Abstract
Local hypoxemia and stasis trigger thrombosis. We have demonstrated previously that in a murine model of normobaric hypoxia pulmonary fibrin deposition is a result of expression of tissue factor, especially in oxygen-deprived mononuclear phagocytes (MPs). We now show that transcription factor early-growth-response gene product (Egr-1) is rapidly activated in hypoxia, both in vitro and in vivo, and is responsible for transcription and expression of tissue factor in hypoxic lung. MPs and HeLa cells subjected to hypoxia (pO2 ≈13 torr) had increased levels of tissue factor transcripts (≈18-fold) and an increased rate of transcription (≈15-fold), based on nuclear run-on analysis. Gel-shift analysis of nuclear extracts from hypoxic MPs and HeLa cells demonstrated increased DNA-binding activity at the serum response region (SRR; −111/+14 bp) of the tissue factor promoter at Egr-1 motifs. Using 32P-labeled Egr consensus oligonucleotide, we observed induction of DNA-binding activity in nuclear extracts from hypoxic lung and HeLa cells because of activation of Egr-1, by means of supershift analysis. Transient transfection of HeLa cells with chimeric plasmids containing wild-type or mutant SRR from the tissue factor promoter showed that intact Sp1 sites are necessary for basal promoter activity, whereas the integrity of Egr-1 sites was required for hypoxia-enhanced expression. A central role for Egr-1 in hypoxia-mediated tissue factor expression was confirmed by experiments with homozygous Egr-1 null mice; wild-type mice subjected to oxygen deprivation expressed tissue factor and showed fibrin deposition, but hypoxic homozygous Egr-1 null mice displayed neither tissue factor nor fibrin. These data delineate a novel biology for hypoxia-induced fibrin deposition, in which oxygen deprivation-induced activation of Egr-1, resulting in expression of tissue factor, has an unexpected and central role.
Keywords: thrombosis/monocyte/transcription
Venous thrombosis, long associated with local hypoxemia and stasis, has a substantial morbidity and mortality, affecting 2–4 persons per 1,000 each year (1–4). Many studies have addressed means of preventing propagation of nascent thrombi, by inhibiting the contribution of platelets and/or plasma coagulation proteins, or by dissolving preformed fibrin, by using fibrinolytic agents. Much less attention has focused on mechanisms underlying the genesis of thrombi, such as that which occurs in stasis, accompanying limb immobilization, by far the most frequent thrombotic disorder in otherwise healthy individuals (1, 2). Almost 20 years ago, brief immobilization of canine extremities was shown to depress blood oxygen tension in the deep venous circulation to virtually undetectable levels (3, 4). A particularly susceptible locus for thrombus formation in this condition was the parietal aspect of the venous valve pocket, where oxygen tension fell off most quickly, strongly suggesting a link between hypoxemia and activation of coagulation (5).
Although multiple metabolic and hemodynamic factors contribute to the dysfunction of ischemic vasculature, oxygen deprivation is the common denominator. We therefore have sought to analyze how it contributes to triggering of procoagulant events in hypoxemic vasculature. Previously, we found that in a hypoxic environment, fibrin deposition occurred in pulmonary vasculature of otherwise normal mice, and that this was a result of de novo expression of tissue factor, especially in oxygen-deprived mononuclear phagocytes (6). We now have observed that hypoxia causes increased transcription of the tissue factor gene mediated by activation of the transcription factor early-growth-response gene product (Egr-1). Hypoxic mononuclear phagocytes (MPs) and HeLa cells showed increased tissue factor mRNA and increased rates of transcription, as compared with normoxic controls. Diminished levels of oxygen caused enhanced nuclear-binding activity for Egr-1 sites in the tissue factor promoter, as well as consensus Egr oligonucleotide. Based on transient transfection analysis, Egr-1 motifs in the tissue factor promoter were critical for hypoxia-mediated transcription of the tissue factor gene. Experiments in homozygous Egr-1 null mice displayed parallel suppression of tissue factor induction and fibrin formation, compared with wild-type controls, consistent with a central role for Egr-1. We propose that activation of Egr-1 is responsible for tissue factor expression in hypoxemic vasculature, thereby triggering a procoagulant pathway eventuating in fibrin formation.
METHODS
Cell Culture and in Vitro Analysis of Tissue Factor Gene Expression.
MPs were harvested from human peripheral blood by differential centrifugation (Histopaque 1077, Sigma), and plated in RPMI 1640 medium with fetal bovine serum (10%) for 48 hr (6). Experiments were also performed with the rat macrophage line (NR8383) obtained from ATCC and grown in F12K medium supplemented with heat-inactivated fetal bovine serum (15%). HeLa cells, obtained from ATCC, were grown in DMEM supplemented with fetal bovine serum (10%). Serum-starved cells (24 hr) were subjected to hypoxia after replacement of the growth medium with fresh medium preequilibrated with the hypoxic gas mixture (6). The balance of the atmosphere was made up of carbon dioxide (pCO2 ≈ 40 torr) and water vapor (≈47 torr), both of which were kept constant, via an automated valve system. Oxygen content of medium bathing the cells was maintained between 12 and 14 torr. Cell viability was maintained throughout experiments performed under the above hypoxic conditions up to the longest time point.
Northern Analysis.
Cultured MPs or HeLa cells were deprived of serum for 24 hr and then subjected to normoxia or hypoxia for 4 hr followed by harvesting of RNA. Total RNA (30 μg/lane), isolated by using TRIzol Reagent (GIBCO/BRL) from MPs, HeLa cells, or murine lung, was subjected to electrophoresis in 0.8% agarose-formaldehyde gels and transferred to Duralon-UV membranes (Stratagene). Membranes were hybridized with human or mouse tissue factor cDNA fragments (6), the latter labeled by using [α-32P]dCTP (3,000 μCi/mmol, DuPont), and then were exposed to Kodak Biomax film at −80°C. Membranes were stripped and rehybridized with human β-actin cDNA probe as a control for RNA loading.
Nuclear Run-On Analysis.
Cultured MPs or HeLa cells were deprived of serum for 24 hr and then subjected to normoxia or hypoxia for 4 hr. Nuclei were isolated and in vitro transcription was performed as described (7).
Gel-Shift Assays.
Nuclear extracts were prepared from MPs, HeLa cells, or murine lung after exposure to hypoxia by the procedure of Dignam et al. (8). Electrophoretic mobility-shift assay (EMSA) was performed by using oligonucleotide probes from the serum response region (SRR) of the tissue factor gene, denoted R1 (−114/−89), R2 (−96/−66), and R3 (−77/−46) (9) or consensus oligonucleotide probes for AP-1 (Santa Cruz Biotechnology), HIF-1 [5′-GCCCTACGTGCTGTCTCA-3′; (10)], Sp1 (Santa Cruz Biotechnology), nuclear factor interleukin 6 (NF-IL-6) (7), or Egr (Santa Cruz Biotechnology). In each case, double-stranded oligonucleotide probes were 5′ end-labeled with [32P]ATP (3,000 Ci/mmol) by using T4 polynucleotide kinase and standard procedures (11). Binding reactions were performed as described (7). Where indicated, antiserum to Egr-1 or nonimmune IgG was incubated with nuclear extract at room temperature for 1 hr, and then the procedure was as described (14). Samples (5 μg of protein in each lane) were loaded directly onto nondenaturing polyacrylamide/bisacrylamide (6%) gels prepared in 0.5× TBE (45 mM Tris/45 mM boric acid/1 mM EDTA). The gels were pre-run for 20 min before samples were loaded. Electrophoresis was performed at room temperature for 1.5–2 hr at 200 V. For competition studies, a 100-fold molar excess of one of the above unlabeled probes was employed.
Transient Transfection.
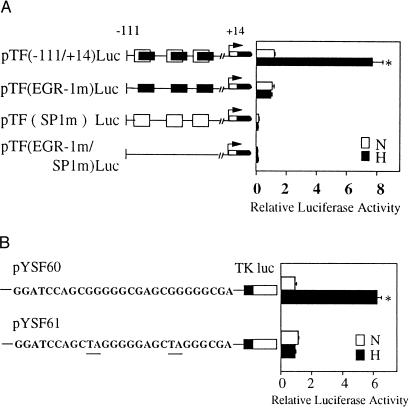
HeLa cells were cotransfected by using lipofectamine reagent (GIBCO). Supercoiled plasmid DNAs were prepared by Endofree Plasmid Maxikit (Qiagen, Chatsworth, CA). The constructs pTF(−111/+14)Luc, pTF(EGR-1m)Luc, pTF(SP1m)Luc, and pTF(EGR-1m/SP1 m)Luc were prepared as described previously (9). To make constructs pYSF60 and pYSF61, the two 27-bp oligonucleotides shown in Fig. 3B were ligated to the basal thymidine kinase promoter (7) and the luciferase reporter gene (from pGL3; Promega). Briefly, the day before transfection, exponentially growing cells were harvested by trypsinization, replated at a density of 4 × 105 per 35-mm well in DMEM containing fetal bovine serum (2%), and cultures were incubated at 37°C in a humidified atmosphere containing 5% CO2 until they reached 50–80% confluence. DNA–lipofectamine complexes were prepared by combining one of the above constructs (1 μg) and pCMV-β-galactosidase DNA (1 μg), as well as 12 μl of lipofectamine reagent in 0.2 ml of serum-free OPTI-MEM1 Reduced Serum Medium (GIBCO/BRL), respectively, followed by incubation at room temperature for 45 min. Then, the complexes were mixed with 1 ml of OPTI-MEM1. After 6 hr of transfection, cultures were washed twice with balanced salt solution (PBS), and then the medium was changed to DMEM containing 2% fetal bovine serum. After another 30 hr at 37°C, medium from cultures was aspirated, cells were washed with balanced salt solution, and serum-free DMEM was added. Cells were incubated for 6 hr at 37°C with 5% CO2 in normoxia or hypoxia. After the incubation period, cell extracts were prepared by using Reporter Lysis Buffer (Promega). Luciferase activity was determined by using the Luciferase Assay System (Promega) and Lumat LB9501 (Wallac, Gaithersburg MD). pCMV-β-galactosidase (CLONTECH) was used as an internal control for efficiency of transfection. Luciferase activity was normalized based on β-galactosidase activity in the same well; this is termed relative luciferase activity (x axis in Fig. 3).
Figure 3.
Hypoxia-inducible tissue factor expression results from transcriptional activation at Egr-1 sites. (A) Transient cotransfection of HeLa cells was performed by using either pTF(−111/+14)Luc, pTF(EGR-1 m)Luc, pTF(SP1 m)Luc, or pTF(EGR-1 m/SP1 m)Luc, and pCMV-β-galactosidase. Cultures were transfected with each of the indicated constructs by using the lipofectamine procedure (GIBCO), and then cells were exposed to normoxia (N) or hypoxia (H) for 5 hr. Luciferase and β-galactosidase activity were then determined. Relative luciferase activity is luciferase activity normalized for β-galactosidase activity. (B) Transient transfection of HeLa cells using pYSF60 (consensus Egr wild-type sequence) or pYSF61 (mutationally inactivated Egr sequence) and pCMV-β-galactosidase by the same procedure described above. Results shown are representative of a minimum of four experiments.
In Vivo Studies of Tissue Factor Expression and Fibrin Deposition.
Homozygous Egr-1 knockout mice (12, 13) or nontransgenic littermate controls were exposed to hypoxia by using an environmental chamber designed for small animals (5). After the indicated time in hypoxia, tissue was fixed in formalin and processed for immunohistochemistry as described (14). Immunoblotting for fibrin epitopes, using an antibody to fibrin γ-γ chain crosslinks (15), and localization tissue factor antigen in tissue sections, by immunohistochemical techniques (6), was performed as described. Fibrin immunostaining was performed by using the same method and antibody prepared to fibrin γ-γ chain crosslinks.
RESULTS
Role of Egr-1 in Hypoxia-Induced Tissue Factor Expression in Cultured Mononuclear Phagocytes and HeLa Cells.
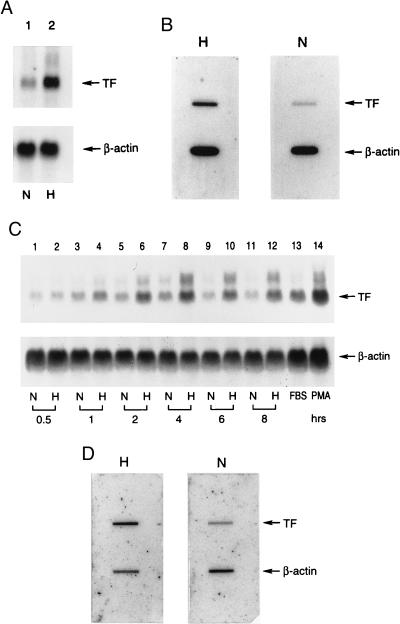
Because hypoxic induction of tissue factor mRNA has been observed in a cultured mutant hepatoma cell line deficient in hypoxia-inducible factor-1 (HIF-1)-β (16), implicating a mechanism independent of HIF-1 (17), a novel pathway was likely to underlie oxygen deprivation-mediated regulation of tissue factor expression. Tissue factor up-regulation occurred prominently in MPs in hypoxic lung, and human peripheral blood-derived mononuclear phagocytes in culture also showed enhanced tissue factor levels on exposure to hypoxia. Up-regulation of tissue factor mRNA (≈18-fold based on densitometry) was observed in hypoxic MPs (Fig. 1A) as a result of, in large part, de novo transcription as shown by nuclear run-on analysis (Fig. 1B; ≈15-fold increase in tissue factor transcripts). To analyze further mechanisms of transcriptional activation, a cell system other than blood monocytes had to be identified, because human monocytes do not proliferate and established monocyte cell lines did not reproducibly show hypoxia-mediated up-regulation of tissue factor. HeLa cells subjected to oxygen deprivation displayed increased tissue factor mRNA, maximal within 4 hr (Fig. 1C), principally reflecting increased transcription (Fig. 1D; ≈10-fold increase in tissue factor).
Figure 1.
Effect of hypoxia on tissue factor gene expression in mononuclear phagocytes (A and B) and HeLa cells (C and D). (A and C) Northern analysis for tissue factor transcripts. Human peripheral blood mononuclear phagocytes (≈106 cells, A) or HeLa cells (≈0.5 × 106 cells, C) in serum-free medium were subjected to normoxia (N) or hypoxia (H; pO2 ≈ 12–14 torr) for the indicated times (HeLa) or for 4 hr (mononuclear phagocytes). Northern analysis was performed by loading 30 μg/lane of total RNA and using 32P-labeled cDNA for human tissue factor (Upper) or human β-actin (Lower). FBS and PMA denote cultures exposed to fetal bovine serum (20%) or phorbol myristate acetate (50 ng/ml), respectively, for 1 hr in each case. (B and D) Nuclear run-on analysis for the rate of tissue factor transcription. Mononuclear phagocytes (≈107 cells, B) or HeLa cells (≈106 cells, D) were subjected to normoxia or hypoxia (as above) for 4 hr, nuclei were harvested, labeled by incubation with [α-32P]dUTP for 1 hr, the RNA was isolated, and the same amount of radioactivity was hybridized with cDNA probes for tissue factor or β-actin, the latter already immobilized on nitrocellulose membranes. Results shown are representative of a minimum of four experiments.
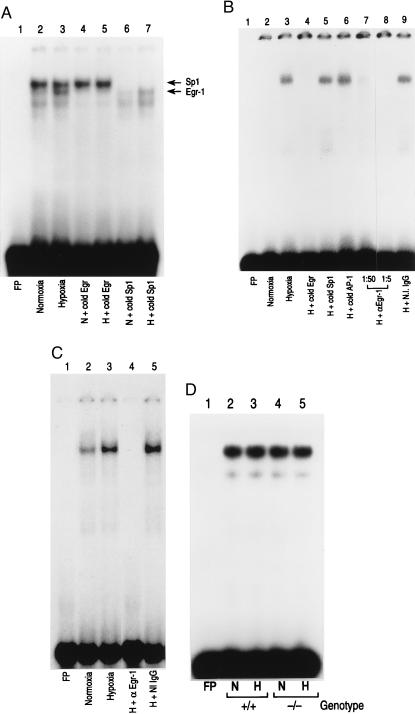
Recent studies have highlighted a role for Egr-1 in vascular injury (18, 19), as well as the contribution of a trio of 12 bp overlapping Egr-1 and Sp1 DNA-binding sites in the SRR of the tissue factor promoter (−111/+14 bp) in regulation of tissue factor expression (9). Oligonucleotide probes spanning each of the Egr-1/Sp1 DNA-binding sites, termed R1, R2, and R3 (9), were 32P-labeled and employed in EMSA with nuclear extracts from hypoxic/normoxic HeLa cells and monocytes. Rat MP and HeLa nuclear extracts (data with HeLa cells are not shown) from both normoxic and hypoxic cultures showed a strong constitutive band (labeled Sp1 in Fig. 2A, lanes 2 and 3; results with the R2 probe are shown) whose appearance was blocked by inclusion of excess unlabeled Sp1 probe (lanes 6 and 7), but not by Egr oligonucleotide (lanes 4 and 5). Only hypoxic cultures showed a new gel-shift band (Fig. 2A, lane 3; this hypoxia-inducible band is labeled Egr-1) whose appearance was blocked by excess unlabeled Egr probe (Fig. 2A, lane 5), but not by Sp1 oligonucleotide (Fig. 2A, lane 7). Because of the greater intensity of the proximal constitutive Sp1 bands relative to the inducible Egr band, it was difficult to accurately judge competition with unlabeled Sp1 oligonucleotide for Egr DNA-binding activity with probes containing overlapping Sp1 and Egr sites. For this reason, experiments also were performed separately by using consensus Sp1 and Egr probes (see below). Competition experiments demonstrated specificity of the observed protein–DNA binding complexes, because excess unlabeled oligonucleotide probes corresponding to consensus sequences for AP-1, HIF-1, and NF-IL-6 had no effect on the observed gel-shift bands (data not shown). Similar results were obtained when nuclear extracts of hypoxic MPs and HeLa cells were studied with 32P-labeled R1 and R3 probes (data not shown). Because of these comparable observations with the two cell types, further in vitro studies were performed in the more conveniently manipulated HeLa cells.
Figure 2.
EMSA of tissue factor promoter DNA-binding motifs: effect of hypoxia. (A) EMSA using R2 from the tissue factor promoter. Rat mononuclear phagocytes (≈107, A) were exposed to normoxia (N) or hypoxia (H; pO2 ≈ 12–14 torr) for 45 min, nuclear extracts were prepared, and EMSA was performed using 32P-labeled oligonucleotide for R2 (−96/−66 bp; ref. 9). Each lane received 10 μg/lane of total nuclear extract protein. The arrows indicate bands corresponding to Egr and Sp1 (A). Note that the unlabeled lower band observed in lanes 2, 3, and 7 may represent Sp3 (38). A 100-fold excess of the indicated unlabeled (cold) oligonucleotide with a consensus sequence of Sp1 or Egr was added. (B) EMSA using consensus oligonucleotide probe Egr. Nuclear extracts were harvested from normoxic/hypoxic HeLa cells and EMSA was performed with the indicated 32P-labeled oligonucleotide probe. Excess unlabeled Sp1, AP-1, or Egr oligonucleotide was added to certain lanes, and either rabbit anti-Egr-1 IgG (1:5 and 1:50 dilution) or nonimmune IgG (1:5) was added to other lanes. (C) EMSA using consensus oligonucleotide for Egr, as above, and nuclear extracts from normoxic/hypoxic murine lung. Mice were subjected to hypoxia (≈6% oxygen; ref. 6) for 45 min, lungs were rapidly harvested, and nuclear extracts were prepared. EMSA was performed as above, and, where indicated, anti-Egr-1 IgG or nonimmune IgG (Santa Cruz Biotechnology) was added. (D) EMSA using consensus oligonucleotide for Sp-1 and nuclear extracts from normoxic/hypoxic murine lung was performed using the same conditions as above (+/+, wild-type mice; −/−, Egr-1 null mice). Results shown are representative of a minimum of four experiments.
To facilitate analysis of the protein–DNA complexes binding to the R2 probe, EMSA was performed by using 32P-labeled consensus oligonucleotides for only Sp1 or Egr. EMSA using 32P-labeled consensus Egr probe showed a gel-shift band in nuclear extracts from hypoxic (Fig. 2B, lane 3), but not from normoxic, HeLa cells (Fig. 2B, lane 2). Addition of excess unlabeled Egr probe blocked appearance of the hypoxia-induced gel-shift band (Fig. 2B, lane 4), but oligonucleotides corresponding to Sp1 or AP-1 were without effect (Fig. 2B, lanes 5 and 6). Inclusion of antibody to Egr-1 in the reaction mixture blocked appearance of the hypoxia-induced gel-shift band observed with the Egr probe, depending on the amount of antibody added (Fig. 2B, lanes 7 and 8), whereas nonimmune IgG was without effect (Fig. 2B, lane 9). Consistent with these data, EMSA of nuclear extracts from hypoxic murine lung (harvested within 45 min of oxygen deprivation) similarly displayed an enhancement of DNA-binding activity for the Egr probe (Fig. 2C, lanes 2 and 3), and the latter gel-shift band disappeared on addition of antibody to Egr-1, but not with nonimmune IgG (Fig. 2C, lanes 4 and 5).
Quite different results were obtained when nuclear extracts from hypoxic cells and lung were evaluated for DNA-binding activity for the Sp1 consensus oligonucleotide. With 32P-Sp1 probe, a prominent band of equal intensity was observed with nuclear extracts from both normoxic and hypoxic HeLa cells (data not shown). Gel-shift analysis of nuclear extracts from hypoxic wild-type murine lung using the Sp-1 probe showed no change in intensity of the band when comparing samples from hypoxic and normoxic tissue (Fig. 2D, +/+). A similar pattern of Sp-1 DNA-binding activity in hypoxic and normoxic lung was shown in nuclear extracts from Egr-1 null mice (Fig. 2D, −/−).
To better determine the role of Egr-1 in driving tissue factor transcription in HeLa cells subjected to hypoxia, transient transfection was performed with chimeric promoter–reporter constructs spanning the SRR (−111/+14 bp) (Fig. 3A). As an internal control for efficiency of transfection, cotransfection was performed with pCMV-β-galactosidase. Results are shown as the fold-increase in luciferase activity normalized for β-galactosidase activity (see Methods). Increased expression of the luciferase reporter was observed in hypoxic cultures after transfection with wild-type SRR in which both Sp1 and Egr-1 sites were intact (Fig. 3A; ≈6-fold), compared with normoxic controls. A mutant construct in which all three Egr-1 sites in the SRR were inactivated showed no increase in luciferase activity when transfected cultures were exposed to hypoxia (Fig. 3A). Consistent with a central role for Sp1 in basal tissue factor expression (9), mutational inactivation of all three Sp1 sites blocked both basal and hypoxia enhanceable expression of luciferase (Fig. 3A). Similarly, a construct in which both Sp1 and Egr-1 sites were inactivated (Fig. 3A) showed expression similar to that with the construct in which Sp1 was inactivated. These data led us to further dissect the role of Egr-1 by making a construct composed of the Egr consensus DNA-binding motif, the basal thymidine kinase promoter (−80/+18), and luciferase. Two constructs were prepared: pYSF60, corresponding to wild-type Egr consensus site, and pYSF61, corresponding to a mutationally inactivated Egr consensus site (Fig. 3B). Transient transfection of these constructs into HeLa cells demonstrated hypoxic enhancement of reporter expression only in the presence of the intact Egr DNA-binding site, but not with the mutationally inactivated Egr construct.
Role of Egr-1 in Pulmonary Tissue Factor Expression and Fibrin Deposition in a Murine Model of Hypoxia/Hypoxemia.
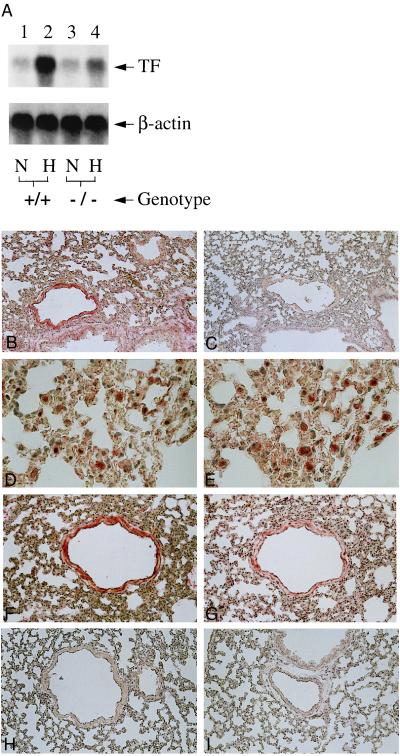
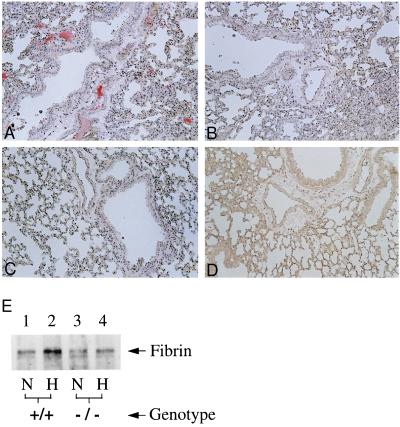
To move from an association of Egr-1 with enhanced tissue factor expression in hypoxia to a cause–effect relationship, we studied Egr-1 null mice (12, 13, 20). Our analysis was performed in Egr-1 null mice and nontransgenic controls, and representative results are shown. Exposure of female Egr-1 knockout mice to hypoxia resulted in a severely blunted increase in tissue factor mRNA in lung: compared with an ≈20-fold increase in tissue factor transcripts in wild-type mice subjected to hypoxia (versus normoxic controls), there was only an ≈2-fold increase in tissue factor mRNA in Egr-1 null mice (Fig. 4A). Abundant tissue factor antigen was present in lungs of wild-type mice subjected to hypoxia (Fig. 4B, versus wild-type normoxic controls in Fig. 4C). Immunohistologic studies were performed on animals to detect tissue factor or fibrin antigen (see below) by using Egr-1 null mice and nontransgenic littermates. For each animal, sections from every lobe of one lung were analyzed; sections in which there were pulmonary vessels of comparable size were compared, and representative results are shown. Tissue factor antigen (Fig. 4 D and F) was colocalized with mononuclear phagocytes (Fig. 4E; identified using the F4/80 antibody) and smooth muscle cells (Fig. 4G; identified using antibody to smooth muscle α-actin). In contrast, tissue factor was virtually undetectable in lungs from hypoxic and normoxic Egr-1 null mice (Fig. 4 H and I, respectively). Consistent with a central role for hypoxia-induced tissue factor expression in fibrin formation/deposition in the vasculature (6), immunostaining of hypoxic lung with antibody to γ-γ chain dimers (15) showed intravascular fibrin in samples from wild-type mice (Fig. 5A), whereas there was no detectable fibrin in samples from normoxic wild-type controls (Fig. 5B). In contrast, no fibrin epitopes were observed in either hypoxic or normoxic Egr-1 null mice (Fig. 5 C and D, respectively). To more quantitatively assess fibrin deposition in lung, immunoblotting with antibody to γ-γ chain crosslinks was performed on lung tissue extracts (6): a strong immunoreactive band was detected in samples from wild-type hypoxic mice, but only a faint band was seen in samples from normoxic controls (Fig. 5E, lanes 1 and 2). Only a very weak band corresponding to fibrin neoepitope was observed in either normoxic or hypoxic samples from Egr-1 null mice (Fig. 5E, lanes 3 and 4).
Figure 4.
Hypoxia-mediated induction of tissue factor expression in murine lung: Egr-1 null mice show reduced tissue factor mRNA (A) and antigen (B–I). (A) Mice (wild-type, +/+, or homozygous null mice, −/−) were subjected to normoxia (N) or hypoxia (H; 6% oxygen), lungs were rapidly harvested, total RNA was prepared, and Northern analysis (30 μg/lane of total RNA) was performed with 32P-labeled cDNA for mouse tissue factor (Upper) or β-actin (Lower). (B–I) Immunohistochemical analysis for tissue factor antigen was performed on lung tissue from mice exposed to normoxia or hypoxia. (B and C) Hypoxic and normoxic wild-type mouse lung, respectively, stained with antitissue factor IgG. (D and E) Higher-power micrograph of hypoxic wild-type mouse lung stained with anti-tissue factor IgG (D) with an adjacent section stained with antibody to F4/80 to detect mononuclear phagocytes (E). (F and G) Higher-power micrograph of hypoxic wild-type mouse lung stained with anti-tissue factor IgG (F) with an adjacent section stained with antibody to smooth muscle α-actin (G). (H and I) Hypoxic and normoxic egr-1 −/− mouse stained with anti-tissue factor IgG. [×200 (B and C, F–I) and ×600 (D and E).] Results shown are representative of a minimum of three experiments.
Figure 5.
Hypoxia-mediated induction of fibrin deposition in murine lung: egr-1 null mice show no increase in fibrin deposition. Wild-type or homozygous Egr-1 null mice were subjected to hypoxia as above. (A–D) Immunostaining was performed with antibody to fibrin γ-γ chain crosslinks (15). (A) Hypoxic wild-type murine lung. (B) Normoxic wild-type murine lung. (C) Hypoxic Egr-1 −/− murine lung. (D) Normoxic Egr-1 −/− murine lung. (×200.) (E) Immunoblotting (5) using the antibody to γ-γ chain crosslinks (15) was performed on plasmin-treated extracts of murine lung harvested from normoxic (N)/hypoxic (H) wild-type or Egr-1 −/− mice. Results shown are representative of a minimum of three experiments.
DISCUSSION
Mononuclear phagocytes subjected to hypoxia/hypoxemia display enhanced tissue factor expression in vitro and in vivo. These observations suggest a novel scenario for hypoxia-induced thrombosis associated with diminished blood flow or frank stasis. Blood monocytes become arrested (6) and activated at sites of hypoxemic vascular injury potentially because of signals emitted by hypoxic endothelium. Perturbation of mononuclear phagocytes in the oxygen-depleted environment causes expression of tissue factor, allowing coagulation to be initiated, and activated products of the procoagulant pathway accumulate in static portions of the vascular tree. This biology of thrombosis is especially relevant to venous valve pockets, which are frequently subject to stasis and are a well known nidus for clot formation (1–4). A contribution of endothelium may be at the level of expression of chemotactic activity; monocyte chemotactic protein-1 (JE/MCP-1) is expressed by hypoxic endothelial cells (21). Thus, we speculate that the biology of hypoxia-mediated thrombosis assigns monocytes a central role to effect a primordial response causing isolation of an ischemic area from the circulation by a protective fibrin clot.
In addition to providing insights into a key mechanism underlying hypoxia-induced thrombosis, our studies have demonstrated a pathophysiologically relevant model of hypoxia-associated gene regulation independent of HIF-1 (17). The role of Egr-1 in physiologic and pathophysiologic (22) processes has been clarified by observations in Egr-1 null mice (12 and 13). Whereas Egr-1 was postulated to be an essential factor in macrophage differentiation (23) and multiple other basic cellular homeostatic events (22), based on in vitro studies, analysis of Egr-1 null mice has not borne out many of these predictions (12, 13, 20). Rather, the phenotype of Egr-1 knockout mice becomes evident only after environmental stress: female homozygote knockout mice are sterile because of luteinizing hormone (LH)-β deficiency, an effect mediated at the level of LH-β transcription (12), and we have shown here that Egr-1 is necessary for tissue factor induction in the setting of environmental oxygen deprivation. The potential significance of Egr-1 activation in hypoxia concerns the range of potential target genes and cellular functions thereby stimulated by these gene products. Of these, we previously have observed enhanced expression of intercellular adhesion molecule-1 in hypoxemic vasculature (24), where it could have an important role in arresting leukocytes in ischemic tissues. These considerations suggest that there may be important contrasts between hypoxia-induced adaptive responses subject to regulation by HIF-1 (17, 25, 26) and those that we think to be under control of Egr-1. Many studies have shown that HIF-1 regulates expression of genes involved in the metabolic adjustment to oxygen deprivation [glycolytic enzymes (27–30) and the noninsulin-dependent glucose transporter, GLUT-1 (31)], angiogenesis (vascular endothelial growth factor; ref. 32), and vasomotor control (nitric-oxide synthase and heme oxygenase; refs. 33 and 34). Egr-1-mediated gene expression appears to orchestrate thromobogenesis and, potentially, cell–cell interactions and inflammatory mechanisms (18, 35–37), suggesting activation of a distinct type of host response mechanisms in the hypoxemic milieu. The range of Egr-1 target genes activated by hypoxia and their consequences for the host response to oxygen deprivation provide a new approach to the biology of thrombosis and effector mechanisms triggered in ischemia.
Acknowledgments
Dr. Gabriel Godman provided valuable suggestions during the course of this work and preparation of the manuscript. This work was supported by grants from the U.S. Public Health Service (HL35246 to W.K.; HL42507, HL55397, and HD13063C to D.S.) and the Surgical Research Fund.
ABBREVIATIONS
- EGR-1
early-growth-response gene product
- MP
mononuclear phagocyte
- EMSA
electrophoretic mobility-shift assay
- SRR
serum response region
- HIF-1
hypoxia-inducible factor-1
References
- 1.Geerts W, Code K, Jay R, Chen E, Szalai J. N Engl J Med. 1994;331:1601–1606. doi: 10.1056/NEJM199412153312401. [DOI] [PubMed] [Google Scholar]
- 2.Anderson F, Wheeler H, Goldberg R, Hosmer D, Patwardhan N, Jovanovic B, Forcier A, Dalen J. Arch Intern Med. 1991;151:933–938. [PubMed] [Google Scholar]
- 3.Malone P, Morris C. J Pathol. 1978;125:119–129. doi: 10.1002/path.1711250302. [DOI] [PubMed] [Google Scholar]
- 4.Hamer J, Malone P, Silver I. Br J Surg. 1981;68:166–170. doi: 10.1002/bjs.1800680308. [DOI] [PubMed] [Google Scholar]
- 5.Malone P. Med Hypotheses. 1977;5:189–201. doi: 10.1016/0306-9877(77)90005-6. [DOI] [PubMed] [Google Scholar]
- 6.Lawson C, Laio H, Yan S-D, Yan S-F, Sobel J, Kisiel W, Stern D, Pinsky D. J Clin Invest. 1997;99:1729–1738. doi: 10.1172/JCI119337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan S-F, Tritto I, Pinsky D, Liao H, Huang J, Fuller G, Brett J, May L, Stern D. J Biol Chem. 1995;270:11463–11471. doi: 10.1074/jbc.270.19.11463. [DOI] [PubMed] [Google Scholar]
- 8.Dignam J, Lebovitz R, Roeder R. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui M-Z, Parry G, Oeth P, Larson H, Smith M, Huang R-P, Adamson E, Mackman N. J Biol Chem. 1996;271:2731–2739. doi: 10.1074/jbc.271.5.2731. [DOI] [PubMed] [Google Scholar]
- 10.Semenza G, Wang G. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 12.Lee S, Sadovsky Y, Swirnoff A, Polish J, Goda P, Gavrilina G, Milbrandt J. Science. 1996;273:1219–1221. doi: 10.1126/science.273.5279.1219. [DOI] [PubMed] [Google Scholar]
- 13.Lee S, Wang Y, Milbrandt J. Mol Cell Biol. 1996;16:4566–4572. doi: 10.1128/mcb.16.8.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan S-F, Mendelsohn M, Zou Y S, Gao Y, Naka Y, Yan S-D, Pinsky D, Stern D. J Biol Chem. 1997;272:4287–4294. doi: 10.1074/jbc.272.7.4287. [DOI] [PubMed] [Google Scholar]
- 15.Lahiri B, Koehn J, Canfield R, Birken S, Lewis J. Thromb Res. 1981;23:103–112. doi: 10.1016/0049-3848(81)90243-7. [DOI] [PubMed] [Google Scholar]
- 16.O’Rourke J, Pugh C, Bartlett S, Ratcliffe P. Eur J Biochem. 1996;241:403–410. doi: 10.1111/j.1432-1033.1996.00403.x. [DOI] [PubMed] [Google Scholar]
- 17.Guillemin K, Krasnow M. Cell. 1997;89:9–12. doi: 10.1016/s0092-8674(00)80176-2. [DOI] [PubMed] [Google Scholar]
- 18.Khachigian L, Lindner V, Williams A, Collins T. Science. 1996;271:1427–1431. doi: 10.1126/science.271.5254.1427. [DOI] [PubMed] [Google Scholar]
- 19.Khachigian L, Williams A, Collins T. J Biol Chem. 1995;270:27679–27686. doi: 10.1074/jbc.270.46.27679. [DOI] [PubMed] [Google Scholar]
- 20.Lee S, Tourtellotte L, Wesselschmidt R, Milbrandt J. J Biol Chem. 1995;270:9971–9977. doi: 10.1074/jbc.270.17.9971. [DOI] [PubMed] [Google Scholar]
- 21.Karakurum M, Shreeniwas R, Chen J, Sunouchi J, Hamilton T, Anderson M, Kuwabara K, Rot A, Nowygrod R, Stern D. J Clin Invest. 1994;93:1564–1570. doi: 10.1172/JCI117135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gashler A, Sukhatme V. Prog Nucleic Acid Res Mol Biol. 1995;50:191–224. doi: 10.1016/s0079-6603(08)60815-6. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen H, Hoffman-Liebermann B, Liebermann D. Cell. 1993;72:197–209. doi: 10.1016/0092-8674(93)90660-i. [DOI] [PubMed] [Google Scholar]
- 24.Shreeniwas R, Koga S, Pinsky D, Kaiser E, Brett J, Wolitzky B, Norton C, Plocinski J, Benjamin W, Burns D, Goldstein A, Stern D. J Clin Invest. 1992;90:2333–2339. doi: 10.1172/JCI116122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semenza G. Trends Cardiovasc Med. 1996;6:151–158. doi: 10.1016/1050-1738(96)00039-4. [DOI] [PubMed] [Google Scholar]
- 26.Ratcliffe P, Ebert B, Firth J, Gleadle J, Maxwell P, Nagao M, O’Rourke J, Pugh C, Wood S. Kidney Intl. 1997;51:514–526. doi: 10.1038/ki.1997.72. [DOI] [PubMed] [Google Scholar]
- 27.Semenza G, Roth P, Fang H-M, Wang G. J Biol Chem. 1994;269:23757–23763. [PubMed] [Google Scholar]
- 28.Firth J, Ebert B, Pugh C, Ratcliffe P. Proc Natl Acad Sci USA. 1994;91:6496–6500. doi: 10.1073/pnas.91.14.6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Firth J, Ebert B, Ratcliffe P. J Biol Chem. 1995;270:21021–21027. doi: 10.1074/jbc.270.36.21021. [DOI] [PubMed] [Google Scholar]
- 30.Ebert B, Gleadle J, O’Rourke J, Bartlett S, Poulton J, Ratcliffe P. Biochem J. 1996;313:809–814. doi: 10.1042/bj3130809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebert B, Firth J, Ratcliffe P. J Biol Chem. 1995;270:29083–29089. doi: 10.1074/jbc.270.49.29083. [DOI] [PubMed] [Google Scholar]
- 32.Shweiki D, Itin A, Soffer D, Keshet E. Nature (London) 1992;359:843–845. doi: 10.1038/359843a0. [DOI] [PubMed] [Google Scholar]
- 33.McQuillan L, Leung G, Marsden P, Kostyk S, Kourembanas S. Am J Physiol. 1994;267:H1921–H1927. doi: 10.1152/ajpheart.1994.267.5.H1921. [DOI] [PubMed] [Google Scholar]
- 34.Morita T, Perrella M, Lee M-E, Kourembanas S. Proc Natl Acad Sci USA. 1995;92:1475–1479. doi: 10.1073/pnas.92.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maltzman J, Carman J, Monroe J. J Exp Med. 1996;183:1747–1759. doi: 10.1084/jem.183.4.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skerka C, Decker E, Zipfel P. J Biol Chem. 1995;270:22500–22506. doi: 10.1074/jbc.270.38.22500. [DOI] [PubMed] [Google Scholar]
- 37.Kramer B, Meichle A, Hensel G, Charnay P, Kronke M. Biochim Biophys Acta. 1994;1219:413–421. doi: 10.1016/0167-4781(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 38.Hagen G, Muller S, Beato M, Suske G. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]