1::gfp is expressed in the main body hypodermis (hyp7) until the L3 stage and in the ventral nerve cord (VNC) until the early-L4 stage of development (Abrahante et al., 2003; Lin et al., 2003). Previous work has demonstrated that hbl-1 down-regulation in epidermal and neural tissues is dependent on the hbl-1 3’UTR (Abrahante et al., 2003; Lin et al., 2003), similar to Drosophila hunchback. When the heterologous unc-54 3’UTR was substituted for the native hbl-1 3’UTR, proper down-regulation was abrogated and GFP expression was maintained in both the hyp7 and the VNC in the adult stage (Lin et al., 2003). Potential cis-regulatory elements in the hbl-1 3’UTR have been identified, including complementary sites for the miRNAs let-7 (LCS) and lin-4 (LCE), and their family members (Lin et al., 2003). The 3’UTR mediated regulation of hbl-1 in the hypodermis at the L2/L3 stage requires the let-7 family microRNAs, mir-48, mir-84 and mir-241 (Abbott et al., 2005), while let-7 and lin-4 are needed for hbl-1 3’UTR reporter down-regulation in the ventral nerve cord (VNC) at the L4 stage (Lin et al., 2003). Therefore, hbl-1 plays a role in defining both the early (L2/L3) and late (L4/Adult) transitions (Abbott et al., 2005; Abrahante et al., 2003; Lin et al., 2003). When the let-7 family members are mutated, hbl-1 continues to be partially down-regulated in both the hypodermis and the VNC (Lin et al., 2003), suggesting that additional factors are required for the regulation of hbl-1. However, protein co-factors that function with the let-7 family to mediate hbl-1 regulation remain unidentified.
There are many intriguing molecular parallels between the Drosophila spatial patterning genes and C. elegans temporal patterning genes. In Drosophila, pattern formation in the early embryo is established through the spatially restricted expression of a complicated network of transcription factors, including hunchback (hb) (Irish et al., 1989; Wu et al., 2001). In C. elegans, hbl-1 is highly expressed during early larval stages and is down-regulated over time in a variety of tissues (Abbott et al., 2005; Abrahante et al., 2003; Fay et al., 1999; Lin et al., 2003) and is thus regulated along a temporal rather than a spatial gradient. In Drosophila, the transcription factor kruppel is a direct target of hunchback and acts in a cascade of multiple transcription factors to ultimately establish segmentation patterning of the fly (Wu et al., 2001). The C. elegans kruppel homologue, lin-29, is analogously regulated along a temporal gradient with its highest expression peaking in the later larval stages (Bettinger et al., 1996). Interestingly, in addition to their functions in body plan formation, hunchback and kruppel have been shown to play a temporal role in establishing neuroblast identity during cortical layer formation in Drosophila (Isshiki et al., 2001). Finally, LIN-41 and Drosophila Brat share high structural similarities and belong to the same RBCC-NHL family of translational regulatory proteins (Slack and Ruvkun, 1998). The possibility exists that additional Drosophila spatial patterning homologs may function to regulate developmental timing in C. elegans. We hypothesized that one or more of the C. elegans pumilio homologs may interact with the late-stage timing factors, lin-41 and hbl-1, to regulate the stem cell-like proliferation and differentiation of seam cells at L/A transition.
Here, we report that the C. elegans pumilio homolog, puf-9, acts to control the 3’UTR mediated expression of hbl-1 in the hypodermis and the ventral nerve cord (VNC). We demonstrate that this repression requires a region of the hbl-1 3’UTR that contains potential Puf protein binding sites (NRE) and let-7 family complementary sites (LCS), suggesting that Pumilios and miRNAs may cooperate at the 3’UTRs of target genes to control their translation. We show that puf-9 genetically interacts with known regulators of the L/A switch, including the let-7 miRNA and two let-7 target genes, lin-41 and hbl-1. Our data reveals that puf-9 is required for seam cell morphology and adult cuticle formation, expanding a somatic role for the Puf family proteins in C. elegans development.
Materials and methods
Strain maintenance and RNAi
C. elegans strains were maintained on NGM plates containing E. coli OP50 bacterial lawns. Experiments were performed at 20°C, unless otherwise indicated. Gene knock-down was achieved through RNAi by feeding using previously published reagents and protocols (Kamath et al., 2003; Timmons et al., 2001). All RNAi experiments were done by plating the indicated worm strain onto plates containing E. coli HT115(DE3) bacteria lawns expressing the appropriate dsRNA. RNAi constructs for fbf-2, puf-3, puf-5, puf-7, puf-8, puf-9, nos-1, nos-2, and nos-3 (Subramaniam and Seydoux, 1999) contained genomic fragments of coding sequence cloned into the MCS of pL4440 RNAi vector (Timmons et al., 2001). RNAi constructs for fbf-2 and puf-7 also knock-down fbf-1 and puf-6 levels, respectively (K. Subramaniam, personal communication). For double and triple-RNAi experiments, bacteria containing each single RNAi construct were diluted with an equal number of cells (determined by optical density at 600nm) containing the indicated RNAi plasmid or an control empty vector (pL4440) RNAi plasmid. In the puf-9(RNAi) analyses, late L4 stage animals were exposed to RNAi to allow for depletion of maternal puf-9 during oogenesis. The puf-9 depleted progeny were then scored at the adult stage for the presence of alae and additional defects. For let-7(n2853ts);puf-9(RNAi) alae and survival analysis, a population of let-7(n2853ts) animals were synchronized as starved L1s, exposed to puf-9(RNAi) and scored for alae or survival at the young adult stage. The puf-9(ok1136) deletion strain was obtained from the C. elegans Gene Knockout Consortium and was back-crossed to wild type (N2) animals four times prior to mutant characterization.
puf-9::gfp Construct Design and Expression Analysis
PCR was used to generate a 1.9 kb genomic fragment from the WO6B11 cosmid (Genebank accession number U39854) including the predicted puf-9 promoter, using primers PUF9A and PUF9GFP. Primers GFPAK9 and GFP2C were used to amplify a 1.8 kb fragment containing gfp followed by the unc-54 3’UTR from pPD95.75 vector. The puf-9 promoter PCR product was then fused in frame to gfp::unc-54 3’UTR using overlap extension PCR using primers PUF9B and GFP2C to create MJC20. The resulting PCR products for MJC20 were co-injected directly into N2 animals at a concentration of 50ng/ul along with the pRF4 (rol-6) marker at 100ng/ul. Individually isolated lines were established and used for puf-9::gfp expression analysis.
X-gal assay and transgenic lines
For all plasmids with designation “pSJA#” and “pMJC#”: PCR fragments were amplified from genomic DNA, digested with SacII and NcoI and ligated into the unc-54 3’UTR of the B29 vector (Reinhart et al, 2000) cut with the same restriction enzymes. Transgenic animals were created by co-injecting the hbl-1 3’UTR constructs (pSJA2, pSJA3, pSJA4, pSJA5, pMJC2, pMJC12 and pFS1038) at 5 ng/ul and the pKP13 (goa-1::gfp) marker at 50 ng/ul into wild type (N2) animals. Refer to published methods for pFS1038 (Lin et al., 2003). puf-9(ok1136) mutant animals were crossed into the existing ZaEx6 strain, containing the pFS1038 hbl-1 3’UTR construct in a wild type background. Independent lines, carrying the extrachromosomal arrays, were tested for β-galactosidase expression. Wild type lines expressing the control reporter construct (pFS1038) were assayed in parallel with either the puf-9(ok1136) mutant background or wild type animals expressing the truncated reporter constructs for each separate experimental trial. The average of the individual trials is shown.
The inserted hbl-1 3’UTR sequence for pSJA2 was amplified using primers F13D11(16) and SJA2; pSJA3 was amplified using primers F13D11(16) and SJA3; pSJA4 was amplified using primers F13D11(16) and SJA4; and pSJA5 was amplified using primers SJA2 and SJA5. pMJC2 was created by overlap extension PCR using primers F13D11(16) and ONS2, and F13D11(17) and ONS1. X-gal staining of animals expressing the lacZ transgene was performed as described (Vella et al., 2004a). All primers were synthesized by the Keck Facility, Yale University Medical School (see Supplemental Fig. 9 for oligonucleotide sequences).
Quantification of hbl-1::gfp expression
GFP levels were quantified in BW1932 and puf-9(ok1136);BW1932 lines using the Carl Zeiss AxioVision Release 4.4 program. All fluorescence images used for GFP quantitation were taken at 40x objective and at 750 ms exposure. From these images, multiple representative VNC neuron cell bodies were selected within a single animal and an average Mean GFP value (MeanG) was recorded for each individual animal. The single animal average MeanG values were then used to calculate an average MeanG for the L1 or adult stages for each background.
Results and Discussion
Mutations in puf-9 enhance let-7 lethal and heterochronic alae phenotypes
Based on the parallels between the Drosophila spatial patterning genes and the genes known to be involved in the C. elegans heterochronic pathway, we investigated a possible post-embryonic role for the C. elegans homologs of nanos (nos) and pumilio (fbf/puf) in regulating developmental timing and epidermal stem cell development. In C. elegans, hbl-1 regulation through its 3’UTR is, in part, dependent upon let-7 miRNA family members (Abbott et al., 2005; Abrahante et al., 2003; Lin et al., 2003). Therefore, we reasoned that if the puf or nos homologs regulated hbl-1, similar to Drosophila hunchback, then they might also genetically interact with the miRNA let-7. We utilized the let-7(n2853ts) strain that had been shown previously to display 100% lethality due to vulval bursting at the non-permissive temperatures of 20°C and 25°C, while a significant number of animals are able to survive through adulthood at 15°C (Reinhart et al., 2000). A candidate RNAi screen knocking down each of the C. elegans pumilio, fbf/puf, and nanos, nos, homologs, identified puf-9 as being able to enhance the vulva bursting phenotype seen in let-7(n2853ts) mutant animals (Fig. 1A and Supplemental Fig. 1A). For instance, at the permissive temperature, only 12% of let-7(n2853ts) animals survived when exposed to puf-9(RNAi) compared to 87% survival in let-7(n2853) animals on mock RNAi (Fig. 1A). None of the other puf homologs affected the survival rate of let-7(n2853ts) mutants (Supplemental Fig. 1A). While this analysis does not rule out a role for other Puf proteins in the heterochronic pathway, we decided to concentrate our efforts on the characterization of puf-9.
Figure 1.
puf-9 depletion enhances the lethal bursting and heterochronic phenotypes of let-7(ts) mutants. (A) let-7(n2853ts) animals exposed to puf-9(RNAi) show an increase in the number of animals that die due to bursting at the adult stage (n=533) compared to let-7(n2853ts) animals grown on a control mock RNAi (n=1114). puf-9(ok1136) mutants (n=746) and wild type animals on puf-9(RNAi) (n=483) showed similar survival rates as let-7(n2853ts) animals on mock RNAi. 100% survival rate was seen in wild type animals grown on mock RNAi (n=652). (B) A cartoon of the intron/exon structure of the puf-9 gene (contained on the W06B11.2 cosmid). The relative position of the region deleted in the puf-9(ok1136) allele is boxed and shaded. The Pum-HD region is indicated within the ok1136 deletion region. The puf-9(ok1136) deletion removes 1581 nt beginning 154 nt into exon 4 and ending 77 nt into exon 8 (nt 5727 to 7309 in the W06B11.2 cosmid sequence). This deletion ends 632 nt upstream of the predicted stop codon and causes a frame-shift to create a premature stop codon 29 nt downstream of the deletion break. (C-E) Depletion of let-7 and puf-9 gene products results in a similar vulval bursting phenotype. Nomarski DIC images of a wild type adult (C, vulva is marked with an arrow), an adult stage let-7(n2853ts) animal with a burst vulva (D), and a puf-9(ok1136) adult animal showing a bursting phenotype similar to let-7(lf) mutants (E). (F) puf-9(RNAi) enhances the lack of adult alae phenotype of let-7(lf) mutants. When grown on puf-9(RNAi), less than half of the let-7(n2853ts) animals displayed any alae at the young adult stage (n=40) compared to wild type animals exposed to puf-9(RNAi) (n=27) or let-7(n2853ts) mutants on mock RNAi (n=61).
In addition to bursting, let-7(n2853ts) animals also display a retarded developmental phenotype where the lateral hypodermal seam cells fail to secrete alae at the young adult stage (Reinhart et al., 2000). Because the bursting phenotype of let-7(n2853) animals may not be directly related to their heterochronic phenotypes, we tested for enhancement of the retarded alae phenotype of let-7 mutant animals on puf-9(RNAi). In a let-7(lf) background, parental depletion of puf-9 during oocyte formation resulted in an almost complete adult lethality in their progeny (Fig. 1A). Therefore, we exposed let-7(n2853ts) animals to puf-9(RNAi) post-embryonically, and scored these animals for alae at the young adult stage. Consistent with the increased bursting data, puf-9(RNAi) also enhanced the delayed adult alae defect of let-7(n2853ts) mutants (Fig. 1F), while puf-9(RNAi) of wild type animals and let-7(n2853ts) mutants on mock RNAi displayed a similar number of adult stage animals lacking alae (Fig. 1F). Due to the dramatic enhancement of let-7(lf) phenotypes by puf-9(RNAi), we conclude that puf-9 plays an important role in tissues affected by let-7 regulation.
The findings from our RNAi screen suggested that puf-9 acted independently of the nanos homologs, nos-1, nos-2 and nos-3, to affect let-7 mutant bursting induced lethality (Supplemental Figs. 1A and 1B). This was confirmed by the observation that let-7(n2853ts) animals exposed to RNAi simultaneously targeting the three nos genes, phenocopied let-7 mutants alone for adult survival under all conditions tested (Supplemental Fig. 1B). Furthermore, let-7 mutants on RNAi targeting puf-9 in combination with the three nos genes, phenocopied let-7(n2853ts);puf-9(RNAi) animals (Supplemental Fig. 1B). These data suggest that the nos genes are not involved in the puf-9 directed enhancement of the let-7 mutant bursting phenotype. However our analysis does not rule out a subtle role for the nos genes. There are many examples of Puf proteins acting independently of a Nanos protein. For instance, Drosophila Pumilio regulates a second mRNA target, bicoid, in the syncitial embryo in a Nanos independent manner (Gamberi et al., 2002).
Previous work has shown that expression of at least three of the C. elegans Pufs, puf-8, fbf-1 and fbf-2, are primarily detected in the germline (Subramaniam and Seydoux, 2003; Zhang et al., 1997). Of the characterized Puf genes in C. elegans, puf-8 is the most closely related to puf-9 (Wickens et al., 2002). In contrast to puf-8, puf-9 levels were not affected in mutants that lack a germline (Subramaniam and Seydoux, 2003). puf-9 is thus unique among the C. elegans Puf genes in that it appears to be predominantly expressed in somatic tissues. Therefore, it is not surprising that puf-9 would be the main, if not the only, C. elegans Puf gene to show an interaction with let-7 in the hypodermis. The enhancement of the let-7 phenotypes by puf-9 and the phenotypic similarities between the two individual mutant backgrounds (see below) suggests that puf-9 could be acting in the same pathway as let-7 and that these two regulatory factors may function in a similar manner to specify hypodermal fates.
puf-9 null mutants display let-7-like vulval bursting phenotypes
To better understand the function of puf-9 and to confirm the results seen with puf-9(RNAi), we obtained the puf-9(ok1136) deletion allele from the C. elegans Gene Knockout Consortium. The puf-9(ok1136) deletion removes the entire conserved Pum-HD (Fig. 1B). Of the nine C. elegans Pumilio proteins, PUF-9 is the most similar to Drosophila Pumilio (Spassov and Jurecic, 2003; Wickens et al., 2002) and based on the importance of the Pum-HD for the function of Drosophila Pumilio, we predict that the ok1136 deletion mutant is a molecular null (see Supplemental Figure 2 for the sequence conservation of the PUF-9 Pum-HD). Interestingly, when puf-9 was reduced through the puf-9(ok1136) deletion or through puf-9(RNAi) exposure, a similar level of lethality was seen as with let-7(n2853ts) mutants on mock RNAi (Fig. 1A). Of the animals that died, the phenotypes observed for both let-7 (Reinhart et al., 2000) and puf-9 mutants were essentially identical at the gross level, with severe hemorrhaging (bursting) through the vulva (Fig. 1D and 1E). puf-9(ok1136) homozygotes showed abnormalities similar to those seen in puf-9(RNAi) animals in all assays performed, indicating that puf-9 gene levels were effectively reduced upon exposure to puf-9(RNAi).
puf-9 is required for proper adult cuticle formation and seam cell fusion
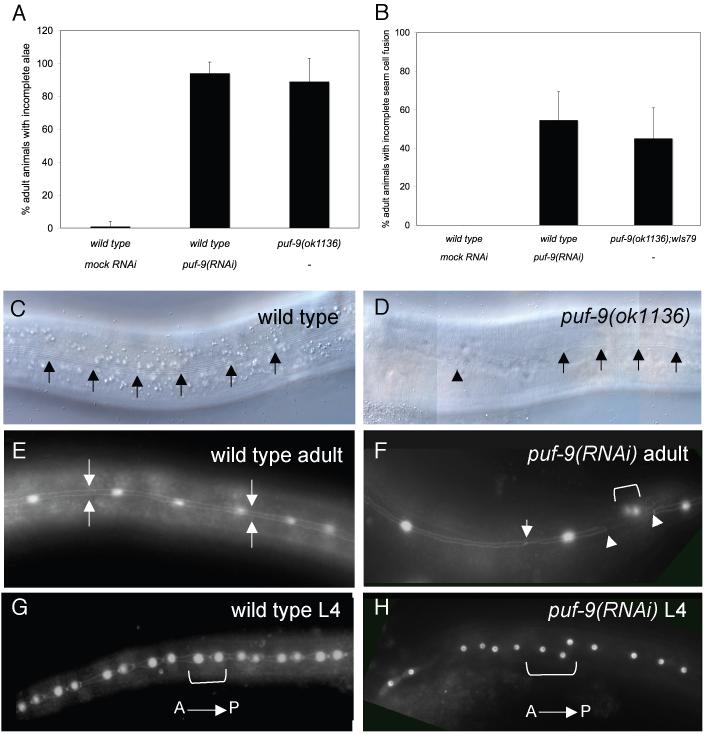
We next examined the effects of puf-9 depletion on hypodermal development, independent of the let-7 mutant background. We observed that puf-9 mutants exhibited an almost completely penetrant alae defect where both puf-9(ok1136) and puf-9(RNAi) animals showed alae breaks at the adult stage (Fig.2A). In the majority of puf-9 mutant animals, alae structures were generally rough in appearance and cover only a few cell lengths at the head and tail of an individual animal (Fig. 2D and Supplemental Fig. 3B). Occasionally degenerative alae covered one half of the length of the animal and were only rarely observed at the cells over the mid-section of the animal. The lack of completely formed adult alae in puf-9(ok1136) mutant animals is reminiscent of let-7 mutants (Reinhart et al., 2000), however, their individual mutant alae phenotypes differ slightly. In order to determine the underlying cause for the alae defects observed, we utilized a wIs79 reporter strain that has gfp marking the seam cell borders (adherens junctions) and seam cell nuclei (Lin et al., 2003). This marker allows us to visualize seam cell exit from the cell cycle as well as the extent and timing of seam cell fusion. When the wIs79 strain was crossed into puf-9(ok1136) mutants or was exposed to puf-9(RNAi) we found that the number of seam cells in puf-9 mutants was comparable to wild type at all larval stages (data not shown). Thus, this analysis did not reveal a distinct reiteration of L4 seam cell divisions as is often noted in late-acting retarded mutants such as let-7 and lin-29 (Ambros, 1989; Reinhart et al., 2000). Additionally, at the L/A transition in a wild type animal, the seam cells fuse to form a linear syncytium (Fig. 2E). We observed that the timing of the initiation of seam cell fusion appeared altered in a subset of animals deficient for puf-9. For example, in puf-9(ok1136) and puf-9(RNAi) animals, approximately half of the animals scored displayed incomplete seam cell fusion at the L/A transition (Figs. 2B and 2F).
Figure 2.
puf-9 deletion mutants display abnormal alae and seam cell fusion phenotypes. (A) puf-9(ok1136) mutants (n=88) and puf-9(RNAi) animals (n=157) show incomplete alae at the adult stage (89% and 94%, respectively) compared to 1% in wild type animals (n=129) (C,D) Nomarski DIC images of adult stage alae in a wild type animal (C, alae marked with arrows) and an adult stage puf-9(ok1136) animal showing incomplete alae (D, alae marked with arrows and the alae break marked with an arrow head). (B, E-H) Seam cell analysis of wIs79 animals carrying a gfp reporter marking the seam cell nuclei and adherens junctions. (B) Approximately half of the wIs79 animals exposed to puf-9(RNAi) (n=96) or in a puf-9(ok1136) mutant background (n=21) showed incomplete seam cell fusion at the young adult stage compared to wIs79 on mock RNAi (n=63). (E-H) GFP images of wIs79;puf-9(RNAi) animals. Fusion defects included gaps along the length of the seam cell boundary syncytium (F, gap edges marked by arrow heads) and single seam cell nuclei separated from the rest of the syncytium (F, the anterior boundary of a single unfused seam cell is marked by the arrow), indicating that a number of seam cells failed to properly fuse to the seam cell syncytium. puf-9(RNAi) animals also show seam cell nuclei that are abnormally spaced (F, set of nuclei between arrow heads and H, nuclei within bracket) and are often spatially displaced relative to one another (H, bracketed nuclei). (E) Wild type adult seam cell fusion (seam cell syncytium between arrows) and (G) anterior-posterior seam cell nuclei orientation (a seam cell pair is in the bracket). Anterior is to the left and ventral down in all images.
We also observed a large number of puf-9(RNAi) adults with abnormally spaced seam cell nuclei (Fig. 2F) and instances where the seam nuclei were positioned next to each other in a dorsal-to-ventral orientation (Fig. 2H) rather than the typical anterior-to-posterior orientation (Fig. 2G), indicating that perhaps seam cell divisions in puf-9(RNAi) animals occur in the wrong axial plane. The seam cell defects are consistent with our previous observation that the seam cells fail to correctly secrete alae at the adult stage. In fact, in most cases observed, sections of an animal that lacked alae corresponded to regions of abnormal fusion and seam cell spacing (data not shown).
To determine if the phenotypes observed in puf-9 mutants were due to altered let-7 expression, we compared let-7 miRNA levels in wild type and puf-9(ok1136) young adult animals. By northern analysis, let-7 levels were not reduced in a puf-9(ok1136) mutant background (Supplemental Fig. 4), suggesting that puf-9 does not function through let-7 miRNA processing and most likely acts synergistically with let-7 for example, to control downstream targets.
The alae and fusion defects seen in puf-9 mutants suggest that puf-9 activity is required for the proper timing of seam cell fusion and for the seam cells to differentiate and attain adult fates, but not for exit from the cell cycle. These results support the previously expressed idea that seam cell fusion, cell cycle exit, and alae formation are independently controlled developmental events (Banerjee et al., 2005; Lin et al., 2003). This is also consistent with our observations that removal of puf-9 activity resulted in a failure of some of the seam cells to fuse at the correct time, but did not show an observable reiteration of larval cell divisions in the hypodermal seam cells, as is seen in let-7 and lin-29 retarded loss-of-function mutants (Ambros, 1989; Papp et al., 1991; Reinhart et al., 2000). While a puf-9 mutation does not cause classic retarded heterochronic phenotypes on its own, it is possible that puf-9 still modulates the activity of genes in the heterochronic pathway.
puf-9 interacts genetically with the let-7 targets, lin-41 and hbl-1
To further establish a role for puf-9 in adult cuticle formation, we tested the ability of puf-9 to interact with additional heterochronic genes involved in the in the L/A switch. We hypothesized that if puf-9 was acting in a similar manner to let-7, then puf-9 might also interact genetically with the known let-7 downstream targets, lin-41 and hbl-1, in the hypodermis.
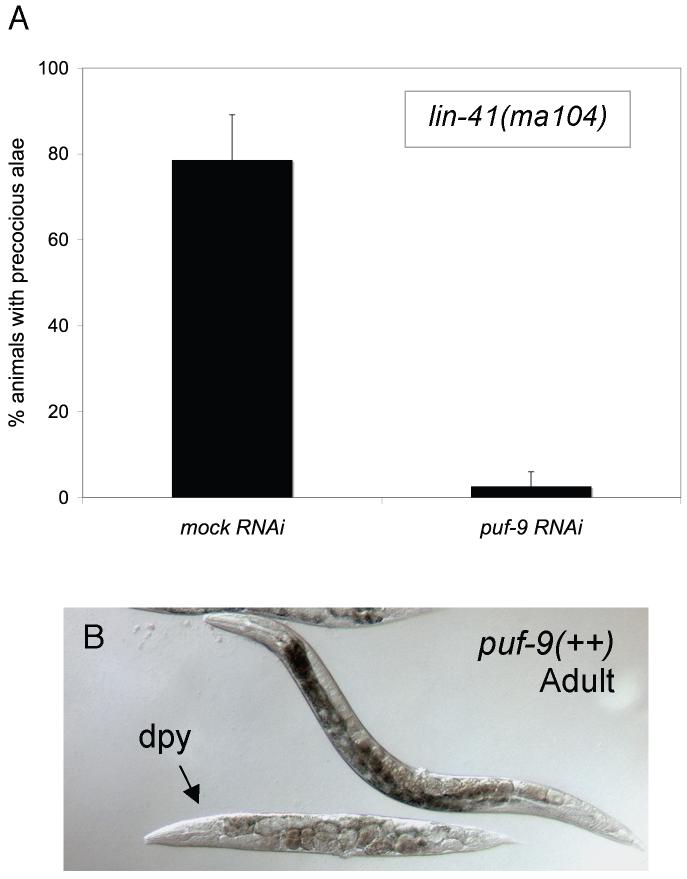
lin-41(RNAi) and the strong loss-of-function (ma104) allele display a partially penetrant alae defect in which alae are precociously observed at the L4 stage (Pasquinelli et al., 2000). Upon exposure to puf-9(RNAi), which was indistinguishable from puf-9(ok1136) in our previous work, the lin-41(ma104) precocious alae defect was completely suppressed (Fig. 3A). Therefore, the lin-41 precocious alae phenotype in the hypodermis requires a wild type copy of puf-9, suggesting that puf-9 acts downstream of, or in parallel to, lin-41 in the heterochronic pathway. Additionally, lin-41(RNAi) was unable to suppress puf-9(ok1136) adult alae defects (data not shown). This further supports our hypothesis that puf-9 is downstream of lin-41.
Figure 3.
puf-9(RNAi) by feeding suppresses lin-41(lf) precocious alae. (A) lin-41(ma104) animals exposed to puf-9(RNAi) (n=35) showed an almost complete reduction in the number of animals that displayed alae in the early L4 stage compared to animals on mock RNAi (n=21). (B) Full length puf-9::gfp over-expressing animals showed dumpy (Dpy) phenotypes reminiscent of lin-41 and hbl-1 loss-of-function mutants. The transgenic animal in (B) is indicated by an arrow.
Furthermore, animals carrying a full-length puf-9::gfp translational fusion construct often displayed a potential over-expression phenotype where the animals have a dumpy (Dpy) appearance (Fig. 3B). This Dpy phenotype is a characteristic of precocious heterochronic mutants, such as lin-41 and hbl-1 loss-of-function mutants, which could again signify that puf-9 is functioning in an opposing manner to both lin-41 and hbl-1, and acting similarly to let-7.
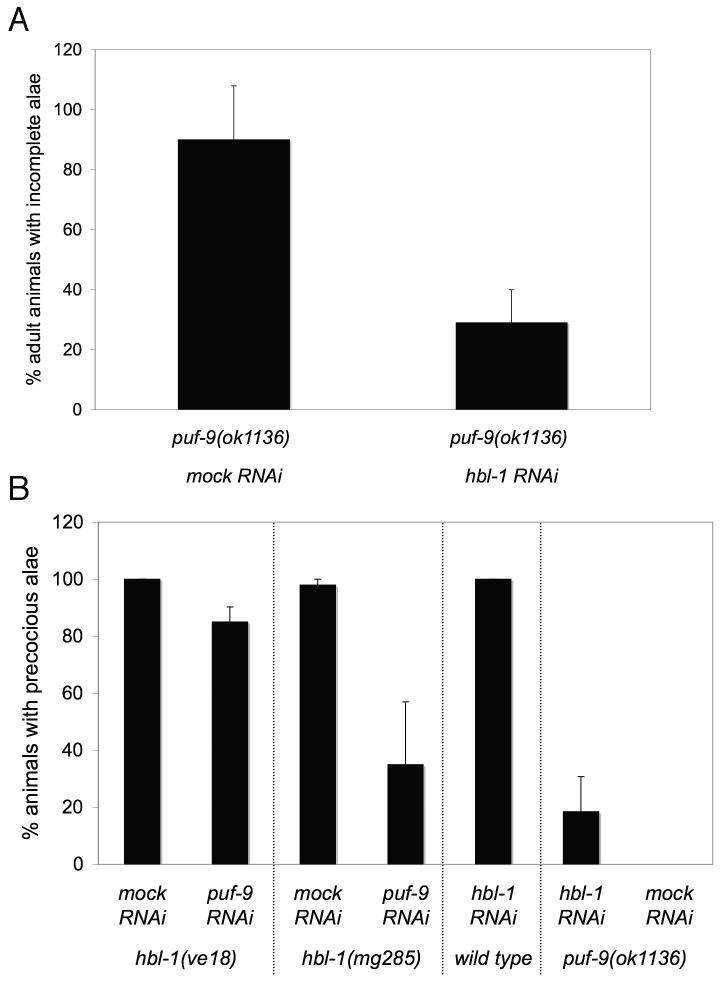
Previous reports showed that lin-41 and hbl-1 gene products can partially substitute for one another to regulate the L/A transition (Abrahante et al., 2003; Lin et al., 2003). Therefore, we tested our hypothesis that puf-9 could also act opposite to hbl-1 genetically, by looking at the effect of hbl-1 depletion on the adult alae phenotype of puf-9 mutants. We observed that hbl-1(RNAi) was able to suppress the adult alae phenotype of puf-9(ok1136) mutants (Fig. 4A). Thus, the puf-9 adult alae defects require a wild type copy of hbl-1. The reciprocal RNAi experiment showed that the majority of hbl-1(ve18) mutant animals (Abrahante et al., 2003) exposed to puf-9(RNAi) continued to display adult type alae in the early L4 stage characteristic of strong hbl-1(lf) mutants (left data set, Fig. 4B). Thus, puf-9(lf) is unable to suppress hbl-1(ve18) for the precocious alae defect, suggesting that hbl-1 acts downstream of, or in parallel to, puf-9 in the heterochronic pathway. This is consistent with our hypothesis that puf-9 acts to negatively regulate hbl-1 in a manner similar to let-7.
Figure 4.
hbl-1(RNAi) suppresses puf-9(ok1136) adult alae phenotypes while puf-9(RNAi) is unable to suppress a strong loss-of-function hbl-1 allele. (A) hbl-1(RNAi) suppresses the puf-9(ok1136) adult alae phenotypes. The number of puf-9(ok1136) animals displaying incomplete alae was decreased on hbl-1(RNAi) (n=55) compared to puf-9(ok1136) mutants alone (n=59). (B, left data sets) puf-9 is unable to suppress the hbl-1(ve18) precocious alae phenotype. hbl-1(ve18) mutants on mock RNAi (n=76) show a complete penetrance of precocious alae in the early L4 stage and nearly all hbl-1(ve18); puf-9(RNAi) animals (n=96) continued to display this precocious alae defect. (B, middle and right data sets) The precocious alae phenotypes of a weak hbl-1 allele and hbl-1(RNAi) are partially suppressed by removal of puf-9. hbl-1(mg285) animals exposed to puf-9(RNAi) (n=133) show a reduction in the number of animals displaying precocious alae at the early L4 stage, compared with mock RNAi (n=77). Similarly, puf-9(ok1136) mutants on hbl-1(RNAi) (n=75) suppressed the precocious alae seen with hbl-1(RNAi) exposure in a wild type background (n=69). puf-9(ok1136) mutant animals did not show any precocious alae on their own when fed mock RNAi (n=43). Error bars indicate standard deviations.
When the hbl-1(mg285) allele, or hbl-1(RNAi) by feeding was used to reduce hbl-1 levels, we found that a subsequent loss of puf-9 was able to partially suppress the formation of precocious alae (Fig. 4B). One explanation for the observed mutual suppression is that in a weak hbl-1 allele, loss of PUF-9 activity leads to overexpression of residual hbl-1 and thus restores partially active hbl-1 levels. It is possible that in the absence of PUF-9, overexpression of residual hbl-1 was able to prevent alae formation in a portion of the animals. When wild type animals are exposed to hbl-1(RNAi) during embryogenesis, a high degree of embryonic and larval lethality is observed (Fay et al., 1999). Therefore, it was necessary in our analysis to expose animals to hbl-1(RNAi) at the first larval stage, which could also allow for residual hbl-1 during development. Another possible explanation for the observed partial suppression of hbl-1(lf) preocious alae is that puf-9 likely suppresses additional targets, other than hbl-1, involved in promoting larval cell fates at the L/A switch. Therefore, in the hbl-1(lf);puf-9 double mutant, de-repression of additional puf-9 targets may be able to substitute for the loss of hbl-1 and, along with any residual hbl-1, could result in the failure to secrete alae and the observed suppression of the hbl-1 precocious alae phenotype. It is also possible that hbl-1 negatively feeds back on puf-9 activity or expression.
In order to determine epistatic relationships it is ideal to use null alleles. However, the puf-9 genetic interaction analysis described here was performed using a combination of genetic mutants and RNAi for several reasons. First, lin-41 null animals display a completely sterile phenotype, thus making this strain difficult to generate and maintain. Second, true hbl-1(null) alleles confer an embryonic lethal phenotype (Fay et al., 1999), which would make larval analysis of hbl-1(null) mutants impossible. Therefore strong loss-of-function alleles were used in combination with RNAi to carry out semi-epistatic genetic analysis with puf-9. Thus, due to the variability of RNAi and non-null alleles, interpretation of the genetic interactions presented here must take into consideration that residual gene activity could remain and that the proposed genetic pathway order may have alternative interpretations.
Taken together, our genetic data suggests a relationship among let-7, lin-41, hbl-1 and puf-9, where puf-9 negatively regulates hbl-1 and is itself negatively regulated by lin-41. Our observation that the bursting and alae phenotypes of let-7;puf-9 double mutants is more severe than either let-7 or puf-9 single mutants suggests that puf-9 acts in a parallel pathway to reinforce let-7 activity and to perhaps regulate downstream genes in the hypodermal seam cells. The placement of puf-9 upstream of hbl-1 brings up the interesting possibility that puf-9 may directly regulate hbl-1 in the heterochronic pathway.
A wild type copy of puf-9 is required for regulation of a hbl-1 3’UTR reporter in the seam cells
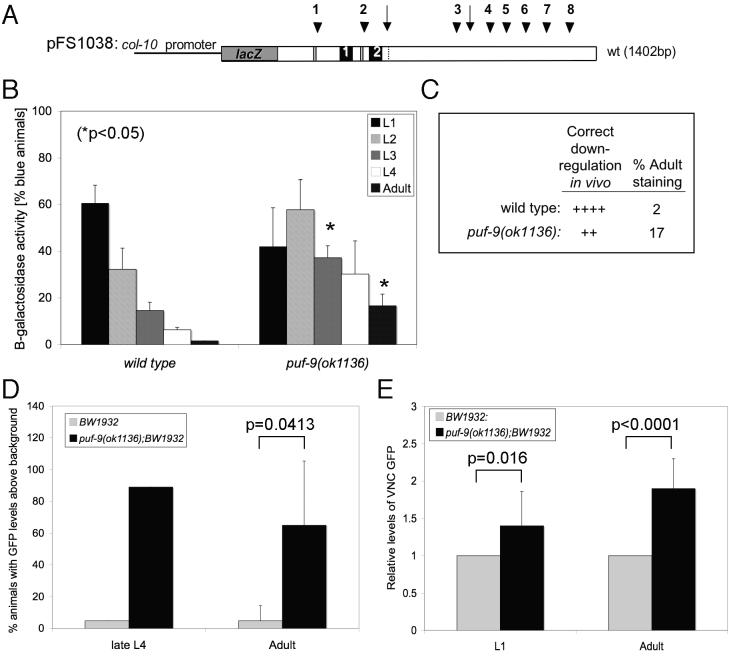
To test our hypothesis that hbl-1 is down-stream of and negatively regulated by puf-9 in the heterochronic pathway, we investigated whether the 3’UTR-mediated regulation of hbl-1 is dependent on a wild type copy of puf-9. A heterologous lacZ reporter gene, driven by the hypodermally expressed col-10 promoter and containing the full-length hbl-1 3’UTR (pFS1038, Fig. 5A) (Lin et al., 2003), was crossed into puf-9(ok1136). In a wild type background the hbl-1 3’UTR directs down-regulation of the reporter gene beginning in the L3 stage and continues down-regulation at later stages of development, such that β-galactosidase staining is almost completely undetectable by the adult stage (left data set, Fig. 5B and 5C) (Lin et al., 2003). In the puf-9(ok1136) mutant background, hbl-1 down-regulation was incomplete, such that puf-9(ok1136) mutant animals retained significant β-galactosidase activity at the adult stage (right data set, Fig. 5B and 5C). These data show that a wild type copy of puf-9 is necessary for the hbl-1 3’UTR directed regulation of a lacZ reporter gene in the hypodermis and suggests that PUF-9 likely regulates hbl-1 in vivo. The observed remaining partial regulation indicates that additional trans-factors are required to work with PUF-9 to fully regulate hbl-1 at the adult stage. We can conclude that trans-factors required for hbl-1 3’UTR regulation, including PUF-9, are present and active in the seam cells during the later larval and adult stages.
Figure 5.
A wild type copy of puf-9 is required for hbl-1 3’UTR reporter down-regulation in the hypodermis and the neurons. (A) A diagram of the heterologous reporter gene (pFS1038) used to assay hbl-1 3’UTR-directed regulation in the hypodermis, consisting of the wild type hbl-1 3’UTR fused downstream of the Escherichia coli lacZ gene and driven by the hypodermally expressed col-10 promoter (not to scale). (B, C) puf-9(ok1136) mutant animals carrying the pFS1038 reporter showed maintenance of β-galactosidase activity in the adult stage (17%), while the same construct in wild type animals showed expected levels of reporter gene down-regulation (2%) at the adult stage. Three independent staining trials were performed. The number of lines scored for each strain are, wild type: 1 line and puf-9(ok1136): 5 lines, n > 270 for all stages scored. For statistical analysis, an unpaired Student’s t test was performed between corresponding stages for the wild type and puf-9(ok1136) backgrounds. Error bars indicate standard deviations. (C) The correct down-regulation for each construct was compared to wild type and assigned values as follows: (++++) 0%−5% of animals with hypodermal expression at the adult stage; (+++) 5%−10%; (++) 10%−20%; (+) 20%−60%; (−) >60%. (D, E) A wild type copy of puf-9 is required for adult stage gfp::hbl-1 3’UTR reporter down-regulation in the VNC. (D) Late L4 (n=18) and adult stage (n=67) puf-9(ok1136) mutants showed an increase in the number of animals with bright HBL-1/GFP in the VNC compared to the same stages in wild type animals (L4, n=21 and adult, n=63). Wild type and puf-9(ok1136) animals were scored in three independent trials for adult GFP expression, and one trial for L4 GFP expression. (E) Quantification of wild type and puf-9(ok1136) GFP levels in the VNC was performed for a sample of L1 and adult stage animals. puf-9(ok1136) mutants showed a significant up-regulation of GFP at the L1 (n=8) and adult stages (n=12) compared to wild type animals (L1, n=4 and adult, n=10). The levels of GFP expression were calculated by normalizing the quantity of average GFP signal in puf-9(ok1136) mutants against the average GFP signal in wild type animals (for L1 and adult stages). Statistics were performed using an unpaired Student’s t test.
puf-9 is required for hbl-1 down-regulation in the VNC
To test the hypothesis that PUF-9 is also required in tissues where hbl-1 expression is known to be regulated at the post-transcriptional level (Abrahante et al., 2003; Fay et al., 1999; Lin et al., 2003), we utilized the BW1932 strain, which carries an integrated array consisting of the hbl-1 promoter driving expression of the first 133 amino acids of HBL-1 and a GFP reporter, followed by the hbl-1 3’UTR (Fay et al., 1999). The wild type GFP expression pattern in this strain showed a distinct temporal expression in the VNC. Beginning in the early L1 stage, the GFP signal progressively decreased until it was essentially undetectable by the late L4 stage (Abrahante et al., 2003; Lin et al., 2003). A failure to properly down-regulate hbl-1 expression in the VNC at the late L4 and adult stages was observed in a puf-9(ok1136) background where the majority of late L4 and adult stage animals expressed detectable GFP levels (Figs. 5D and Supplemental Fig. 5). Further quantitation of GFP levels in individual neurons from wild type and puf-9(ok1136) animals confirmed de-repression of HBL-1/GFP in puf-9(ok1136) adults (Fig. 5E). While VNC GFP expression was essentially the same in the puf-9(ok1136) and wild type backgrounds at the L1 stage, puf-9 mutant animals showed a significant increase in the relative amount of GFP signal in the VNC compared to wild type at the adult stage (Fig. 5E). The failure to appropriately down-regulate hbl-1::gfp demonstrates that a wild type copy of puf-9 is required in a subset of the neurons, as well as the hypodermis, for the correct 3’UTR mediated regulation of hbl-1.
Interestingly, Drosophila hunchback (hb) is temporally expressed during cortical laminar formation and is required to promote first born neuron fates (Cleary and Doe, 2006; Isshiki et al., 2001). Mis-expression of Hb in later born neuroblasts can drive the inappropriate transformation of neurons to the first born fate (Isshiki et al., 2001). Therefore, the appropriate expression of Hb is essential for proper neuron identity and cortical layering in Drosophila. However, the factors required for hb regulation during neurogenesis have not been identified. Our work could provide evidence for a conserved mechanism for hunchback regulation by Pumilio in neural tissues.
hbl-1 requires multiple cis-sequences in its 3’UTR for proper regulation
The dependence on PUF-9 for the proper 3’UTR-mediated regulation of hbl-1 brings up the interesting possibility that PUF-9 could bind directly to sequences within the hbl-1 3’UTR. Previous work identified many putative cis-regulatory elements in the hbl-1 3’UTR, including potential binding sites for the miRNAs let-7 (LCS) and lin-4 (LCE), and their family members (Abrahante et al., 2003; Lin et al., 2003). Additionally, the hbl-1 3’UTR contains sequences that resemble Nanos response elements (NREs) (Supplemental Fig. 6), which are known binding sites for Pumilio in Drosophila (Murata and Wharton, 1995; Wharton and Struhl, 1991), and represent potential Puf family binding sites.
We observed that a highly conserved 169 nt region of the hbl-1 3’UTR, containing two putative NRE sites (termed NRE1 and NRE2) flanking a predicted LCS and closely preceded by a potential LCE (Supplemental Fig. 6), which we called the “NRE region” was sufficient to form PUF-9 dependent complexes in vitro (Supplemental Fig. 7). This molecular data suggests that the NRE-region of the hbl-1 3’UTR binds, either directly or in a complex, to PUF-9 in vivo.
In order to determine the importance of the NRE region on hbl-1 3’UTR down-regulation in the hypodermis, we generated various deletions of the hbl-1 3’UTR (Fig. 6A) from the pFS1038 construct described above (Fig. 5A), and injected the constructs into wild type animals. Multiple independent lines were generated for each deletion construct and were scored for β-galactosidase activity in the hypodermis at all post-embryonic stages (Fig. 6B). We observed that inclusion of the intact NRE region (pSJA2 and pSJA5), containing the two NRE-like sequences and the LCS2 and LCE1 sites, lead to substantial reporter down-regulation, suggesting that a combination of NRE1, NRE2, and/or LCS 2 are able to mediate partial regulation of the hbl-1 3’UTR reporter gene at the adult stage. Removal of the majority of the hbl-1 3’UTR, such that only LCS1 remained (pSJA4), resulted in almost complete loss of reporter gene down-regulation, indicating that the LCS1 site and upstream sequences are not sufficient to direct regulation of the lacZ reporter in the hypodermis. The partial de-repression observed in pSJA2 and pSJA5 constructs indicates that sequences downstream of NRE2 are also necessary, in addition to the NRE region, for full hbl-1 3’UTR repression. This idea is supported by the staining pattern seen when just the NRE region by itself is deleted (pMJC2) where we observed that keeping the 3’UTR intact, outside of the NRE region, resulted in some adult stage down-regulation of the reporter.
Figure 6.
hbl-1 is regulated by multiple sequences, including the NRE region, in its 3’UTR. (A) Schematic of the hbl-1 3’UTR deletion constructs used to determine the necessary regulatory region of the hbl-1 3’UTR. Each construct shares a common col-10 promoter and contains the indicated portions of the Caenorhabditis elegans hbl-1 3’UTR fused to the Escherichia coli lacZ reporter gene. LCS are designated by arrowheads, with LCS 1 and LCS 2 specified by the double lines. LCE are shown as arrows, with LCE 1 designated as a dashed line. NRE1 and NRE2 are shown as black, numbered boxes within the construct. The values for correct down-regulation were assigned as in Figure 5. pFS1038 is the full-length reporter construct and contains the wild type 1402 bp hbl-1 3’UTR. (A, B) The deletion constructs all showed statistically significant up-regulation at the adult stage compared to wild type (students t test). The number of lines scored for each construct in (B) are pFS1038: 1 line (4 trials), pSJA2: 3 lines, pSJA3: 4 lines, pSJA4: 2 lines, pSJA5: 3 lines, and pMJC2: 4 lines, n > 50 for all stages scored. Error bars indicate standard deviations.
We have subsequently identified multiple miRNA complementary sites (LCS and LCE), as well as additional conserved NRE-like sequences downstream of the defined NRE region (Supplemental Fig 6A). Any number of these sites could be required in addition to the NRE region for complete down-regulation of hbl-1 at the adult stage. It is possible that PUF-9 and a let-7 family member are able to bind at multiple regulatory elements along the length of the hbl-1 3’UTR to mediate wild type down-regulation in vivo. It is also possible that other factors can partially substitute for puf-9 to regulate targets, such as hbl-1, in the seam cells.
Our hypothesis that sequences outside of the NRE region are important for hbl-1 regulation during development was further supported by the observation that mutating the individual NRE1 and NRE2 sequences resulted in levels of staining that were not significantly changed from wild type (Supplemental Fig. 8B). While there appeared to be a trend toward mis-regulation of the reporter at the adult stage in pMJC12 lines, the variability of staining between trials does not allow us to definitively conclude that there is significant adult up-regulation in the mutated transgenes.
Taken together, our data suggests that PUF-9 may regulate hbl-1 by binding to a region of the hbl-1 3’UTR that contains modified Nanos Response Elements (NRE1 and NRE2) and a let-7 family complementary site (LCS2). However, it is unclear whether PUF-9 binds directly to sequences in the NRE region, in vivo. It is possible that PUF-9 binds a combination of sites throughout the hbl-1 3’UTR to regulate hbl-1 expression in different tissue types. Given that let-7 and puf-9 individually repress hbl-1 expression via its 3’UTR (Abrahante et al., 2003; Lin et al., 2003), the genetic enhancement of let-7 by puf-9(RNAi), and the close association of LCS and NRE-like sequences in the hbl-1 3’UTR, it is intriguing to speculate that post-transcriptional hbl-1 regulation is achieved through cooperative binding of PUF-9 and a let-7 family member.
puf-9 is expressed in the hypodermis and the neurons
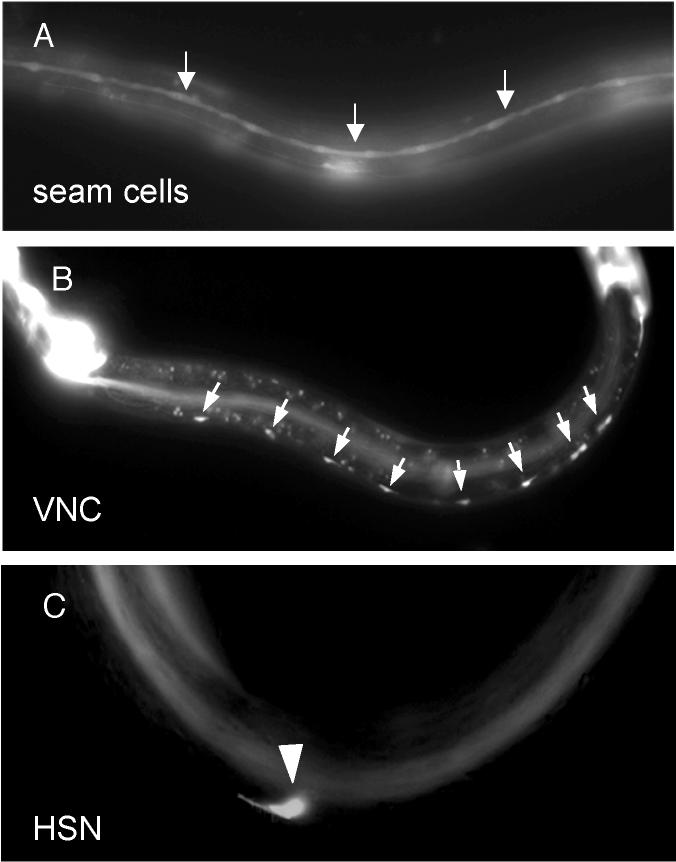
To see if puf-9 is expressed in tissues where hbl-1 is also expressed, we analyzed the post-embryonic expression pattern of a puf-9::gfp transcriptional fusion reporter. Promoter activity with distinct GFP expression in lateral hypodermal seam cells (Fig. 7A, arrows) and in the non-seam cell hypodermis (Fig. 7A, diffuse gfp) was observed throughout development. The observed hypodermal GFP expression pattern is consistent with the noted seam cell and alae defects in puf-9 mutants, supporting a function for puf-9 in these tissues. Additionally, expression was noted in many neurons, including the ventral nerve cord (VNC) (Figure 7B, arrows) and the hermaphrodite specific neuron (HSN) (Figure 7C, arrow head). GFP was also observed in various somatic gonad tissues including the anchor cell in larval stages and adult vulval muscle cells, the distal tip cells, a subset of the vulval precursor cells, uterine cells, and spermatheca (not shown). PUF-9/GFP translational fusion lines, containing the entire PUF-9 coding region, recapitulated the expression pattern seen in the transcriptional fusion lines described above, including in the hypodermis and neurons (not shown).
Fig. 7.
puf-9::gfp is expressed in the hypodermis and a subset of the neurons. Animals carrying a puf-9::gfp transcriptional fusion (MJC20) showed larval GFP expression in the lateral hypodermal seam cells (A, arrows) and non-seam cell hypodermis (A, diffuse staining). Additionally, GFP expression was noted in various neural cells including the ventral neural cord (VNC) (B, cell bodies marked by arrows), and the hermaphrodite specific neurons (HSN) (C, marked by an arrowhead).
The promoter driven expression pattern of puf-9 supports our hypothesis that PUF-9 regulates hbl-1 in both the hypodermis and the VNC. The puf-9::gfp constructs above do not appear to be temporally regulated. However, we cannot rule out possible post-transcriptional temporal regulation of PUF-9 expression.
Conclusions and implications
Puf mediated RNA regulation has been co-opted by many different organisms during evolution, illustrating the importance of this highly adaptable family of gene regulators. However, despite their widespread abundance, the molecular mechanisms by which Puf proteins regulate their target RNAs are still largely unknown. The work presented in this study could elucidate a general mechanism of translation regulation by Puf proteins, relevant to both the temporal and spatial control of gene expression.
Puf proteins have an evolutionarily conserved function in promoting stem cell proliferation and self-renewal (Crittenden et al., 2002; Wickens et al., 2002). For example, in the adult fly, a wild type copy of pumilio (pum) is required to maintain a population of germline stem cells (GSCs) and in Pum deficient animals, the GSCs will differentiate after a few rounds of division, leading to small or empty ovary phenotypes (Forbes and Lehmann, 1998). In pumilio mutant fly larvae, germline progenitor cells, called pole cells, fail to migrate to the presumptive gonad, exit their proliferative state, and prematurely reinitiate mitosis (Asaoka-Taguchi et al., 1999; Gilboa and Lehmann, 2004). Our work shows that in C. elegans, puf-9 may promote adult hypodermal fates by acting to repress genes, such as hbl-1, which prevent cell differentiation and promote continued cell divisions. Therefore, this work may give further insight into the role of Puf proteins during the regulation of stem cell fate determination.
The transcription factor hbl-1 is a candidate target of PUF-9 for a variety of reasons. First, hbl-1 is the homolog of Drosophila hunchback (Fay et al., 1999), which is a known target of repression by Pumilio in Drosophila. Second, hbl-1 and puf-9 expression coincides in many tissues, including the hypodermis (specifically the seam cells), the VNC and the HSN (Abrahante et al., 2003; Fay et al., 1999; Lin et al., 2003). Third, our genetic evidence and reporter gene assays indicate that PUF-9 negatively regulates hbl-1 in the hypodermis and the VNC. Finally, PUF-9 is able to interact in vitro with a region of the hbl-1 3’UTR containing potential Puf protein family binding sites.
In C. elegans there are four let-7 family members, including let-7, mir-48, mir-84, and mir-241 (Lim et al., 2003). However, since hbl-1::gfp is down-regulated by the L2 stage in the hypodermis (Abrahante et al., 2003; Fay et al., 1999; Lin et al., 2003) and let-7 RNA is not expressed until the L3 stage (Abbott et al., 2005; Esquela-Kerscher et al., 2005; Johnson et al., 2005; Johnson et al., 2003; Reinhart et al., 2000), other factors most likely contribute to the early regulation of hbl-1. In support of this, data from Lin et al. (2003) indicate that loss of let-7 had no detectable effect on early hbl-1 expression in the hypodermis. Therefore, it is possible that puf-9 may act with mir-48, mir-84, or mir-241 at early larval stages to repress a subset of RNA targets, including hbl-1, to control adult seam fates. At later stages, puf-9 may act with let-7 to regulate hbl-1 and additional targets required for the larval-to-adult transition.
Our genetic enhancement data supports the idea that let-7 and puf-9 work together to promote adult fates during C. elegans development. let-7 is highly conserved in both invertebrate and vertebrate organisms (Pasquinelli et al., 2000) and multiple let-7 homologs have been found in humans (Lim et al., 2003), underscoring the evolutionary importance of this regulatory molecule. Due to their high abundance and evolutionary conservation, the importance of miRNAs as translational regulators is apparent. However, the binding partners and exact mechanism by which miRNAs act to regulate their mRNA targets is still unclear. Therefore, finding binding partners that work with miRNAs may give insight into the mechanism by which these regulatory RNAs act on their gene targets. Our data suggests that the important translational regulatory families, Pumilios and microRNAs, may cooperate at the 3’UTRs of target genes to control their translation.
Supplementary Material
puf-9(RNAi) enhances the bursting phenotype of let-7(n2853ts) mutants. (A) Single and combinatorial RNAi depletion of each of the nos and puf genes led to the observation that the knockdown of only puf-9 enhanced the vulva bursting phenotype seen in let-7(n2853ts) mutant animals. None of the other nos or puf genes tested interacted with let-7 mutants in this assay and gave similar levels of survival as let-7 animals on mock RNAi at all temperatures tested (n > 150 animals scored for all RNAi constructs tested). (B) The nos genes do not contribute to puf-9 enhancement of let-7(n2853ts) bursting phenotypes. let-7(n2853ts) animals grown on RNAi simultaneously depleting puf-9 and nos-1, nos-2, and nos-3 (collectively designated as nos) show similar adult survival compared to let-7(n2853ts) animals grown on puf-9(RNAi) alone. Further, let-7 animals exposed to RNAi for the three nos genes gave similar levels of adult survival as let-7(n2853ts) animals on mock RNAi, at the permissive temperature. (n > 240 animals scored for all RNAi constructs tested).
The puf-9 gene product shares high similarity with other Puf proteins and is most closely related to Drosophila Pumilio. (A) A cartoon showing the PUF-9 C-terminal Pumilio homology domain (Pum-HD) (indicated in yellow) in relation to the full-length 703 aa protein product. (B) A pile-up of the C-terminal RNA binding domain of PUF-9 compared to representative Puf proteins from flies, humans, and C. elegans. The eight Pum-HD repeats are numbered in parenthesis above the repeats and identical residues are shaded in black. Sequence homology was determined by ClustalW in the MegAlign program, using default settings. (C) Phylogenetic dendogram of Puf family proteins, including PUF-9, Drosophila Pumilio, Human PumH1, and the C. elegans PUF-8 and FBF-1. PUF-9 is more similar to Drosophila Pumilio, and Human PUMH1 than to the other C. elegans Puf proteins, PUF-8 and FBF-1, as determined by Clustal W analysis in the Megalign program using the full-length amino acid sequences.
puf-9 deletion mutants display abnormal adult alae phenotypes. High resolution images of the pictures shown in Fig. 2. Nomarski DIC images of adult stage alae in a wild type animal (A, alae marked with arrows) and an adult stage puf-9(ok1136) animal showing incomplete alae (B, alae marked with arrows and the alae break marked with arrow head).
By Northern analysis, let-7 miRNA levels are not decreased in a puf-9(ok1136) mutant background. Lane 1, wild type; Lane 2, puf-9(ok1136). Phosphoimager signal intensities were determined using ImageQuant software. The puf-9(ok1136) signal is represented as a relative value compared to wild type for both let-7 and U6 probes. The same blot was stripped, following let-7 probe hybridization, and re-probed with U6 as a loading control for total RNA and Ethidium bromide staining is shown for the same gel, done prior to transfer.
puf-9 mutants showed a higher number of animals with GFP above background levels at all stages scored compared to wild type. For each stage, GFP brightness was scored on a scale of 1 to 5, with Level 1 = background and Level 5 = brightest expression. The values shown represent the percentage of animals with an assigned level of GFP expression in the VNC for each developmental stage scored. Wild type (BW1932) animals are grouped on the top half of the chart and puf-9(ok1136);BW1932 animals are grouped on the bottom half.
The hbl-1 3’UTR contains many cis-sequences that are conserved with a divergent nematode species. (A) A diagram of conserved potential regulatory cis-sequences within the hbl-1 3’UTR. let-7 family member complementary sites (LCS) are marked by arrow heads, and lin-4 complementary sites (LCE) are shown as arrows. Putative Puf family binding sites (NRE) are represented by blue boxes. Sequences are given as approximate locations along the 3’UTR and are not drawn to scale. The detailed region shows a sequence alignment of a portion of the hbl-1 3’UTR containing a let-7 family complementary site (LCS2) closely flanked by two modified NRE-like sequences (NRE1 and NRE2, large blue boxes). This region of the hbl-1 3’UTR shows high conservation with C. briggsae (C. elegans sequence on top and C. briggsae sequence on bottom) with identical nucleotides shown in the middle line. The assigned NRE1 and NRE2 sequences are shaded in blue with the Box A underlined in red and Box B outlined in red. Non-canonical Box A and Box B sequences are indicated by an asterisk. Note that the sequence is represented as DNA. (B) The canonical Drosophila hunchback NRE site consists of a bipartite 36 nucleotide sequence, composed of two conserved regions (Box A and Box B) separated by a divergent 6 nucleotide spacer. The minimal canonical Box A and Box B sequences are shown.
A labeled sense RNA probe containing the NRE-region of the hbl-1 3’UTR was able to direct a shift when incubated with cell extract from wild type (N2) animals, but not puf-9(RNAi) depleted extract. Gel retardation assays. 32P-labeled hbl-1 3’UTR RNA was incubated with 0, 2, or 4 ug of total protein from wild type or puf-9(ok1136) mixed stage animals. Three different complexes were noted and are indicated by brackets; arrow, free RNA. Complexes 1 and 2 were visibly reduced when the same hbl-1 3’UTR probe was incubated with extract from wild type animals exposed to puf-9(RNAi).
Sequences other than NRE1 and NRE2 can facilitate hbl-1 3’UTR down-regulation. (A) pMJC12 has three nucleotides each in NRE 1 and NRE 2 mutated within the full hbl-1 3’UTR. The sequences of the NRE 1 and NRE 2 Box B sites (underlined in the enlarged sequence region) are shown surrounding the intervening LCS 2 site (boxed area). The ‘UGU’ in each NRE Box B sequence was mutated to ‘AAA’ (the mutated residues are indicated by red stars). (B) Mutating the NRE sites in the context of the 3’UTR (pMJC12) showed a trend toward increased hypodermal staining at the later developmental stages compared to wild type. However, the error bars are large and make interpretation of this data inconclusive. The amount of reporter expression at each stage (L3-adult) was normalized relative to the combined L1/L2 stages for each individual construct. The number of lines scored for each construct in (B) are pFS1038: 1 line (3 trials), pMJC12: 2 lines. Error bars indicate standard deviations.
Sequences of the oligonucleotide primers used to create the various plasmids and PCR products used in this work.
Acknowledgements
Grant Information: NIH: GM62594 (to FJS) and NIH: GM64701 (to FJS).
We thank A. Esquela-Kerscher and M. Boehm for critical reading of this manuscript. We also thank the C. elegans Gene Knockout Consortium and the Caenorhabditis Genetics Center for strains. The nos and puf RNAi bacteria strains were generously given to us by K. Subramaniam and G. Seydoux. The W06B11 cosmid was kindly provided by Dr. Alan Coulson at the Wellcome Trust Sanger Institute. We also thank Sharon Moulis and Stephanie Arnoldi for plasmid construction. This work was supported by grants from the National Institutes of Health to FJS (GM62594 and GM64701).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, Horvitz HR, Ambros V. The let-7 MicroRNA Family Members mir-48, mir-84, and mir-241 Function Together to Regulate Developmental Timing in Caenorhabditis elegans. Dev Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahante JE, Daul AL, Li M, Volk ML, Tennessen JM, Miller EA, Rougvie AE. The Caenorhabditis elegans hunchback-like gene lin-57/hbl-1 controls developmental time and is regulated by microRNAs. Dev Cell. 2003;4:625–637. doi: 10.1016/s1534-5807(03)00127-8. [DOI] [PubMed] [Google Scholar]
- Ambros V. A hierarchy of regulatory genes controls a larva-to-adult developmental switch in C. elegans. Cell. 1989;57:49–57. doi: 10.1016/0092-8674(89)90171-2. [DOI] [PubMed] [Google Scholar]
- Ambros V, Horvitz HR. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat Cell Biol. 1999;1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Kwok A, Lin SY, Slack FJ. Developmental timing in C. elegans is regulated by kin-20 and tim-1, homologs of core circadian clock genes. Dev Cell. 2005;8:287–295. doi: 10.1016/j.devcel.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Banerjee D, Slack F. Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays. 2002;24:119–129. doi: 10.1002/bies.10046. [DOI] [PubMed] [Google Scholar]
- Bettinger JC, Lee K, Rougvie AE. Stage-specific accumulation of the terminal differentiation factor LIN-29 during Caenorhabditis elegans development. Development. 1996;122:2517–2527. doi: 10.1242/dev.122.8.2517. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-Step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cleary MD, Doe CQ. Regulation of neuroblast competence: multiple temporal identity factors specify distinct neuronal fates within a single early competence window. Genes Dev. 2006;20:429–434. doi: 10.1101/gad.1382206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden SL, Bernstein DS, Bachorik JL, Thompson BE, Gallegos M, Petcherski AG, Moulder G, Barstead R, Wickens M, Kimble J. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature. 2002;417:660–663. doi: 10.1038/nature754. [DOI] [PubMed] [Google Scholar]
- Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Johnson SM, Bai L, Saito K, Partridge J, Reinert KL, Slack FJ. Post-embryonic expression of C. elegans microRNAs belonging to the lin-4 and let-7 families in the hypodermis and the reproductive system. Dev Dyn. 2005;234:868–877. doi: 10.1002/dvdy.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay DS, Stanley HM, Han M, Wood WB. A Caenorhabditis elegans homologue of hunchback is required for late stages of development but not early embryonic patterning. Dev Biol. 1999;205:240–253. doi: 10.1006/dbio.1998.9096. [DOI] [PubMed] [Google Scholar]
- Forbes A, Lehmann R. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development. 1998;125:679–690. doi: 10.1242/dev.125.4.679. [DOI] [PubMed] [Google Scholar]
- Gamberi C, Peterson DS, He L, Gottlieb E. An anterior function for the Drosophila posterior determinant Pumilio. Development. 2002;129:2699–2710. doi: 10.1242/dev.129.11.2699. [DOI] [PubMed] [Google Scholar]
- Gilboa L, Lehmann R. Repression of primordial germ cell differentiation parallels germ line stem cell maintenance. Curr Biol. 2004;14:981–986. doi: 10.1016/j.cub.2004.05.049. [DOI] [PubMed] [Google Scholar]
- Irish V, Lehmann R, Akam M. The Drosophila posterior-group gene nanos functions by repressing hunchback activity. Nature. 1989;338:646–648. doi: 10.1038/338646a0. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnson SM, Lin SY, Slack FJ. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, Welchman DP, Zipperlen P, Ahringer J. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Lamont LB, Crittenden SL, Bernstein D, Wickens M, Kimble J. FBF-1 and FBF-2 regulate the size of the mitotic region in the C. elegans germline. Dev Cell. 2004;7:697–707. doi: 10.1016/j.devcel.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Weinstein EG, Abdelhakim A, Yekta S, Rhoades MW, Burge CB, Bartel DP. The microRNAs of Caenorhabditis elegans. Genes Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SY, Johnson SM, Abraham M, Vella MC, Pasquinelli A, Gamberi C, Gottlieb E, Slack FJ. The C elegans hunchback homolog, hbl-1, controls temporal patterning and is a probable microRNA target. Dev Cell. 2003;4:639–650. doi: 10.1016/s1534-5807(03)00124-2. [DOI] [PubMed] [Google Scholar]
- Murata Y, Wharton RP. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell. 1995;80:747–756. doi: 10.1016/0092-8674(95)90353-4. [DOI] [PubMed] [Google Scholar]
- Papp A, Rougvie AE, Ambros V. Molecular cloning of lin-29, a heterochronic gene required for the differentiation of hypodermal cells and the cessation of molting in C.elegans. Nucleic Acids Res. 1991;19:623–630. doi: 10.1093/nar/19.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Basson M, Liu Z, Ambros V, Horvitz HR, Ruvkun G. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–669. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- Slack FJ, Ruvkun G. A novel repeat domain that is often associated with RING finger and B-box motifs. Trends Biochem Sci. 1998;23:474–475. doi: 10.1016/s0968-0004(98)01299-7. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Wharton RP. Drosophila Brain Tumor is a translational repressor. Genes Dev. 2001;15:762–773. doi: 10.1101/gad.870801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassov DS, Jurecic R. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life. 2003;55:359–366. doi: 10.1080/15216540310001603093. [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development. 1999;126:4861–4871. doi: 10.1242/dev.126.21.4861. [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G. Dedifferentiation of primary spermatocytes into germ cell tumors in C. elegans lacking the pumilio-like protein PUF-8. Curr Biol. 2003;13:134–139. doi: 10.1016/s0960-9822(03)00005-8. [DOI] [PubMed] [Google Scholar]
- Tautz D. Regulation of the Drosophila segmentation gene hunchback by two maternal morphogenetic centres. Nature. 1988;332:281–284. doi: 10.1038/332281a0. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3'UTR. Genes Dev. 2004a;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella MC, Reinert K, Slack FJ. Architecture of a validated microRNA::target interaction. Chem Biol. 2004b;11:1619–1623. doi: 10.1016/j.chembiol.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell. 2001;7:855–865. doi: 10.1016/s1097-2765(01)00229-5. [DOI] [PubMed] [Google Scholar]
- Wharton RP, Struhl G. RNA regulatory elements mediate control of Drosophila body pattern by the posterior morphogen nanos. Cell. 1991;67:955–967. doi: 10.1016/0092-8674(91)90368-9. [DOI] [PubMed] [Google Scholar]
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- Wu X, Vasisht V, Kosman D, Reinitz J, Small S. Thoracic patterning by the Drosophila gap gene hunchback. Dev Biol. 2001;237:79–92. doi: 10.1006/dbio.2001.0355. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Williamson JR, Lehmann R. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. Rna. 1997;3:1421–1433. [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gallegos M, Puoti A, Durkin E, Fields S, Kimble J, Wickens MP. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature. 1997;390:477–484. doi: 10.1038/37297. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
puf-9(RNAi) enhances the bursting phenotype of let-7(n2853ts) mutants. (A) Single and combinatorial RNAi depletion of each of the nos and puf genes led to the observation that the knockdown of only puf-9 enhanced the vulva bursting phenotype seen in let-7(n2853ts) mutant animals. None of the other nos or puf genes tested interacted with let-7 mutants in this assay and gave similar levels of survival as let-7 animals on mock RNAi at all temperatures tested (n > 150 animals scored for all RNAi constructs tested). (B) The nos genes do not contribute to puf-9 enhancement of let-7(n2853ts) bursting phenotypes. let-7(n2853ts) animals grown on RNAi simultaneously depleting puf-9 and nos-1, nos-2, and nos-3 (collectively designated as nos) show similar adult survival compared to let-7(n2853ts) animals grown on puf-9(RNAi) alone. Further, let-7 animals exposed to RNAi for the three nos genes gave similar levels of adult survival as let-7(n2853ts) animals on mock RNAi, at the permissive temperature. (n > 240 animals scored for all RNAi constructs tested).
The puf-9 gene product shares high similarity with other Puf proteins and is most closely related to Drosophila Pumilio. (A) A cartoon showing the PUF-9 C-terminal Pumilio homology domain (Pum-HD) (indicated in yellow) in relation to the full-length 703 aa protein product. (B) A pile-up of the C-terminal RNA binding domain of PUF-9 compared to representative Puf proteins from flies, humans, and C. elegans. The eight Pum-HD repeats are numbered in parenthesis above the repeats and identical residues are shaded in black. Sequence homology was determined by ClustalW in the MegAlign program, using default settings. (C) Phylogenetic dendogram of Puf family proteins, including PUF-9, Drosophila Pumilio, Human PumH1, and the C. elegans PUF-8 and FBF-1. PUF-9 is more similar to Drosophila Pumilio, and Human PUMH1 than to the other C. elegans Puf proteins, PUF-8 and FBF-1, as determined by Clustal W analysis in the Megalign program using the full-length amino acid sequences.
puf-9 deletion mutants display abnormal adult alae phenotypes. High resolution images of the pictures shown in Fig. 2. Nomarski DIC images of adult stage alae in a wild type animal (A, alae marked with arrows) and an adult stage puf-9(ok1136) animal showing incomplete alae (B, alae marked with arrows and the alae break marked with arrow head).
By Northern analysis, let-7 miRNA levels are not decreased in a puf-9(ok1136) mutant background. Lane 1, wild type; Lane 2, puf-9(ok1136). Phosphoimager signal intensities were determined using ImageQuant software. The puf-9(ok1136) signal is represented as a relative value compared to wild type for both let-7 and U6 probes. The same blot was stripped, following let-7 probe hybridization, and re-probed with U6 as a loading control for total RNA and Ethidium bromide staining is shown for the same gel, done prior to transfer.
puf-9 mutants showed a higher number of animals with GFP above background levels at all stages scored compared to wild type. For each stage, GFP brightness was scored on a scale of 1 to 5, with Level 1 = background and Level 5 = brightest expression. The values shown represent the percentage of animals with an assigned level of GFP expression in the VNC for each developmental stage scored. Wild type (BW1932) animals are grouped on the top half of the chart and puf-9(ok1136);BW1932 animals are grouped on the bottom half.
The hbl-1 3’UTR contains many cis-sequences that are conserved with a divergent nematode species. (A) A diagram of conserved potential regulatory cis-sequences within the hbl-1 3’UTR. let-7 family member complementary sites (LCS) are marked by arrow heads, and lin-4 complementary sites (LCE) are shown as arrows. Putative Puf family binding sites (NRE) are represented by blue boxes. Sequences are given as approximate locations along the 3’UTR and are not drawn to scale. The detailed region shows a sequence alignment of a portion of the hbl-1 3’UTR containing a let-7 family complementary site (LCS2) closely flanked by two modified NRE-like sequences (NRE1 and NRE2, large blue boxes). This region of the hbl-1 3’UTR shows high conservation with C. briggsae (C. elegans sequence on top and C. briggsae sequence on bottom) with identical nucleotides shown in the middle line. The assigned NRE1 and NRE2 sequences are shaded in blue with the Box A underlined in red and Box B outlined in red. Non-canonical Box A and Box B sequences are indicated by an asterisk. Note that the sequence is represented as DNA. (B) The canonical Drosophila hunchback NRE site consists of a bipartite 36 nucleotide sequence, composed of two conserved regions (Box A and Box B) separated by a divergent 6 nucleotide spacer. The minimal canonical Box A and Box B sequences are shown.
A labeled sense RNA probe containing the NRE-region of the hbl-1 3’UTR was able to direct a shift when incubated with cell extract from wild type (N2) animals, but not puf-9(RNAi) depleted extract. Gel retardation assays. 32P-labeled hbl-1 3’UTR RNA was incubated with 0, 2, or 4 ug of total protein from wild type or puf-9(ok1136) mixed stage animals. Three different complexes were noted and are indicated by brackets; arrow, free RNA. Complexes 1 and 2 were visibly reduced when the same hbl-1 3’UTR probe was incubated with extract from wild type animals exposed to puf-9(RNAi).
Sequences other than NRE1 and NRE2 can facilitate hbl-1 3’UTR down-regulation. (A) pMJC12 has three nucleotides each in NRE 1 and NRE 2 mutated within the full hbl-1 3’UTR. The sequences of the NRE 1 and NRE 2 Box B sites (underlined in the enlarged sequence region) are shown surrounding the intervening LCS 2 site (boxed area). The ‘UGU’ in each NRE Box B sequence was mutated to ‘AAA’ (the mutated residues are indicated by red stars). (B) Mutating the NRE sites in the context of the 3’UTR (pMJC12) showed a trend toward increased hypodermal staining at the later developmental stages compared to wild type. However, the error bars are large and make interpretation of this data inconclusive. The amount of reporter expression at each stage (L3-adult) was normalized relative to the combined L1/L2 stages for each individual construct. The number of lines scored for each construct in (B) are pFS1038: 1 line (3 trials), pMJC12: 2 lines. Error bars indicate standard deviations.
Sequences of the oligonucleotide primers used to create the various plasmids and PCR products used in this work.