Abstract
Postsynaptic density protein 95 (PSD-95), a specialized scaffold protein with multiple protein interaction domains, forms the backbone of an extensive postsynaptic protein complex that organizes receptors and signal transduction molecules at the synaptic contact zone. Large, detergent-insoluble PSD-95-based postsynaptic complexes can be affinity-purified from conventional PSD fractions using magnetic beads coated with a PSD-95 antibody. In the present study purified PSD-95 complexes were analyzed by LC/MS/MS. A semiquantitative measure of the relative abundances of proteins in the purified PSD-95 complexes and the parent PSD fraction was estimated based on the cumulative ion current intensities of corresponding peptides. The affinity-purified preparation was largely depleted of presynaptic proteins, spectrin, intermediate filaments, and other contaminants prominent in the parent PSD fraction. We identified 525 of the proteins previously reported in parent PSD fractions, but only 288 of these were detected after affinity purification. We discuss 26 proteins that are major components in the PSD-95 complex based upon abundance ranking and affinity co-purification with PSD-95. This subset represents a minimal list of constituent proteins of the PSD-95 complex and includes, in addition to the specialized scaffolds and N-methyl-d-aspartate (NMDA) receptors, an abundance of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) receptors, small G-protein regulators, cell adhesion molecules, and hypothetical proteins. The identification of two Arf regulators, BRAG1 and BRAG2b, as co-purifying components of the complex implies pivotal functions in spine plasticity such as the reorganization of the actin cytoskeleton and insertion and retrieval of proteins to and from the plasma membrane. Another co-purifying protein (Q8BZM2) with two sterile α motif domains may represent a novel structural core element of the PSD.
The postsynaptic density (PSD)1 is a disk-shaped protein complex lining the postsynaptic membrane. In a recent study its total mass was estimated to be around 1 million kDa (1). The function of this massive protein complex appears to be anchoring and organizing postsynaptic neurotransmitter receptors and corresponding signaling molecules at the active zone. Thus, it is expected that the extent and type of the postsynaptic response to neurotransmitter release will largely depend on the molecular composition and organization of the PSD.
The first tentative identification of PSD components was done in the late 1970s when several laboratories developed the methodology to isolate PSD fractions and started analyzing them by biochemical methods (2, 3). The general strategy, still applied today, was treatment of synaptosomal fractions with detergents that solubilize membranes but leave the PSD relatively intact and subsequent separation of membrane-free PSDs by further centrifugation. Analysis of PSD fractions continued to reveal additional putative PSD components in later years (4) and gained new momentum with the introduction of mass spectrometric techniques (5–14). The development of two-hybrid screens and other methods to determine binding partners of already identified components added important complementary approaches (for a review, see Ref. 15).
The global proteomics analysis of isolated PSDs remains a crucial first step in the elucidation of the molecular structure of the PSD. Indeed the strategy constitutes a relatively simple and convenient way for the identification of hundreds of proteins in a single run: one of the most recent studies identified a total of 1264 proteins (13). Also unlike immunological approaches, the strategy is not based on any a priori notion of PSD constituents and therefore can reveal hitherto unsuspected elements. A recent study (16) integrated data from seven proteomics studies (5–9, 17) and other literature on the analysis of PSD fractions. Collins et al. (16) report that altogether 1124 proteins were identified in these seven studies. However, 58% of the proteins were detected in only one study, raising the possibility of a high rate of false positives. The authors also compiled a “consensus PSD” list of 467 proteins that were identified in at least two of the studies, thus reducing the probability of false positives linked to individual protocols.
Although the 467 proteins in the consensus PSD list have a better probability of being genuine PSD components, detection in multiple studies does not necessarily prove that they are. In fact, there seem to be only a handful of proteins that were identified in all seven of the studies (Ref. 16 and Supplemental Tables S2 and S3 in Ref. 16), and this high consensus group includes, in addition to the expected PSD components such as PSD-95, homer, and CaMKII, likely contaminants such as synapsins and intermediate filaments. Moreover because the probability of detection of a protein in a mixture increases with its relative abundance, it is likely that the contaminants in this group are among the most abundant in PSD fractions.
Detection of the same contaminants in all proteomics studies suggests a systemic contamination problem in PSD fractions in general. The most widely applied protocol for the preparation of PSD fractions is the one originally developed by Carlin et al. (18) and uses the relatively mild detergent Triton X-100. Because Triton X-100 is mild, it appears to cause minimal loss of protein from the PSD as judged from morphological criteria. On the other hand, many proteins that are present in the synaptosomal fraction but are not part of the PSD also appear to be resistant to Triton X-100. Indeed electron microscopy (EM) analyses of PSD fractions prepared by this method reveal various particulate contaminants including filamentous material and other protein complexes (19, 20). Some of the filamentous materials that are not associated with the PSD have been identified as neurofilaments and spectrin (20).
Altogether these considerations indicate the need for a purer preparation as a basis for the identification of PSD components. The use of a stronger detergent such as N-laurylsarcosinate (21) eliminates certain contaminants but also appears to dissociate some genuine PSD elements as well. In addition, certain contaminants such as the so-called “CaMKII clusters” that are resistant to the detergent become enriched in the N-laurylsarcosinate-derived PSD fraction (19). On the other hand, because most particulate contaminants in the PSD fraction are membrane-free protein complexes like the PSDs themselves, they are expected to be of the same density as PSDs, excluding the possibility of further fractionation by conventional centrifugation-based techniques.
Affinity-based separation techniques targeting ubiquitous PSD components constitute an orthogonal approach for the isolation of postsynaptic protein complexes, a strategy expected to avoid the problem of detergent-insoluble particulate material contamination prevalent in conventional density-based PSD preparations. Using this approach the isolation of NMDA receptor and PSD-95 complexes from whole cell deoxycholate extracts has been reported (22, 23). However, because these preparations are from detergent-solubilized starting material, isolated complexes tend to be small. Indeed the molecular mass of NMDA receptor complexes was estimated to be around 2000 kDa, which is about 3 orders of magnitude smaller than the whole PSD complex. Thus, the preparation presumably contains receptor complexes that become dissociated from the bigger PSD complex upon treatment with the relatively strong detergent deoxycholate. In addition, because whole cell extracts are used as starting material, complexes that originate from extrasynaptic/intracellular pools are expected to co-purify with complexes from the PSD. These may include extrasynaptic receptors and transport packages.
We evaluated an affinity-based strategy for the isolation of large PSD-95-containing complexes from the Triton X-100-derived PSD fraction (20). The strategy aims to minimize the contribution from extrasynaptic/intracellular PSD-95 pools by starting with a PSD-enriched fraction from which such non-synaptic complexes have been largely eliminated. Importantly by using magnetic beads coated with a PSD-95 antibody, large, insoluble complexes are separated without the use of additional detergents to solubilize particulates.
Postsynaptic PSD-95 complexes are likely to represent the bulk of the PSD observed by EM in intact cells. Indeed PSD-95 is able to interact with a large number of proteins through its three PDZ domains, an SH3 domain, and a guanylate kinase domain and also can multimerize to form an extended scaffold (for a review, see Ref. 15). Moreover proteins that bind PSD-95 in turn interact with yet other postsynaptic components, thus extending the network. The complexes isolated using PSD-95 antibody-coated magnetic beads resemble in situ PSDs (20), suggesting that most if not all of the original components have been retained. These considerations suggested that analysis of the isolated PSD-95 complexes would identify most if not all of the components of the in situ PSD.
In the present study, isolated PSD-95 complexes and the parent PSD fraction were analyzed in parallel by liquid chromatography coupled to tandem mass spectrometry. For each fraction, identified proteins were ranked according to cumulative ion current intensities of corresponding peptides as a semiquantitative measure of relative abundance. Comparison of the ranks of proteins in the parent and affinity-purified preparations allowed identification of those that co-purify with PSD-95 and, importantly, those that are likely contaminants.
MATERIALS AND METHODS
Preparation of the PSD Fraction and Isolation of PSD-95 Complex
The PSD fraction was prepared essentially by the method of Carlin et al. (18) with modifications as described previously (19). Brains from adult Sprague-Dawley rats were custom collected by Pel Freez Biologicals (Rogers, AR), frozen immediately in liquid nitrogen, and shipped on dry ice. Brains were thawed by a 1-min immersion in isotonic sucrose at 37 °C and dissected on ice to remove white matter, and cerebral cortices were then rapidly homogenized. A synaptosomal fraction was obtained and treated with 0.5% Triton X-100. Detergent-insoluble pellets were collected, and a PSD-enriched fraction was separated by sucrose density centrifugation. This fraction was extracted once more with 0.5% Triton X-100, 75 mm KCl.
Isolation of PSD-95 complexes was as described previously (20) with some modifications. Dynabeads (M-450 coated with goat anti-mouse IgG) were obtained from Dynal (Oslo, Norway) and were further coated with a monoclonal PSD-95 antibody (MA1– 046; ABR-Affinity BioReagents, Inc., Golden, CO). PSD fraction (200 μg of protein) was resuspended in 10 ml of 2% BSA (of 99% purity to minimize contamination of the preparation by the blocker), 0.01% Tween 20 in phosphate-buffered saline using a probe sonicator. Sonication (3 × 1 min with ∼3 min cooling intervals in ice) was done at the lowest power setting (<5 watts) taking care not to warm the samples. Resuspended PSD fraction (200 μg) was incubated with antibody-coated magnetic beads (2 × 108 beads) in a final volume of 10 ml for 2 h at 4 °C with continuous rotation. Unbound material was removed, and the beads were washed three times with 2% BSA, 0.01% Tween 20 in phosphate-buffered saline and five times with 0.01% Tween 20 in phosphate-buffered saline for a total of ∼3 h.
Fractionation and Mass Spectrometric Analysis of Preparations
The parent PSD fraction or washed beads with attached complexes were treated with SDS-containing electrophoresis sample buffer and were incubated in boiling water for 5 min. Solubilized affinity-purified samples corresponding to the yield from ∼100 μg of starting PSD fraction were loaded onto individual gel lanes. Gels were stained with Coomassie Brilliant Blue to visualize bands for imaging and then sectioned from the top of the gel to the bottom in 2-mm increments, and the pieces were placed in clean Eppendorf tubes. In-gel tryptic digestion was performed after two washes with 50% acetonitrile in 100 mm ammonium bicarbonate, and dehydration of gel slices was performed with the addition of 100% acetonitrile. Disulfides were reduced with 45 mm dithiothreitol in 50 mm ammonium bicarbonate and alkylated with 100 mm iodoacetamide in 50 mm ammonium bicarbonate. Gel pieces were again dehydrated with 100% acetonitrile. Trypsin (260 ng/gel piece) was added and incubated overnight at room temperature. Peptides were extracted from the gel pieces by subsequent additions of 30% acetonitrile in 0.1% TFA and 80% acetonitrile in 0.1% TFA. Samples were dried, redissolved in 0.1% formic acid, and injected into a Shimadzu HPLC system coupled to a Thermo Finnigan LCQ Classic instrument (24). HPLC separation was performed on a New Objectives Pico-frit column filled with BetaBasic C18. A linear gradient was developed from 10 to 60% B (A, 5% acetonitrile, 95% aqueous 0.1% formic acid; B, 80% acetonitrile, 20% aqueous 0.1% formic acid) at a rate of 1.5%/min. Data were collected continuously for 60 min, selecting the three most intense ions (exceeding 3 × 106 intensity units) in an MS survey scan to subsequent MS/MS analyses using collisionally induced dissociation. Selected precursors were analyzed for two MS/MS cycles and then excluded for redundant analyses for a 90-s interval. Thermo Finnigan Excalibur 2.0 utility extract_msn was used to retrieve peak lists without any added smoothing or signal to noise ratio criteria. The recorded MS/MS files were searched with the Mascot search engine 2.1.04 (Matrix Sciences, London, UK) against the Swiss-Prot database (Sp_Trembl_122406.fas) with the limitation of mammalian species for protein database records; precursor ion mass tolerance, 2.0; fragment ion mass tolerance, 0.8; methionine oxidation and carbamidomethylation of cysteine allowed; and trypsin specificity with one missed cleavage allowed.
Mascot-assigned peptides with Ion Score exceeding their Identity Score were then grouped using the software tool DBParser 2.0 (25), and reports were generated for each lane from the concatenated Mascot files. DBParser 2.0 was also used to compare data from multiple lanes at both the peptide and protein levels. DBParser 3.0 was used to extract ion current intensity values for peptides with Mascot Ion Scores greater than Identity Scores (DBParser 3.0 is further described in Supplement 1).
RESULTS
PSD-95 complexes were affinity-purified from the conventional PSD fraction using magnetic beads coated with an antibody against the core PSD protein PSD-95. Parent and affinity-purified samples were separated by one-dimensional SDS gel electrophoresis and analyzed in parallel. The control sample (Fig. 1, Lane 3) shows no visible band except the secondary antibody used to coat the magnetic beads, indicating the absence of nonspecific binding. The protein profiles of the parent PSD fraction (Fig. 1, Lane 1) and of the affinity-purified PSD-95 complex (Fig. 1, Lane 2) are different, indicating removal of several proteins during purification.
Fig. 1. Coomassie blue-stained SDS gel (7.5% acrylamide).
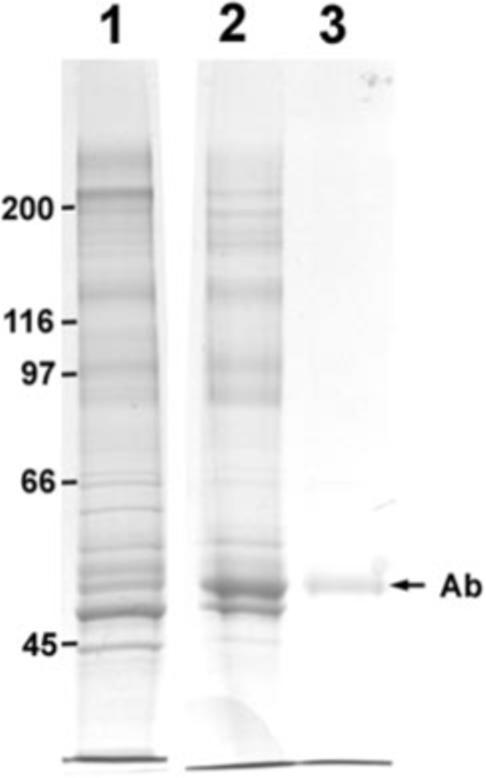
Lane 1, parent PSD fraction; Lane 2, PSD-95 complex affinity-purified with antibody-coated magnetic beads; Lane 3, control, eluant when beads coated with secondary antibody only were incubated with PSD fraction. The position of the antibody (Ab) heavy chain is shown with an arrow. Lane 2 contains PSD-95 antibody and secondary antibody, whereas Lane 3 contains secondary antibody only. Positions of molecular mass standards (in kDa) are indicated on the left.
Gel lanes in their entirety were cut into 40 fractions, digested with trypsin, and analyzed by LC/MS/MS. The resulting mass spectra were assigned probable peptide sequences using the Mascot search engine (Matrix Sciences). For these studies, the Swiss-Prot mammalian protein reference library was selected, and peptides with Mascot Ion Scores exceeding their Identity Scores were analyzed and grouped using DBParser 2.0. This software applies parsimony analysis to reduce the thousands of identified peptides to a minimal protein list (25). Only proteins from the minimal protein list were considered for further evaluation (Tables I–IV). The results from the two preparations include protein assignments from homologous mammalian proteins other than rat as indicated in the tables of results.
Table I.
50 top ranking of proteins in the affinity-purified PSD-95 complex Proteins are ranked in descending order of summed ion current intensity. For complete peptide sequences and statistical confidence scores, refer to Supplemental Table 1.
| Affinity purification rank | Parent rank | Protein (family) common name | UniProt KB/Swiss-Prot entry | Unique peptides |
|---|---|---|---|---|
| 1 | 1 | CaMKII | KCC2A_RAT | 9 |
| 2 | 4 | PSD-95 | DLG4_RAT | 25 |
| 3 | 3 | β-Tubulin | TBB5_RAT | 16 |
| 4 | 7 | SynGAP | SYGP1_RAT | 24 |
| 5 | 2 | α-Tubulin | Q5XIF6_RAT | 10 |
| 6 | 10 | PSD-93 | DLG2_RAT | 16 |
| 7 | 27 | GluR2 | GRIA2_RAT | 24 |
| 8 | 6 | Actin | ACTB_RAT | 6 |
| 9 | 23 | Shank3 | SHAN3_RAT | 25 |
| 10 | 22 | BRAG1 (KIAA0522) | O60275_HUMAN | 20 |
| 11 | 17 | Hypothetical | Q8BZM2_MOUSE | 9 |
| 12 | 48 | GluR3 | GRIA3_RAT | 13 |
| 13 | 35 | Homer | HOME1_RAT | 10 |
| 14 | 19 | Shank1 | SHAN1_RAT | 24 |
| 15 | 26 | Shank2 | SHAN2_RAT | 18 |
| 16 | 14 | IRSp53 | BAIP2_RAT | 12 |
| 17 | 49 | BRAG2b | Q6DN90_HUMAN | 4 |
| 18 | 61 | GluR1 | GRIA1_RAT | 7 |
| 19 | 24 | Densin 180 | LRRC7_RAT | 13 |
| 20 | 13 | α-Catenin | Q5R416_PONPY | 13 |
| 21 | 20 | Glutamine synthetase | GLNA_RAT | 7 |
| 22 | 65 | SAPAP4 | DLGP4_RAT | 4 |
| 23 | 44 | SAPAP1 | DLGP1_RAT | 8 |
| 24 | 29 | ADP/ATP translocase | ADT1_RAT | 6 |
| 25 | 47 | SAPAP3 | DLGP3_MOUSE | 8 |
| 26 | 45 | NMDAR2B | NMDE2_RAT | 13 |
| 27 | 16 | Myosin 10 | MYH10_RAT | 13 |
| 28 | 66 | SAPAP2 | DLGP2_RAT | 6 |
| 29 | 32 | PP1 | PP1A_RAT | 7 |
| 30 | 76 | Neuroligin | NLGN3_RAT | 7 |
| 31 | 87 | Hypothetical (FAM81A) | Q8TBF8_HUMAN | 5 |
| 32 | >93 | GLUR4 | GRIA4_MOUSE | 6 |
| 33 | 84 | Cylindromatosis | Q66H62_RAT | 8 |
| 34 | 30 | PKC-γ | KPCG_RAT | 5 |
| 35 | 12 | SNIP | SNIP_RAT | 6 |
| 36 | 9 | Plectin | PLEC1_RAT | 23 |
| 37 | 25 | Heat shock cognate 71 | HSP7C_RAT | 8 |
| 38 | 62 | NMDAR2A | O08948_RAT | 8 |
| 39 | 37 | SAP97 | DLG1_HUMAN | 4 |
| 40 | 36 | SAP102 | DLG3_RAT | 5 |
| 41 | 92 | Leu-rich repeat ... | LRTM1_HUMAN | 2 |
| 42 | 59 | Hypothetical (FLJ35778) | Q8NA73_HUMAN | 3 |
| 43 | 15 | VDAC1 | VDAC1_RAT | 7 |
| 44 | 38 | Citron | Q9QX19_RAT | 6 |
| 45 | 60 | NMDAR1 | Q62648_RAT | 5 |
| 46 | 58 | TARP | CCG8_RAT | 2 |
| 47 | 18 | α-Actinin | Q6GMN8_RAT | 7 |
| 48 | 28 | G3P | G3P_RAT | 6 |
| 49 | 41 | Kalirin | HAPIP_RAT | 12 |
| 50 | 69 | CPG2 protein | Q63128_RAT | 5 |
Table IV.
Proteins greatly reduced/depleted in the affinity-purified PSD-95 complex Proteins from Table II with a normalized ion current intensity ratio of 0.1 or smaller were included. Certain proteins (not applicable, NA) were abundant in the parent fraction with intensities that would rank them in the top 50 proteins but were not detected after affinity purification. ID, identity; int., intensity; SNIP, SNAP-25-interacting protein.
| Protein (family) name | UniProt KB/Swiss-Prot entry | Change in rank (parent/purified) | Normalized ion current intensity ratio (purified/parent) |
|---|---|---|---|
| Presynaptic proteins | |||
| Bassoon | BSN_RAT | 8/69 | 0.01 |
| SNIP | SNIP_RAT | 12/35 | 0.06 |
| Synapsin 2 | SYN2_RAT | 21/74 | 0.02 |
| ERC protein 2 | ERC2_RAT | 31/71 | 0.04 |
| Synapsin 1 | SYN1_RAT | NA/a | 0.00 |
| Piccolo | PCLO_RAT | NA/a | 0.00 |
| RIM | RIMS_RAT | NA/a | 0.00 |
| Cytoskeletal elements | |||
| α-Spectrin | SPTA2_RAT | 5/59 | 0.01 |
| Plectin | PLEC1_RAT | 9/36 | 0.04 |
| α-Internexin | AINX_RAT | 11/67 | 0.01 |
| β-Spectrin | SPTN2_RAT | NA/a | 0.00 |
| Neurofilament-L | NFL_RAT | NA/a | 0.00 |
| Neurofilament-M | NFM_RAT | NA/a | 0.00 |
| Different organelle/cell type | |||
| VDAC1 (mitochondrial) | VDAC1_RAT | 15/43 | 0.06 |
| VDAC2 (mitochondrial) | VDAC2_RAT | 34/73 | 0.04 |
| Myelin basic protein | MBP_RAT | NA/a | 0.00 |
| Ribosomal proteins | RLA0_RAT | NA/a | 0.00 |
| Other | |||
| α-Actinin | Q6GMN8_RAT | 18/47 | 0.06 |
| Synaptopodin | SYNPO_RAT | NA/a | 0.02 |
| β-Catenin | CTNB1_HUMAN | 33/52 | 0.09 |
| N-cadherin | CADH2_RAT | 42/63 | 0.09 |
Proteins identified with two or more peptides in parent fraction only.
Altogether three sets of samples were analyzed, corresponding to two independent parent and affinity-purified preparations. The concatenated parent (from 133 LC/MS/MS runs) and affinity-purified (125 LC/MS/MS runs) files correspond to summed and integrated analyses. Identified proteins were ranked according to summed ion current intensities of their constituent peptides as a relative abundance index. DBParser 3.0 extracts retention time and peak intensity from raw data based on the precursor mass identified by the search engine as described previously for ion trap LC/MS/MS data (26). The corresponding maximum intensity, peak area, and number of scans for each identified peptide were then calculated from respective selected ion chromatograms recorded during the survey scan cycle (see Supplement 1 for DBParser 3.0 details).
Known contaminants, originating from sources other than brain tissue, including immunoglobulins (antibodies used for affinity purification), serum albumin (blocker), trypsinogen, and human keratin were deleted from the lists. The lists were condensed further by reporting certain related proteins with multiple isoforms as a single family. Families of proteins are represented by their most abundant member. For example, although α-, β-, δ-, and γ-isoforms of CaMKII were identified, CaMKII is reported by a single entry corresponding to its most abundant α-isoform.
Tables I and II list the highest ranking 50 proteins from the simplified lists corresponding to affinity-purified and parent samples, respectively. The tables include only those proteins detected in both fractions and thus allow a fair comparison by rank order. The proteins are listed in descending rank order (highest ion current intensity at the top). Only proteins identified by two or more peptides were included in determining rank order. The second columns in Tables I and II show the rank of the same protein in the other group. Although a number of factors in addition to the abundance of a protein may influence the ion current intensity, these other factors are expected to remain the same for the two fractions so that a change in rank order may be used as an indication of a change in relative abundance. The last columns in Tables I and II indicate the number of peptides identified by MS/MS exclusively associated with the assigned proteins and used for intensity determinations. There is no direct correlation between the number of peptides and total ion current intensity as the number of peptides detectable is a function of both the number of possible tryptic products compatible with mass spectrometric detection and their relative abundance.
Table II.
50 top ranking of proteins in the conventional (parent) PSD fraction Proteins are ranked in descending order of summed ion current intensity. For complete peptide sequences and statistical confidence scores, refer to Supplemental Table 2.
| Parent rank | Affinity purification rank | Protein (family) common name | UniPost KB/Swiss-Prot entry | Unique peptides |
|---|---|---|---|---|
| 1 | 1 | CaMKII | KCC2A_RAT | 11 |
| 2 | 5 | α-Tubulin | Q5XIF6_RAT | 13 |
| 3 | 3 | β-Tubulin | TBB5_RAT | 15 |
| 4 | 2 | PSD-95 | DLG4_RAT | 24 |
| 5 | 59 | α-Spectrin | SPTA2_RAT | 52 |
| 6 | 8 | Actin | ACTB_RAT | 7 |
| 7 | 4 | SynGAP | SYGP1_RAT | 27 |
| 8 | 69 | Bassoon | BSN_RAT | 38 |
| 9 | 36 | Plectin | PLEC1_RAT | 60 |
| 10 | 6 | PSD-93 | DLG2_RAT | 15 |
| 11 | 67 | α-Internexin | AINX_RAT | 11 |
| 12 | 35 | SNIP | SNIP_RAT | 18 |
| 13 | 20 | α-Catenin | Q5R416_PONPY | 19 |
| 14 | 16 | IRSp53 | BAIP2_RAT | 14 |
| 15 | 43 | VDAC1 | VDAC1_RAT | 13 |
| 16 | 27 | Myosin 10 | MYH10_RAT | 22 |
| 17 | 11 | Hypothetical (SAM/PTB) | Q8BZM2_MOUSE | 11 |
| 18 | 47 | α-Actinin | Q6GMN8_RAT | 28 |
| 19 | 14 | Shank1 | SHAN1_RAT | 17 |
| 20 | 21 | Glutamine synthetase | GLNA_RAT | 7 |
| 21 | 74 | Synapsin 2 | SYN2_RAT | 16 |
| 22 | 10 | BRAG1 (KIAA0522) | O60275_HUMAN | 14 |
| 23 | 9 | Shank3 | SHAN3_RAT | 14 |
| 24 | 19 | Densin 180 | LRRC7_RAT | 15 |
| 25 | 37 | Heat shock cognate 71 | HSP7C_RAT | 12 |
| 26 | 15 | Shank2 | SHAN2_RAT | 11 |
| 27 | 7 | GluR2 | GRIA2_RAT | 9 |
| 28 | 48 | G3P | G3P_RAT | 7 |
| 29 | 24 | ADP/ATP translocase | ADT1_RAT | 8 |
| 30 | 34 | PKC-γ | KPCG_RAT | 8 |
| 31 | 71 | ERC protein 2 | ERC2_RAT | 12 |
| 32 | 29 | PP1 | PP1A_RAT | 10 |
| 33 | 52 | β-Catenin | CTNB1_HUMAN | 11 |
| 34 | 73 | VDAC2 | VDAC2_RAT | 8 |
| 35 | 13 | Homer | HOME1_RAT | 9 |
| 36 | 40 | SAP102 | DLG3_RAT | 11 |
| 37 | 39 | SAP97 | DLG1_HUMAN | 3 |
| 38 | 44 | Citron | Q9QX19_RAT | 7 |
| 39 | 60 | GAPDH | Q8K417_SIGHI | 4 |
| 40 | 56 | ArgBP2 | O35413_RAT | 8 |
| 41 | 49 | Kalirin | HAPIP_RAT | 11 |
| 42 | 63 | N-cadherin | CADH2_RAT | 6 |
| 43 | 62 | VDAC3 | Q6GSZ1_RAT | 6 |
| 44 | 23 | SAPAP1 | DLGP1_RAT | 8 |
| 45 | 26 | NMDAR2B | NMDE2_RAT | 7 |
| 46 | 61 | Fructose bisphosphate aldolase | ALDOA_RABIT | 2 |
| 47 | 25 | SAPAP3 | DLGP3_MOUSE | 10 |
| 48 | 12 | GluR3 | GRIA3_RAT | 6 |
| 49 | 17 | BRAG2b | Q6DN90_HUMAN | 6 |
| 50 | 66 | Mitochondrial glutamate carrier | GHC1_MOUSE | 6 |
As a measure of co-purification with PSD-95, the ratios of normalized ion current intensities in the affinity-purified group over that in the parent group were calculated for individual proteins (Tables III and IV, last columns). To compensate for variable total protein contents of samples applied to gels, ion current intensities for individual proteins were normalized with respect to the ion current intensity for PSD-95 in the same sample. For certain proteins, the ratios of normalized ion current intensities were greater than 1 implying that immuno-affinity capture of PSD-95 resulted in the capture of additional specific proteins with higher efficiency than the targeted PSD-95. However, this phenomenon is more likely the result of signal to noise ratio differences between the parent and affinity-purified protein mixtures. Chemical noise from peptides from contaminant proteins masks co-eluting peptides from constitutive proteins present in the parent mixture. Thus, peptides from some proteins are observable only after affinity purification. Comparable amounts of PSD-95 (DLG4) were analyzed in each preparation (24 peptides observed in the parent preparation, summed intensity: 7 × 109; 25 peptides in the affinity-purified, summed intensity: 9 × 109). After removal of interfering peptides from contaminants, signals from peptides from the constituents of the PSD-95 complex were detected more readily. For example, GluR1 (GRIA1_RAT) was characterized by three peptides in 113 MS scans (summed intensity, 2 × 108) in the parent mixture and by seven peptides in 274 MS scans (9 × 108) after affinity purification. The semiquantitative nature of this method assesses relative ranking but cannot be used to infer stoichiometry.
Table III.
Proteins co-purifying with PSD-95 Proteins from Table I (50 top ranking proteins in the affinity-purified preparation) with a normalized ion current intensity ratio greater than 0.5 are included.
| Protein (family) name | UniProt KB/Swiss-Prot entry | Change in rank (parent/affinity-purified) | Normalized ion current intensity ratio (purified/parent) |
|---|---|---|---|
| Glutamate receptors | |||
| GluR2 (AMPAR) | GRIA2_RAT | 27/7 | >1 |
| GluR3 (AMPAR) | GRIA3_RAT | 48/12 | >1 |
| GluR1 (AMPAR) | GRIA1_RAT | 61/18 | >1 |
| GluR4 (AMPAR) | GRIA4_RAT | >93/32 | >1 |
| NMDAR2B | NMDE2_RAT | 45/26 | 0.55 |
| NMDAR2A | O08948_RAT | 62/38 | >1 |
| NMDAR1 | Q62648_RAT | 60/45 | 0.74 |
| Scaffolds | |||
| PSD-95 | DLG4_RAT | 4/2 | 1.00 |
| PSD-93 | DLG2_RAT | 10/6 | 0.67 |
| Shank3 | SHAN3_RAT | 23/9 | >1 |
| Hypothetical (SAM/PTB) | Q8BZM2_MOUSE | 17/11 | 0.62 |
| Homer | HOME1_RAT | 35/13 | >1 |
| Shank2 | SHAN2_RAT | 26/15 | 0.75 |
| SAPAP4 | DLGP4_RAT | 65/22 | >1 |
| SAPAP1 | DLGP1_RAT | 44/23 | 0.59 |
| SAPAP2 | DLGP2_RAT | 66/28 | >1 |
| G-protein regulators | |||
| SynGAP | SYGP1_RAT | 7/4 | 0.64 |
| BRAG1 (KIAA0522) | O60275_HUMAN | 22/10 | >1 |
| BRAG2b | Q6DN90_HUMAN | 49/17 | >1 |
| Other | |||
| Neuroligin | NLGN3_RAT | 76/30 | >1 |
| Hypothetical (FAM81A) | Q8TBF8_HUMAN | 87/31 | >1 |
| Cylindromatosis | Q66H62_RAT | 84/33 | >1 |
| Leu-rich repeat . . . | LRTM1_HUMAN | 92/41 | >1 |
| Hypothetical (FLJ35778) | Q8NA73_HUMAN | 59/42 | 0.73 |
| TARP (γ8) | CCG8_RAT | 58/46 | 0.62 |
| CPG2 protein | Q63128_RAT | 69/50 | 0.95 |
Table III lists those proteins that co-purify with PSD-95 by two criteria presented in the last two columns: change in rank order and normalized ion current intensity ratio (purified/parent). Only the 50 top ranking proteins from the affinity-purified group (Table I) were considered, and the cutoff point for inclusion in the list was a normalized ion current intensity ratio of 0.5. It can be observed that 26 of the 50 top ranking proteins met these criteria and also demonstrated a substantial decrease in rank order (Table III). The 26 proteins shown in Table III are relatively abundant components that are substantially co-purified with PSD-95 and thus most likely represent a minimal list of constitutive PSD-95 complex proteins.
Table IV lists those proteins that are depleted or greatly reduced in the affinity-purified sample with the same indexes as in Table III. For this list, those proteins from Table II (top ranking proteins from the parent group) with normalized ion current intensity ratios of parent/purified below 0.1 were selected. Also five additional top ranking proteins that were detected in the parent group only were included. Of the 21 proteins that show significant reduction/depletion in purified samples, seven are presynaptic elements, and six are cytoskeletal elements.
DISCUSSION
The molecular architecture of the PSD-95 complex is less readily definable than that of the synaptic vesicle as elegantly described by Takamori et al. (27). In the course of that work, the value of immunoaffinity-assisted isolation was recognized in the proteomics analyses of light and heavy synaptic vesicles by Morciano et al. (28). Both heavy and light sucrose density gradient-separated vesicles contain many proteins in common, but the sedimented heavy fraction was contaminated with PSD proteins until separated with anti-SV2 monoclonal antibody-coated magnetic beads. The heavy fraction was then found to consist of synaptic vesicles docked to presynapse plasma membranes but was free of PSD proteins (28). In an analogous manner, we sought to demonstrate that the immunoaffinity-assisted isolation of the PSD-95 complex developed by Vinade et al. (20) provides an isolate more relevant to defining the composition of the PSD-95 complex because it is substantially freer from co-sedimenting components than attainable by sucrose density centrifugation alone.
Because the identification of proteins in the PSD fraction has been the subject of multiple reports, we have reconciled our findings with those of others. Repeated observation of the same proteins by multiple investigators adds certainty to the identified constituents of the PSD fraction. Collins et al. (16) collected and correlated reports from seven laboratories and the literature to assemble a non-redundant set of 1124 accession numbers of proteins. The task of comparing protein identifications is not possible without access to either the original tandem mass spectrometry data or the peptide sequences ascribed to that data. Protein accession numbers and entry names change when protein databases are curated so that differences in published lists of proteins are sometimes due to differences in the versions of the protein reference libraries used for sequence assignment. Consequently we compiled protein sequence fasta files for the entries tabulated by Collins et al. (16) and used these reference sublibraries for Mascot to re-search and identify proteins. The use of the Collins et al. (16) sublibrary (kindly provided by the authors) ensures that peptide sequences recognized can be mapped to the same protein names, eliminating ambiguity of naming conventions or isoform selection. Table V summarizes these results parsed and analyzed using MassSieve software.2
Table V.
Comparison of proteins observed with literature data Bold values are data from the present study. For complete protein information, peptide sequences, and statistical confidence scores, refer to Supplemental Tables 3−6.
| Confidencea | Proteins reportedb | Parent preparation | Affinity purification |
|---|---|---|---|
| 8 | 10 | 10 | 8 |
| 7 | 13 | 13 | 12 |
| 6 | 15 | 15 | 12 |
| 5 | 34 | 30 | 25 |
| 4 | 62 | 56 | 31 |
| 3 | 135 | 91 | 50 |
| 2 | 198 | 112 | 45 |
| 1 | 657 | 192 | 101 |
| Totals | 1124 | 525 | 288 |
Consensus number of studies reporting identification of the same set of proteins.
Number of proteins in the set (Collins et al. (16)).
The proteins identified in our parent preparation are consistent with those reported by others. Of 134 proteins observed by four or more laboratories, we identified peptides from 124 (92%). Of greater importance are differences observed after affinity purification: only 88 (57%) of the 134 proteins observed by four or more laboratories were observed after affinity purification presumably due to removal of contaminating proteins.
In the present study large, affinity-purified PSD-95 complexes derived from the synapse were analyzed, and on the basis of rank order changes with respect to the parent PSD fraction, a subset of the identified proteins were selected as likely to be consistent, integral components of the complex. It should be noted that the 26 proteins included in Table III constitute a minimal list of relatively abundant constituents of the PSD-95 complex. The PSD-95 complex undoubtedly includes additional components. Indeed a much larger number of proteins than those listed in Table III have been identified in the affinity-purified preparation (see Table V and supplemental tables for a complete list of proteins and constituent identified peptides), and many of these appear to be more prominent than in the parent PSD fraction. However, we have chosen to apply stringent criteria (two or more peptides within the top 50 rank order by intensity and normalized ion current ratios greater than 0.5 relative to PSD-95) that would select only for the relatively abundant proteins that co-purify with PSD-95.
A minimal list of constituent proteins of the synaptic PSD-95 complex (Table III) includes specialized PSD scaffolds with multiple protein interaction sites and receptors as well as several proteins likely to be involved in aspects of spine structure and dynamics such as trafficking of receptors, actin dynamics, and cell adhesion. On the other hand, many of the proteins that are greatly reduced or eliminated upon affinity purification are presynaptic or extrasynaptic in origin (Table IV). Effective removal of these known contaminants attests to the success of the affinity-based strategy and establishes the preparation as a good base for the elucidation of the composition of the PSD complex.
Electron microscopy of the affinity-purified preparation shows many particulate structures that look like PSDs (20), suggesting that the isolated PSD-95 complex retains the bulk of the proteins that make up the in situ PSD. However, there are a few proteins, including N-cadherin, catenins, and α-actinin, that have been observed at the contact zone by immuno-EM (29 –31) but whose levels decrease with respect to the parent PSD fraction upon affinity purification. This could be due to the dissociation of some loosely bound components during purification as well as the existence of physically separate complexes that co-localize in situ with the PSD-95 complex. It is interesting to note that all the above mentioned proteins are associated (either directly or indirectly) with the actin cytoskeleton and may be linked to the PSD-95 complex through actin. Partial disassembly of the actin during the purification protocol may have caused their release. Thus, the affinity-purified preparation appears to include the majority but not all of the components of the in situ PSD.
Two measures for affinity co-purification (change in rank order and ratio of normalized ion current intensities) were used to identify integral elements of the PSD-95 complex. The 26 proteins in Table III are thus likely to represent a subset of constituent elements. As discussed below, some of the proteins in the list are expected components of the PSD-95 complex such as specialized PSD scaffolds and NMDA receptors, whereas the involvement of some of the other identified proteins had been somewhat controversial. Definitive stoichiometry of constitutive PSD-95 complex proteins will require either orthogonal techniques such as quantitative Western blotting or stable isotope mass spectrometric standards, and this study refines the list of proteins that should be targeted as well as identifies their proteotypic peptides for synthesis of labeled standards. Most interestingly, the list spotlights certain proteins that had not been generally acknowledged up to now as part of the PSD-95 complex. We discuss below particular proteins in functional groups.
Scaffolds
Most specialized PSD scaffolds are identified as constituents of the PSD-95 complex. Co-purification of PSD-93 with PSD-95, two homologous proteins, each with a guanylate kinase, an SH3, and three PDZ domains (for a review, see Ref. 15), argues against their differential localization in different synapses and indicates that they are part of the same complex. In addition, SAPAPs (SAP90/PSD-95-associated proteins) that bind directly to PSD-95, Shanks that bind to SAPAPs, and homer that binds to Shanks (for a review, see Ref. 15) all co-purify with PSD-95 suggesting that the entire PSD scaffold is preserved in the purified complex.
Shank3, together with PSD-95 and PSD-93, is among the 10 top ranking proteins in the purified fraction. Shanks are thought to multimerize through association of their sterile α motif (SAM) domains (32). Indeed SAM domains that typically spread over ∼70 residues have been described to homo- and hetero-oligomerize (for a review, see Ref. 33). A recent report (34) described large sheets made of stacked helical fibers being formed by the SAM domains of Shank3, which, they suggest, may constitute a core platform for the PSD organization. The present analysis identifies a hypothetical protein (Q8BZM2) with two SAM domains as a constituent of the PSD-95 complex. In analogy to other SAM-containing proteins, this protein is expected to homo- and hetero-oligomerize and thus participate in the formation of the PSD scaffold.
Adhesion Molecules
In contrast to cadherin/catenin, another group of cell adhesion molecules, neuroligins, are identified as constituents of the PSD-95 complex. This finding is in agreement with reports on direct binding of the two proteins (35). In addition, the present analysis reveals the presence of yet another putative adhesion molecule, LRTM1, of the leucine-rich repeat transmembrane protein family in the PSD-95 complex (36).
Cytoskeletal Elements
Cytoskeletal elements make up a major proportion of the proteins in the conventional PSD fraction and were observed to be enriched in the PSD fraction compared with the synaptosome fraction (10). However, with the exception of actin and tubulin most cytoskeletal proteins are substantially reduced/depleted in the affinity-purified PSD-95 complex. Actin constitutes the main cytoskeleton of the dendritic spine and interacts with the PSD in situ (37). The present data, demonstrating a slight tendency for actin to co-purify with PSD-95, is in agreement with the notion that actin is an integral element of the PSD. Another cytoskeletal protein, tubulin, also appears to be present in large quantities in the affinity-purified fraction. This is somewhat surprising because microtubules are rarely observed near the PSD of mature spines. One explanation may be that tubulin is attached to the PSD either in monomeric or a different polymeric form; for example, it has been proposed that NMDA receptors have a preferential affinity for soluble forms of tubulin (for a review, see Ref. 38). On the other hand immuno-EM studies do not support the presence of large quantities of tubulin at the in situ PSD.3 Thus, the possibility remains that at least part of the detected tubulin becomes associated with the PSD-95 complex postmortem when large quantities of depolymerized tubulin become available during homogenization.
In contrast to actin and tubulin, other cytoskeletal elements such as spectrins and intermediate filaments that are prominent in conventional PSD fractions (5, 9, 10) are virtually eliminated upon affinity purification. The removal of spectrin and intermediate filaments suggests that these proteins are not components of the PSD, consistent with the observation of non-associated spectrin filaments and neurofilaments in the parent PSD preparation (20).
Glutamate Receptors and Related Molecules
Glutamate receptors of both NMDA and AMPA types appear to be prominent components of the PSD-95 complex. Our previous work demonstrated by Western immunoblotting the presence of AMPA-type glutamate receptors in the affinity-purified preparation (20). Comparative analysis of the parent and purified fractions in the present study shows that AMPA receptors co-purify with PSD-95. Earlier reports had indicated that NMDA receptor complexes isolated from detergent extracts do not contain AMPA-type receptors (22, 23), an observation that led the authors to conclude that AMPA receptors at the PSD are physically separate from the PSD-95 complex. It is possible that these small NMDA complexes (2000 kDa as compared with 1 million kDa for the PSD) are subcomplexes dissociated from the PSD by the relatively strong detergent (deoxycholate) used. Alternatively they may represent a distinct group of receptors of either synaptic or extrasynaptic origin.
Our observation that AMPA-type receptors are an integral part of the PSD-95 complex is in agreement with accumulated evidence on the regulation of synaptic AMPA receptor levels by PSD-95 (39–42). On the other hand, because direct binding of AMPA receptors to PSD-95 has not been demonstrated, an indirect association through at least one bridging molecule has to be assumed. SAP97 has been detected by Western immunoblotting to be present in affinity-purified complexes (20), but this protein would be able to anchor only those receptors that contain GluR1 subunits. Also the present semiquantitative analysis indicates that SAP97 is not particularly enriched in the PSD-95 complex, whereas GluR1 is. A good candidate for bridging all types of AMPA receptors to the core PSD complex is the TARP (Stargazin) family of proteins, which are reported to bind very strongly to AMPA receptors and also to PSD-95 (43). Our results identify TARP γ8 (CCG8_RAT) as a component of the PSD-95 complex. However, at this stage it is not clear whether the amount of TARP found in the PSD-95 complex is sufficient to support the binding of all AMPA receptors, at least one TARP per five AMPA receptor subunits. Future quantitative studies that assess stoichiometric relationships would clarify that point.
CPG2, a protein that has been proposed to be involved in the constitutive internalization of NMDA- and AMPA-type glutamate receptors and in the activity-induced internalization of AMPA receptors (44) is also a component of the PSD-95 complex. Immuno-EM studies demonstrated that the protein is located laterally and underneath the cytosolic side of the PSD (44). These results together with our data indicating physical attachment of the protein to the PSD-95 complex suggest that CPG2 may be associated as an appendage to the PSD rather than being integrated within the core structure.
CaMKII
The Ca2+-regulated protein kinase CaMKII appears to be the major protein in the PSD-95 complex as judged by its abundance ranking and relative band intensity in Coomassie-stained gels. We had previously demonstrated that part of the CaMKII present in conventional PSD preparations is contributed by contaminating CaMKII clusters, spherical structures ∼100 nm in diameter (19). Thus, it is remarkable that even after the elimination of these CaMKII clusters in affinity-purified samples CaMKII still remains the most abundant protein. However, it should be noted that this may not hold true for in situ PSD-95 complexes at inactive synapses. Indeed the amount of CaMKII associated with the PSD is regulated by synaptic activity. Previous immuno-EM studies have shown that excitatory and ischemia-like conditions cause severalfold increases in CaMKII labeling of the PSD (19, 45, 46). Ischemia-like conditions that prevail during the dissection of brain tissue likewise cause an increase in the CaMKII content of PSD fractions (47). Thus, for the case of CaMKII and certain other variable/transient components of the postsynaptic complex such as GluR1, the abundance is likely to be determined by factors such as the level of neuronal activity and anesthesia prior to sacrifice as well as by postmortem handling.
Small G-protein Regulators
A substrate for CaMKII, the synaptic Ras GTPase-activating protein SynGAP (48) is also among the abundant proteins in the purified PSD-95 complexes. SynGAP can directly bind to PSD-95 (49). Parallel analysis of parent and affinity-purified fractions highlighted the presence of two additional small G-protein regulators of the brefeldin A-resistant ArfGEF (BRAG) family in the PSD-95 complex. One of these (KIAA0522, O60275_HUMAN, IQEC2_HUMAN, gi|62666747|ref|XP_228841.3|) has been classified as BRAG1 (50). A recent study (51) published during the preparation of this manuscript identifies an error in the sequence of KIAA0522 and demonstrates in situ co-localization of BRAG1 and PSD-95. BRAG1 is enriched in the PSD-95 fraction compared with the synaptosome fraction (10, 51). BRAG1 is a 190-kDa protein with a calmodulin binding IQ domain and a sec7 domain, a ∼200-amino acid signature sequence for ArfGEFs that confers guanine nucleotide exchange activity. BRAG1 has been shown to regulate Arf1 (51), and from the sequence similarity, it is probable that it also acts on Arf6 (49).
Surprisingly another member of the same family, BRAG2b (Q6DN90_HUMAN, IQEC1_HUMAN), is also identified in the PSD-95 complex. BRAG1 and BRAG2b are distinguishable proteins and were identified by two entirely different sets of peptides with no overlapping assignments. BRAG2b is a splice isoform of a previously identified ArfGEF (Arf-GEP100 (52) and has been shown to regulate Arf6 (53). Another ArfGEF called SynArfGEF (Q76M68) has recently been reported by Inaba et al. (54) to be enriched in the PSD fraction and is suggested to act on Arf1. SynArfGEF was not observed in this study and differs from both BRAG1 and BRAG2b (63 and 48% homology, respectively).
Arfs are from the Ras family of small G-proteins that regulate membrane trafficking and the actin cytoskeleton. Different types of Arfs appear to be targeted to different subcellular locations (for reviews, see Refs. 55 and 56): Arf1 is preferentially targeted to the Golgi, whereas Arf6 is localized at the plasma membrane and to some extent on endosomes (for reviews, see Refs. 56 and 57). Interestingly Arf6 has been implicated in the insertion of and retrieval of membrane proteins at defined sites as well as in the reorganization of the actin cytoskeleton that results in various forms of membrane remodeling (for a review, see Ref. 57). Considering that synaptic strength is largely regulated by the insertion and retrieval of receptors to and from the postsynaptic contact zone and that actin is a constituent of the cytoskeleton at the spine, the potential relevance of Arf6 to spine structure and function becomes obvious. In fact, two recent studies (58, 59) already reported involvement of Arf6 on spine formation and stability, although with somewhat contradictory conclusions. The finding in the present study that activators of Arf6 are specifically concentrated at the PSD opens the way for the unraveling of new mechanisms for the modification of spine morphology and function.
Acknowledgments
We thank Drs. T. S. Reese and J. Kowalak for advice and helpful discussions throughout this study.
* This work was supported in part by the Intramural Research Programs of the NINDS, National Institutes of Health and the National Institute of Mental Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: PSD, postsynaptic density; NMDA, N-methyl-d-aspartate; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; Arf, ADP-ribosylation factor; SAM, sterile α motif; CaMKII, calcium/calmodulin-dependent protein kinase II; EM, electron microscopy; SAPAP, SAP90/PSD-95-associated protein; TARP, transmembrane AMPA receptor regulatory protein; BRAG, brefeldin A-resistant ArfGEF; GEF, guanine nucleotide exchange factor; GAP, GTPase-activating protein.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
D. Slotta, M. A. McFarland, and S. P. Markey, manuscript in preparation.
J.-H. Tao-Cheng, personal communication.
Supplementary Material
REFERENCES
- 1.Chen X, Vinade L, Leapman RD, Petersen JD, Nakagawa T, Phillips TM, Sheng M, Reese TS. Mass of the postsynaptic density and enumeration of three key molecules. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11551–11556. doi: 10.1073/pnas.0505359102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen RS, Blomberg F, Berzins K, Siekevitz P. The structure of postsynaptic densities isolated from dog cerebral cortex. I. Overall morphology and protein composition. J. Cell Biol. 1977;74:181–203. doi: 10.1083/jcb.74.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matus AI, Taff-Jones DH. Morphology and molecular composition of isolated postsynaptic junctional structures. Proc. R. Soc. Lond. B Biol. Sci. 1978;203:135–151. doi: 10.1098/rspb.1978.0097. [DOI] [PubMed] [Google Scholar]
- 4.Walsh MJ, Kuruc N. The postsynaptic density: constituent and associated proteins characterized by electrophoresis, immunoblotting, and peptide sequencing. J. Neurochem. 1992;59:667–678. doi: 10.1111/j.1471-4159.1992.tb09421.x. [DOI] [PubMed] [Google Scholar]
- 5.Walikonis RS, Jensen ON, Mann M, Provance DWJ, Mercer JA, Kennedy MB. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J. Neurosci. 2000;20:4069–4080. doi: 10.1523/JNEUROSCI.20-11-04069.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jordan BA, Fernholz BD, Boussac M, Xu C, Grigorean G, Ziff EB, Neubert TA. Identification and verification of novel rodent postsynaptic density proteins. Mol. Cell. Proteomics. 2004;3:857–871. doi: 10.1074/mcp.M400045-MCP200. [DOI] [PubMed] [Google Scholar]
- 7.Li KW, Hornshaw MP, Van Der Schors RC, Watson R, Tate S, Casetta B, Jimenez CR, Gouwenberg Y, Gundelfinger ED, Smalla KH, Smit AB. Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J. Biol. Chem. 2004;279:987–1002. doi: 10.1074/jbc.M303116200. [DOI] [PubMed] [Google Scholar]
- 8.Yoshimura Y, Yamauchi Y, Shinkawa T, Taoka M, Donai H, Takahashi N, Isobe T, Yamauchi T. Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J. Neurochem. 2004;88:759–768. doi: 10.1046/j.1471-4159.2003.02136.x. [DOI] [PubMed] [Google Scholar]
- 9.Peng J, Kim MJ, Cheng D, Duong DM, Gygi SP, Sheng M. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J. Biol. Chem. 2004;279:21003–21011. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- 10.Li K, Hornshaw MP, van Minnen J, Smalla KH, Gundelfinger ED, Smit AB. Organelle proteomics of rat synaptic proteins: correlation-profiling by isotope-coded affinity tagging in conjunction with liquid chromatography-tandem mass spectrometry to reveal post-synaptic density specific proteins. J. Proteome Res. 2005;4:725–733. doi: 10.1021/pr049802+. [DOI] [PubMed] [Google Scholar]
- 11.Collins MO, Yu L, Coba MP, Husi H, Campuzano I, Blackstock WP, Choudhary JS, Grant SG. Proteomic analysis of in vivo phosphorylated synaptic proteins. J. Biol. Chem. 2005;280:5972–5982. doi: 10.1074/jbc.M411220200. [DOI] [PubMed] [Google Scholar]
- 12.Trinidad JC, Thalhammer A, Specht CG, Schoepfer R, Burlingame AL. Phosphorylation state of postsynaptic density proteins. J. Neurochem. 2005;92:1306–1316. doi: 10.1111/j.1471-4159.2004.02943.x. [DOI] [PubMed] [Google Scholar]
- 13.Trinidad JC, Specht CG, Thalhammer A, Schoepfer R, Burlingame AL. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteomics. 2006;5:914–922. doi: 10.1074/mcp.T500041-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Cheng D, Hoogenraad CC, Rush J, Ramm E, Schlager MA, Duong DM, Xu P, Wijayawardana SR, Hanfelt J, Nakagawa T, Sheng M, Peng J. Relative and absolute quantification of postsynaptic density proteome isolated from rat forebrain and cerebellum. Mol. Cell. Proteomics. 2006;5:1158–1170. doi: 10.1074/mcp.D500009-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Kim E, Sheng M. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- 16.Collins MO, Husi H, Yu L, Brandon JM, Anderson CN, Blackstock WP, Choudhary JS, Grant SG. Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 2006;97(Suppl 1):16–23. doi: 10.1111/j.1471-4159.2005.03507.x. [DOI] [PubMed] [Google Scholar]
- 17.Satoh K, Takeuchi M, Oda Y, Deguchi-Tawarada M, Sakamoto Y, Matsubara K, Nagasu T, Takai Y. Identification of activity-regulated proteins in the postsynaptic density fraction. Genes Cells. 2002;7:187–197. doi: 10.1046/j.1356-9597.2001.00505.x. [DOI] [PubMed] [Google Scholar]
- 18.Carlin RK, Grab DJ, Cohen RS, Siekevitz P. Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J. Cell Biol. 1980;86:831–845. doi: 10.1083/jcb.86.3.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dosemeci A, Reese TS, Petersen J, Tao-Cheng JH. A novel particulate form of Ca2+/calmodulin-dependent [correction of Ca2+/CaMKII-dependent] protein kinase II in neurons. J. Neurosci. 2000;20:3076–3084. doi: 10.1523/JNEUROSCI.20-09-03076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vinade L, Chang M, Schlief ML, Petersen JD, Reese TS, Tao-Cheng JH, Dosemeci A. Affinity purification of PSD-95-containing postsynaptic complexes. J. Neurochem. 2003;87:1255–1261. doi: 10.1046/j.1471-4159.2003.02091.x. [DOI] [PubMed] [Google Scholar]
- 21.Cho KO, Hunt CA, Kennedy MB. The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron. 1992;9:929–942. doi: 10.1016/0896-6273(92)90245-9. [DOI] [PubMed] [Google Scholar]
- 22.Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat. Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 23.Husi H, Grant SG. Isolation of 2000-kDa complexes of N-methyl-D-aspartate receptor and postsynaptic density 95 from mouse brain. J. Neurochem. 2001;77:281–291. doi: 10.1046/j.1471-4159.2001.t01-1-00248.x. [DOI] [PubMed] [Google Scholar]
- 24.Masuda J, Maynard DM, Nishimura M, Ueda T, Kowalak JA, Markey SP. Fully automated micro- and nanoscale one- or two-dimensional high-performance liquid chromatography system for liquid chromatography-mass spectrometry compatible with non-volatile salts for ion exchange chromatography. J. Chromatogr. A. 2005;1063:57–69. doi: 10.1016/j.chroma.2004.11.084. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Dondeti V, Dezube R, Maynard DM, Geer LY, Epstein J, Chen X, Markey SP, Kowalak JA. DBParser: web-based software for shotgun proteomic data analyses. J. Proteome Res. 2004;3:1002–1008. doi: 10.1021/pr049920x. [DOI] [PubMed] [Google Scholar]
- 26.Chelius D, Bondarenko PV. Quantitative profiling of proteins in complex mixtures using liquid chromatography and mass spectrometry. J. Proteome Res. 2002;1:317–323. doi: 10.1021/pr025517j. [DOI] [PubMed] [Google Scholar]
- 27.Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, Muller SA, Rammner B, Grater F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmuller H, Heuser J, Wieland F, Jahn R. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. doi: 10.1016/j.cell.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 28.Morciano M, Burre J, Corvey C, Karas M, Zimmermann H, Volknandt W. Immunoisolation of two synaptic vesicle pools from synaptosomes: a proteomics analysis. J. Neurochem. 2005;95:1732–1745. doi: 10.1111/j.1471-4159.2005.03506.x. [DOI] [PubMed] [Google Scholar]
- 29.Wyszynski M, Kharazia V, Shanghvi R, Rao A, Beggs AH, Craig AM, Weinberg R, Sheng M. Differential regional expression and ultrastructural localization of α-actinin-2, a putative NMDA receptor-anchoring protein, in rat brain. J. Neurosci. 1998;18:1383–1392. doi: 10.1523/JNEUROSCI.18-04-01383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Petralia RS, Sans N, Wang YX, Wenthold RJ. Ontogeny of postsynaptic density proteins at glutamatergic synapses. Mol. Cell. Neurosci. 2005;29:436–452. doi: 10.1016/j.mcn.2005.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rubio ME, Curcio C, Chauvet N, Bruses JL. Assembly of the N-cadherin complex during synapse formation involves uncoupling of p120-catenin and association with presenilin 1. Mol. Cell. Neurosci. 2005;30:611–623. [PubMed] [Google Scholar]
- 32.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 33.Qiao F, Bowie JU. The many faces of SAM. Sci. STKE. 2005;2005:re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 34.Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR, Salyer D, Gundelfinger ED, Bowie JU. An architectural framework that may lie at the core of the postsynaptic density. Science. 2006;311:531–535. doi: 10.1126/science.1118995. [DOI] [PubMed] [Google Scholar]
- 35.Irie M, Hata Y, Takeuchi M, Ichtchenko K, Toyoda A, Hirao K, Takai Y, Rosahl TW, Sudhof TC. Binding of neuroligins to PSD-95. Science. 1997;277:1511–1515. doi: 10.1126/science.277.5331.1511. [DOI] [PubMed] [Google Scholar]
- 36.Lauren J, Airaksinen MS, Saarma M, Timmusk T. A novel gene family encoding leucine-rich repeat transmembrane proteins differentially expressed in the nervous system. Genomics. 2003;81:411–421. doi: 10.1016/s0888-7543(03)00030-2. [DOI] [PubMed] [Google Scholar]
- 37.Gulley RL, Reese TS. Cytoskeletal organization at the postsynaptic complex. J. Cell Biol. 1981;91:298–302. doi: 10.1083/jcb.91.1.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Rossum D, Hanisch UK. Cytoskeletal dynamics in dendritic spines: direct modulation by glutamate receptors? Trends Neurosci. 1999;22:290–295. doi: 10.1016/s0166-2236(99)01404-6. [DOI] [PubMed] [Google Scholar]
- 39.Migaud M, Charlesworth P, Dempster M, Webster LC, Watabe AM, Makhinson M, He Y, Ramsay MF, Morris RG, Morrison JH, O'Dell TJ, Grant SG. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 1998;396:433–439. doi: 10.1038/24790. [DOI] [PubMed] [Google Scholar]
- 40.El-Husseini AE, Schnell E, Chetkovich DM, Nicoll RA, Bredt DS. PSD-95 involvement in maturation of excitatory synapses. Science. 2000;290:1364–1368. [PubMed] [Google Scholar]
- 41.El-Husseini A-D, Schnell E, Dakoji S, Sweeney N, Zhou Q, Prange O, Gauthier-Campbell C, Aguilera-Moreno A, Nicoll RA, Bredt DS. Synaptic strength regulated by palmitate cycling on PSD-95. Cell. 2002;108:849–863. doi: 10.1016/s0092-8674(02)00683-9. [DOI] [PubMed] [Google Scholar]
- 42.Ehrlich I, Malinow R. Postsynaptic density 95 controls AMPA receptor incorporation during long-term potentiation and experience-driven synaptic plasticity. J. Neurosci. 2004;24:916–927. doi: 10.1523/JNEUROSCI.4733-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dakoji S, Tomita S, Karimzadegan S, Nicoll RA, Bredt DS. Interaction of transmembrane AMPA receptor regulatory proteins with multiple membrane associated guanylate kinases. Neuropharmacology. 2003;45:849–856. doi: 10.1016/s0028-3908(03)00267-3. [DOI] [PubMed] [Google Scholar]
- 44.Cottrell JR, Borok E, Horvath TL, Nedivi E. CPG2: a brain- and synapse-specific protein that regulates the endocytosis of glutamate receptors. Neuron. 2004;44:677–690. doi: 10.1016/j.neuron.2004.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dosemeci A, Tao-Cheng JH, Vinade L, Winters CA, Pozzo-Miller L, Reese TS. Glutamate-induced transient modification of the postsynaptic density. Proc. Natl. Acad. Sci. U. S. A. 2001;98:10428–10432. doi: 10.1073/pnas.181336998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dosemeci A, Vinade L, Winters CA, Reese TS, Tao-Cheng JH. Inhibition of phosphatase activity prolongs NMDA-induced modification of the postsynaptic density. J. Neurocytol. 2002;31:605–612. doi: 10.1023/a:1025735410738. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki T, Okumura-Noji K, Tanaka R, Tada T. Rapid translocation of cytosolic Ca2+/calmodulin-dependent protein kinase II into postsynaptic density after decapitation. J. Neurochem. 1994;63:1529–1537. doi: 10.1046/j.1471-4159.1994.63041529.x. [DOI] [PubMed] [Google Scholar]
- 48.Oh JS, Manzerra P, Kennedy MB. Regulation of the neuron-specific Ras GTPase-activating protein, synGAP, by Ca2+/calmodulin-dependent protein kinase II. J. Biol. Chem. 2004;279:17980–17988. doi: 10.1074/jbc.M314109200. [DOI] [PubMed] [Google Scholar]
- 49.Kim JH, Liao D, Lau LF, Huganir RL. SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron. 1998;20:683–691. doi: 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- 50.Cox R, Mason-Gamer RJ, Jackson CL, Segev N. Phylogenetic analysis of Sec7-domain-containing Arf nucleotide exchangers. Mol. Biol. Cell. 2004;15:1487–1505. doi: 10.1091/mbc.E03-06-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy JA, Jensen ON, Walikonis RS. BRAG1, a Sec7 domain-containing protein, is a component of the postsynaptic density of excitatory synapses. Brain Res. 2006;1120:35–45. doi: 10.1016/j.brainres.2006.08.096. [DOI] [PubMed] [Google Scholar]
- 52.Someya A, Sata M, Takeda K, Pacheco-Rodriguez G, Ferrans VJ, Moss J, Vaughan M. ARF-GEP100, a guanine nucleotide-exchange protein for ADP-ribosylation factor 6. Proc. Natl. Acad. Sci. U. S. A. 2001;98:2413–2418. doi: 10.1073/pnas.051634798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dunphy JL, Moravec R, Ly K, Lasell TK, Melancon P, Casanova JE. The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of beta1 integrins. Curr. Biol. 2006;16:315–320. doi: 10.1016/j.cub.2005.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inaba Y, Tian QB, Okano A, Zhang JP, Sakagami H, Miyazawa S, Li W, Komiyama A, Inokuchi K, Kondo H, Suzuki T. Brain-specific potential guanine nucleotide exchange factor for Arf, synArfGEF (Po), is localized to postsynaptic density. J. Neurochem. 2004;89:1347–1357. doi: 10.1111/j.1471-4159.2004.02440.x. [DOI] [PubMed] [Google Scholar]
- 55.Shin HW, Nakayama K. Guanine nucleotide-exchange factors for arf GTPases: their diverse functions in membrane traffic. J. Biochem. (Tokyo) 2004;136:761–767. doi: 10.1093/jb/mvh185. [DOI] [PubMed] [Google Scholar]
- 56.Donaldson JG, Honda A. Localization and function of Arf family GTPases. Biochem. Soc. Trans. 2005;33:639–642. doi: 10.1042/BST0330639. [DOI] [PubMed] [Google Scholar]
- 57.D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nat. Rev. Mol. Cell. Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 58.Miyazaki H, Yamazaki M, Watanabe H, Maehama T, Yokozeki T, Kanaho Y. The small GTPase ADP-ribosylation factor 6 negatively regulates dendritic spine formation. FEBS Lett. 2005;579:6834–6838. doi: 10.1016/j.febslet.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 59.Choi S, Ko J, Lee JR, Lee HW, Kim K, Chung HS, Kim H, Kim E. ARF6 and EFA6A regulate the development and maintenance of dendritic spines. J. Neurosci. 2006;26:4811–4819. doi: 10.1523/JNEUROSCI.4182-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


