Abstract
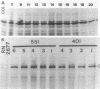
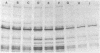
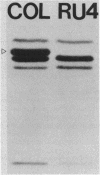
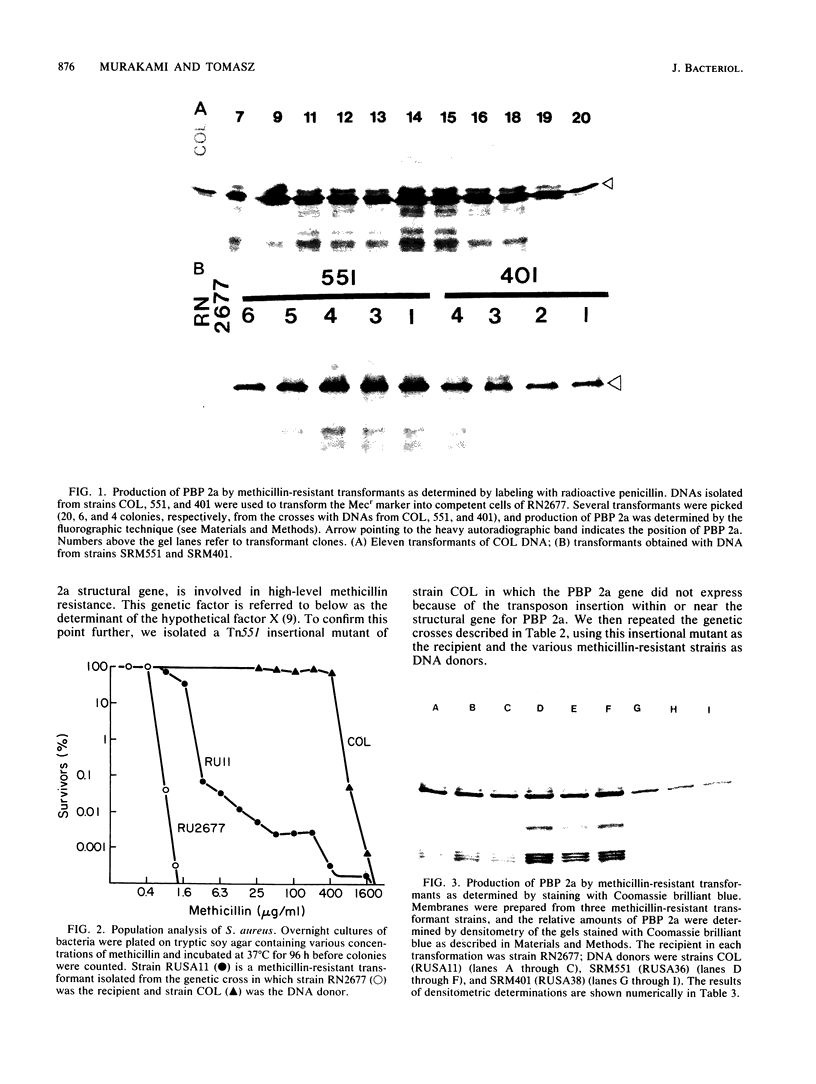
A methicillin-susceptible, novobiocin-resistant strain of Staphylococcus aureus (RN2677; methicillin MIC, 0.8 micrograms/ml) was transformed with DNA prepared from highly and homogeneously methicillin-resistant S. aureus strains (methicillin MIC, greater than or equal to 400 micrograms/ml) or from heterogeneous strains in which the majority of cells had a low level of resistance (methicillin MIC, 6.3 micrograms/ml). All methicillin-resistant transformants showed low and heterogeneous resistance (methicillin MIC, 3.1 micrograms/ml) irrespective of the resistance level of DNA donors. All transformants examined produced normal amounts of the low-affinity penicillin-binding protein (PBP) 2a, and methicillin resistance and the capacity to produce PBP 2a showed the same degree of genetic linkage to the novobiocin resistance marker with both homogeneous and heterogeneous DNA donors. Next, we isolated a methicillin-susceptible mutant from a highly and homogeneously resistant strain which had a Tn551 insertion near or within the PBP 2a gene and thus did not produce PBP 2a. With this mutant used as the recipient, genetic transformation of the methicillin resistance gene was repeated with DNA isolated either from highly and homogeneously resistant strains or from heterogeneous (low-resistance) strains. All transformants obtained expressed high and homogeneous resistance and produced PBP 2a irrespective of the resistance level of the DNA donors. Our findings suggest that (i) the methicillin resistance locus is identical to the structural gene for PBP 2a, (ii) although the ability to produce PBP 2a is essential for resistance, the MICs for the majority of cells are not related to the cellular concentration of PBP 2a, and (iii) high MICs and homogeneous expression of resistance require the products of other distinct genetic elements as well.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J. M., Freitag C. S., Clements J. R., Eisenstein B. I. An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Sep;82(17):5724–5727. doi: 10.1073/pnas.82.17.5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B. Insertional inactivation of staphylococcal methicillin resistance by Tn551. J Bacteriol. 1983 Apr;154(1):479–487. doi: 10.1128/jb.154.1.479-487.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B., Strässle A., Kayser F. H. Characterization of an isogenic set of methicillin-resistant and susceptible mutants of Staphylococcus aureus. Eur J Clin Microbiol. 1986 Dec;5(6):697–701. doi: 10.1007/BF02013308. [DOI] [PubMed] [Google Scholar]
- Brown D. F., Reynolds P. E. Intrinsic resistance to beta-lactam antibiotics in Staphylococcus aureus. FEBS Lett. 1980 Dec 29;122(2):275–278. doi: 10.1016/0014-5793(80)80455-8. [DOI] [PubMed] [Google Scholar]
- Chambers H. F., Hartman B. J., Tomasz A. Increased amounts of a novel penicillin-binding protein in a strain of methicillin-resistant Staphylococcus aureus exposed to nafcillin. J Clin Invest. 1985 Jul;76(1):325–331. doi: 10.1172/JCI111965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyke K. G. Penicillinase production and intrinsic resistance to penicillins in methicillin-resistant cultures of Staphylococcus aureus. J Med Microbiol. 1969 Aug;2(3):261–278. doi: 10.1099/00222615-2-3-261. [DOI] [PubMed] [Google Scholar]
- Georgopapadakou N. H., Smith S. A., Bonner D. P. Penicillin-binding proteins in a Staphylococcus aureus strain resistant to specific beta-lactam antibiotics. Antimicrob Agents Chemother. 1982 Jul;22(1):172–175. doi: 10.1128/aac.22.1.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Expression of methicillin resistance in heterogeneous strains of Staphylococcus aureus. Antimicrob Agents Chemother. 1986 Jan;29(1):85–92. doi: 10.1128/aac.29.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984 May;158(2):513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblum J., Hartman B. J., Novick R. P., Tomasz A. Conversion of a homogeneously methicillin-resistant strain of Staphylococcus aureus to heterogeneous resistance by Tn551-mediated insertional inactivation. Eur J Clin Microbiol. 1986 Dec;5(6):714–718. doi: 10.1007/BF02013311. [DOI] [PubMed] [Google Scholar]
- Kuhl S. A., Pattee P. A., Baldwin J. N. Chromosomal map location of the methicillin resistance determinant in Staphylococcus aureus. J Bacteriol. 1978 Aug;135(2):460–465. doi: 10.1128/jb.135.2.460-465.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Nomura K., Doi M., Yoshida T. Production of low-affinity penicillin-binding protein by low- and high-resistance groups of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1987 Sep;31(9):1307–1311. doi: 10.1128/aac.31.9.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M. T., Hewitt J. H. Methicillin resistance in Staphylococcus aureus. Lancet. 1970 Apr 18;1(7651):800–804. doi: 10.1016/s0140-6736(70)92408-6. [DOI] [PubMed] [Google Scholar]
- Rossi L., Tonin E., Cheng Y. R., Fontana R. Regulation of penicillin-binding protein activity: description of a methicillin-inducible penicillin-binding protein in Staphylococcus aureus. Antimicrob Agents Chemother. 1985 May;27(5):828–831. doi: 10.1128/aac.27.5.828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabath L. D., Wallace S. J. The problems of drug-resistant pathogenic bacteria. Factors influencing methicillin resistance in staphylococci. Ann N Y Acad Sci. 1971 Jun 11;182:258–266. doi: 10.1111/j.1749-6632.1971.tb30662.x. [DOI] [PubMed] [Google Scholar]
- Silverman M., Zieg J., Hilmen M., Simon M. Phase variation in Salmonella: genetic analysis of a recombinational switch. Proc Natl Acad Sci U S A. 1979 Jan;76(1):391–395. doi: 10.1073/pnas.76.1.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. D., Wachi M., Doi M., Ishino F., Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987 Aug 31;221(1):167–171. doi: 10.1016/0014-5793(87)80373-3. [DOI] [PubMed] [Google Scholar]
- Stahl M. L., Pattee P. A. Confirmation of protoplast fusion-derived linkages in Staphylococcus aureus by transformation with protoplast DNA. J Bacteriol. 1983 Apr;154(1):406–412. doi: 10.1128/jb.154.1.406-412.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson N. E., Pattee P. A. Transformation in Staphylococcus aureus: role of bacteriophage and incidence of competence among strains. J Bacteriol. 1977 Feb;129(2):778–788. doi: 10.1128/jb.129.2.778-788.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukata K., Yamashita N., Konno M. Occurrence of a beta-lactam-inducible penicillin-binding protein in methicillin-resistant staphylococci. Antimicrob Agents Chemother. 1985 May;27(5):851–857. doi: 10.1128/aac.27.5.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsui Y., Yokota T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1985 Sep;28(3):397–403. doi: 10.1128/aac.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]