Abstract
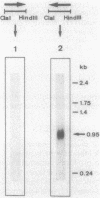
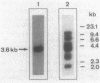
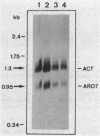
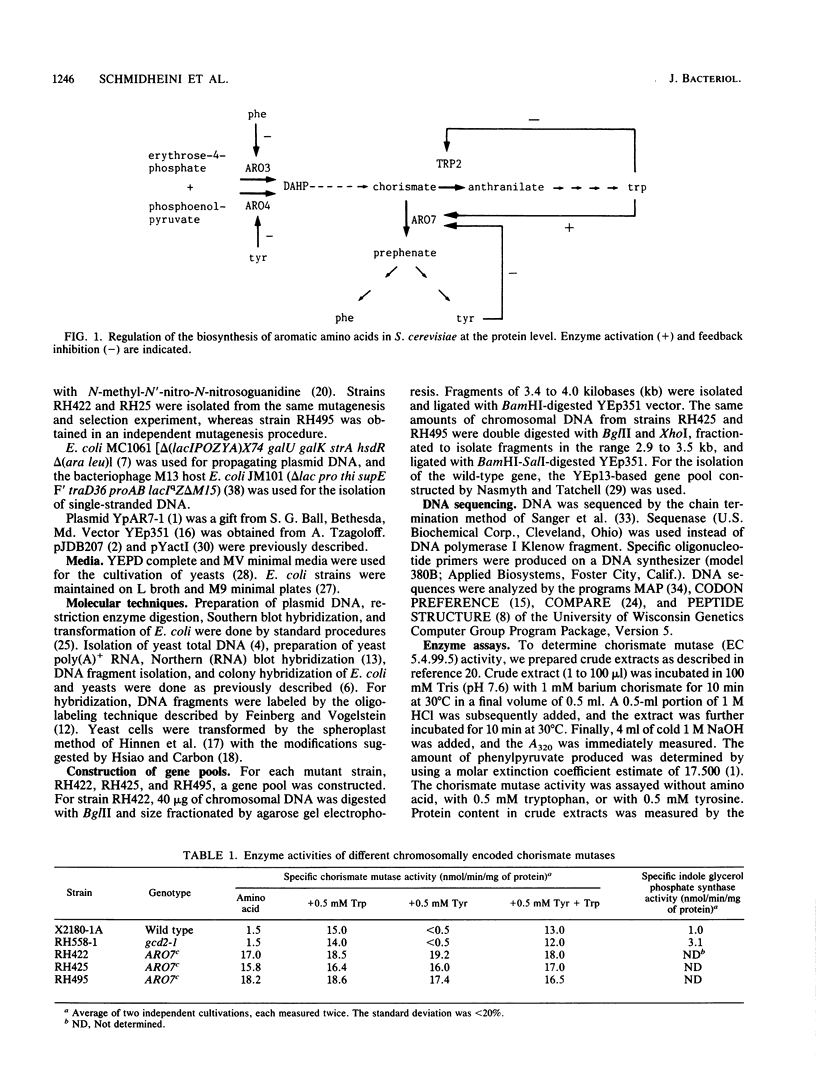
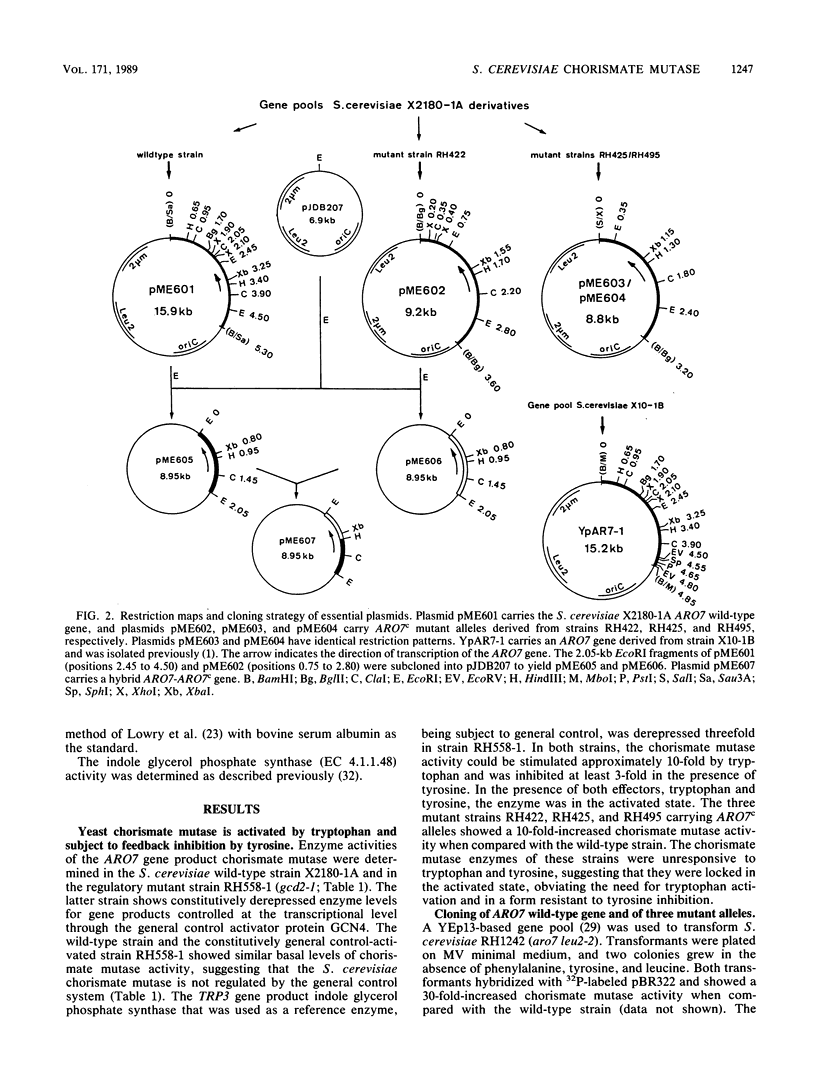
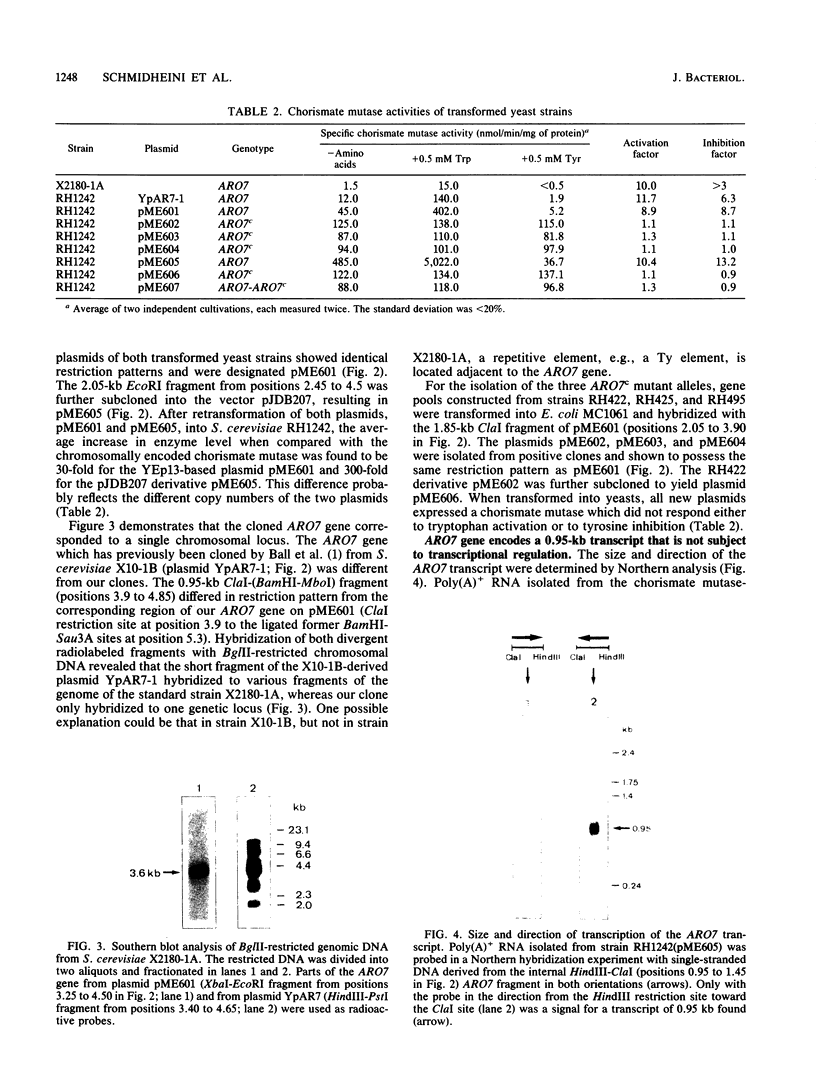
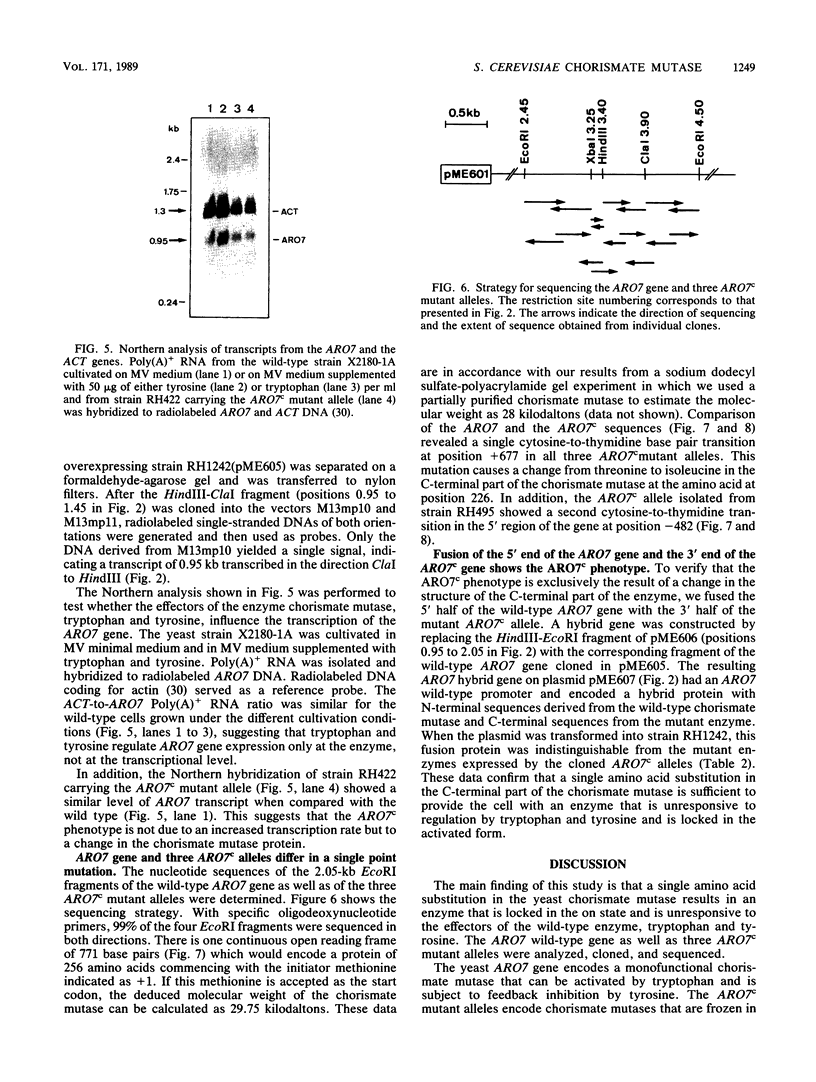
The Saccharomyces cerevisiae ARO7 gene product chorismate mutase, a single-branch-point enzyme in the aromatic amino acid biosynthetic pathway, is activated by tryptophan and subject to feedback inhibition by tyrosine. The ARO7 gene was cloned on a 2.05-kilobase EcoRI fragment. Northern (RNA) analysis revealed a 0.95-kilobase poly(A)+ RNA, and DNA sequencing determined a 771-base-pair open reading frame capable of encoding a protein 256 amino acids. In addition, three mutant alleles of ARO7 were cloned and sequenced. These encoded chorismate mutases which were unresponsive to tyrosine and tryptophan and were locked in the on state, exhibiting a 10-fold-increased basal enzyme activity. A single base pair exchange resulting in a threonine-to-isoleucine amino acid substitution in the C-terminal part of the chorismate mutase was found in all mutant strains. In contrast to other enzymes in this pathway, no significant homology between the monofunctional yeast chorismate mutase and the corresponding domains of the two bifunctional Escherichia coli enzymes was found.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ball S. G., Wickner R. B., Cottarel G., Schaus M., Tirtiaux C. Molecular cloning and characterization of ARO7-OSM2, a single yeast gene necessary for chorismate mutase activity and growth in hypertonic medium. Mol Gen Genet. 1986 Nov;205(2):326–330. doi: 10.1007/BF00430446. [DOI] [PubMed] [Google Scholar]
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Braus G. H., Luger K., Paravicini G., Schmidheini T., Kirschner K., Hütter R. The role of the TRP1 gene in yeast tryptophan biosynthesis. J Biol Chem. 1988 Jun 5;263(16):7868–7875. [PubMed] [Google Scholar]
- Braus G., Furter R., Prantl F., Niederberger P., Hütter R. Arrangement of genes TRP1 and TRP3 of Saccharomyces cerevisiae strains. Arch Microbiol. 1985 Sep;142(4):383–388. doi: 10.1007/BF00491908. [DOI] [PubMed] [Google Scholar]
- Braus G., Paravicini G., Hütter R. A consensus transcription termination sequence in the promoter region is necessary for efficient gene expression of the TRP1 gene of Saccharomyces cerevisiae. Mol Gen Genet. 1988 Jun;212(3):495–504. doi: 10.1007/BF00330855. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Martinez-Arias A., Shapira S. K., Chou J. Beta-galactosidase gene fusions for analyzing gene expression in escherichia coli and yeast. Methods Enzymol. 1983;100:293–308. doi: 10.1016/0076-6879(83)00063-4. [DOI] [PubMed] [Google Scholar]
- Chou P. Y., Fasman G. D. Prediction of the secondary structure of proteins from their amino acid sequence. Adv Enzymol Relat Areas Mol Biol. 1978;47:45–148. doi: 10.1002/9780470122921.ch2. [DOI] [PubMed] [Google Scholar]
- Davidson B. E. Chorismate mutase-prephenate dehydratase from Escherichia coli. Methods Enzymol. 1987;142:432–439. doi: 10.1016/s0076-6879(87)42054-5. [DOI] [PubMed] [Google Scholar]
- Davidson B. E., Hudson G. S. Chorismate mutase-prephenate dehydrogenase from Escherichia coli. Methods Enzymol. 1987;142:440–450. doi: 10.1016/s0076-6879(87)42055-7. [DOI] [PubMed] [Google Scholar]
- Drake J. W., Baltz R. H. The biochemistry of mutagenesis. Annu Rev Biochem. 1976;45:11–37. doi: 10.1146/annurev.bi.45.070176.000303. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. "A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity". Addendum. Anal Biochem. 1984 Feb;137(1):266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Furter R., Paravicini G., Aebi M., Braus G., Prantl F., Niederberger P., Hütter R. The TRP4 gene of Saccharomyces cerevisiae: isolation and structural analysis. Nucleic Acids Res. 1986 Aug 26;14(16):6357–6373. doi: 10.1093/nar/14.16.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Devereux J., Burgess R. R. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görisch H., Lingens F. Chorismate mutase from Streptomyces aureofaciens: a heat-stable enzyme. J Bacteriol. 1973 May;114(2):645–651. doi: 10.1128/jb.114.2.645-651.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J. E., Myers A. M., Koerner T. J., Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986 Sep;2(3):163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C. L., Carbon J. High-frequency transformation of yeast by plasmids containing the cloned yeast ARG4 gene. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3829–3833. doi: 10.1073/pnas.76.8.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G. S., Davidson B. E. Nucleotide sequence and transcription of the phenylalanine and tyrosine operons of Escherichia coli K12. J Mol Biol. 1984 Dec 25;180(4):1023–1051. doi: 10.1016/0022-2836(84)90269-9. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Llewellyn D. J., Daday A., Smith G. D. Evidence for an artificially evolved bifunctional 3-deoxy-D-arabinoheptulosonate-7-phosphate synthase-chorismate mutase in Bacillus subtilis. J Biol Chem. 1980 Mar 10;255(5):2077–2084. [PubMed] [Google Scholar]
- Lorence J. H., Nester E. W. Multiple molecular forms of chorismate mutase in Bacillus subtillis. Biochemistry. 1967 May;6(5):1541–1553. doi: 10.1021/bi00857a041. [DOI] [PubMed] [Google Scholar]
- Maizel J. V., Jr, Lenk R. P. Enhanced graphic matrix analysis of nucleic acid and protein sequences. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7665–7669. doi: 10.1073/pnas.78.12.7665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruya A., O'Connor M. J., Backman K. Genetic separability of the chorismate mutase and prephenate dehydrogenase components of the Escherichia coli tyrA gene product. J Bacteriol. 1987 Oct;169(10):4852–4853. doi: 10.1128/jb.169.10.4852-4853.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miozzari G., Niederberger P., Hütter R. Tryptophan biosynthesis in Saccharomyces cerevisiae: control of the flux through the pathway. J Bacteriol. 1978 Apr;134(1):48–59. doi: 10.1128/jb.134.1.48-59.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K. The structure of transposable yeast mating type loci. Cell. 1980 Mar;19(3):753–764. doi: 10.1016/s0092-8674(80)80051-1. [DOI] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeschger M. P., Berlyn M. K. A simple procedure for localized mutagenesis using nitrosoguanidine. Mol Gen Genet. 1974;134(1):77–83. doi: 10.1007/BF00332814. [DOI] [PubMed] [Google Scholar]
- Prantl F., Strasser A., Aebi M., Furter R., Niederberger P., Kirschner K., Huetter R. Purification and characterization of the indole-3-glycerolphosphate synthase/anthranilate synthase complex of Saccharomyces cerevisiae. Eur J Biochem. 1985 Jan 2;146(1):95–100. doi: 10.1111/j.1432-1033.1985.tb08624.x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder J. L., Blattner F. R. Formal description of a DNA oriented computer language. Nucleic Acids Res. 1982 Jan 11;10(1):69–84. doi: 10.1093/nar/10.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch A., Miozzari J., Hütter R. Regulation of tryptophan biosynthesis in Saccharomyces cerevisiae: mode of action of 5-methyl-tryptophan and 5-methyl-tryptophan-sensitive mutants. J Bacteriol. 1974 Mar;117(3):1131–1140. doi: 10.1128/jb.117.3.1131-1140.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Chen W., Hill D. E., Hope I. A., Oettinger M. A. Constitutive and coordinately regulated transcription of yeast genes: promoter elements, positive and negative regulatory sites, and DNA binding proteins. Cold Spring Harb Symp Quant Biol. 1985;50:489–503. doi: 10.1101/sqb.1985.050.01.061. [DOI] [PubMed] [Google Scholar]
- Teshiba S., Furter R., Niederberger P., Braus G., Paravicini G., Hütter R. Cloning of the ARO3 gene of Saccharomyces cerevisiae and its regulation. Mol Gen Genet. 1986 Nov;205(2):353–357. doi: 10.1007/BF00430450. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zalkin H., Paluh J. L., van Cleemput M., Moye W. S., Yanofsky C. Nucleotide sequence of Saccharomyces cerevisiae genes TRP2 and TRP3 encoding bifunctional anthranilate synthase: indole-3-glycerol phosphate synthase. J Biol Chem. 1984 Mar 25;259(6):3985–3992. [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]