Abstract
The clinicopathological phenotype of the Gerstmann–Sträussler–Scheinker disease (GSS) variant linked to the codon 102 mutation in the prion protein (PrP) gene (GSS P102L) shows a high heterogeneity. This variability also is observed in subjects with the same prion protein gene PRNP haplotype and is independent from the duration of the disease. Immunoblot analysis of brain homogenates from GSS P102L patients showed two major protease-resistant PrP fragments (PrP-res) with molecular masses of ≈21 and 8 kDa, respectively. The 21-kDa fragment, similar to the PrP-res type 1 described in Creutzfeldt–Jakob disease, was found in five of the seven subjects and correlated with the presence of spongiform degeneration and “synaptic” pattern of PrP deposition whereas the 8-kDa fragment, similar to those described in other variants of GSS, was found in all subjects in brain regions showing PrP-positive multicentric amyloid deposits. These data further indicate that the neuropathology of prion diseases largely depends on the type of PrP-res fragment that forms in vivo. Because the formation of PrP-res fragments of 7–8 kDa with ragged N and C termini is not a feature of Creutzfeldt–Jakob disease or fatal familial insomnia but appears to be shared by most GSS subtypes, it may represent a molecular marker for this disorder.
In humans, prion diseases comprise a broad spectrum of clinicopathological variants that usually are classified in four major groups: Creutzfeldt–Jakob disease (CJD), Gerstmann–Sträussler–Scheinker disease (GSS), kuru and fatal familial insomnia (1). Although CJD includes sporadic, familial, and acquired forms, fatal familial insomnia can be either inherited or sporadic, and kuru is acquired, GSS only has been described associated with prion protein gene (PRNP) mutations (1, 2). Among these mutations, a point mutation at codon 102, which results in the substitution of proline to leucine, is by far the most common. Other mutations at codons 105, 117, 145, 198, and 217 and an insert mutation between codons 51 and 91 also have been linked to GSS but are rarer (1, 2). The common phenotypic trait of all GSS variants is the presence of multicentric amyloid plaques composed of prion protein (PrP) fragments (2). Other features, however, show differences among GSS patients. The presentation can be characterized by motor signs or dementia whereas, pathologically, amyloid plaques may or may not be associated with spongiform degeneration or neurofibrillar degeneration (2).
We showed (3–5) that, in CJD and kuru as well as in fatal familial insomnia, PrP protease resistant (PrP-res) comprises glycosylated and N-terminally truncated fragments designated PrP-res type 1 (Mr of unglycosylated form ≈21 kDa) and PrP-res type 2 (Mr of unglycosylated form ≈19 kDa) (3–5). In contrast, purified amyloid preparations from GSS affected subjects carrying the Y145STOP, F198S, A117V, or Q217R mutations were shown to contain PrP-res fragments of 11 and 7 kDa with ragged N and C termini (6, 7). To contribute to the definition of the full spectrum of PrP-res types that forms in human prion diseases, we examined the physicochemical properties of the PrP-res fragments extracted from the affected brain of subjects carrying the P102L mutation, the most common mutation linked to GSS. The study of the PrP-res and its correlation with the phenotype of the disease is particularly appropriate in the P102L GSS subtype because this disease shows an heterogeneous phenotype that is not fully explained by variation in the PRNP genotype (8, 9). We show that, in subjects carrying the P102L mutation who were syngenic at PRNP codon 129 and 219, distinct pathological features of the disease correlate with different patterns of PrP-res fragments.
MATERIALS AND METHODS
Patients.
The seven subjects analyzed were from seven unrelated kindreds. Pedigree, clinical features, and histopathological data for five subjects were reported in detail (8–13). These data, and those obtained from the two previously unreported patients, are summarized in Table 1.
Table 1.
Summary of genetic, clinical, and histopathologic features in the seven P102L GSS patients
| Patient | Codon 129 | Kindred | Sex | Age at onset | Duration (months) | Clinical signs | Pathology |
|---|---|---|---|---|---|---|---|
| 1 | M/M | French | M | 56 | 30 | Ataxia, pyramidal signs, dysarthria, dysphagia, dementia; no PSW on EEG | Moderate spongiosis, gliosis, neuronal loss, amyloid plaques |
| 2 | M/M | Austrian | F | 37 | 34 | Dementia, ataxia, apraxia, dysphagia, myoclonus, seizures, coma; PSW on EEG | Severe spongiosis, gliosis, neuronal loss, amyloid plaques |
| 3 | M/M | English/American | M | 42 | 13 | Severe dementia, ataxia, myoclonus, rigidity, coma; PSW on EEG | Severe spongiosis, gliosis, neuronal loss, amyloid plaques |
| 4 | M/M | Japanese | F | 26 | 51 | Ataxia, dysarthria, pyramidal, dysphagia, hallucinations, dementia; EEG: NA | Severe spongiosis, gliosis, neuronal loss, amyloid plaques |
| 5 | M/M | Italian/American | M | 60 | NA | At terminal stage: severe dementia, bedridden, rigidity; EEG: NA | Severe spongiosis, gliosis, neuronal loss, amyloid plaques |
| 6 | M/M | American | F | 66 | 51 | Ataxia, tremor, dementia, pyramidal signs; no PSW on EEG | Diffuse atrophy, no spongiosis, amyloid plaques |
| 7 | M/M | German | M | 53 | 72 | Ataxia, nystagmus, dysarthria, dementia; no PSW on EEG | Diffuse atrophy, no spongiosis, amyloid plaques |
NA, not available; EEG, electroencephalogram; PSW, periodic sharp waves complexes; M/M, methionine/methionine.
Tissues.
Frozen tissue from the cerebral cortex (n = 7), basal ganglia (n = 2), and cerebellum (n = 3) was obtained. In addition, paraffin blocks of tissue from the cerebral cortex, hippocampus, basal ganglia, thalamus, midbrain, pons, and cerebellum were available in four subjects.
Histopathological Examination.
A brain autopsy was performed in six patients, and a brain biopsy was performed in one (patient 4). A semiquantitative evaluation of spongiosis, neuronal loss, and gliosis was carried out on hematoxylin- and eosin-stained paraffin sections from the above-listed brain regions. Results of histopathologic examination for the other three subjects were obtained from the original pathologic reports and from previous publications.
Molecular Genetic Analysis.
Analysis of PRNP was performed by using genomic DNA isolated from frozen brain tissue according to standard procedures. The presence of the P102L mutation was confirmed in all patients by digestion of amplified DNA with the restriction enzyme DdeI. Additionally, codons 129 and 219 of PRNP were examined by digestion with the restriction endonucleases Nsp I and Mae II (codon 129) and by restriction site-generated PCR (codon 219) (14).
Antibodies.
The following antibodies were used: mouse mAb 3F4, which reacts with PrP residues 109–112 (15), and rabbit antisera to synthetic peptides homologous to human PrP residues 220–231 (Anti-C) and 23–40 (Anti-N) (16).
PrP Immunocytochemistry.
Paraffin sections were processed for PrP immunostaining after hydrolytic autoclaving as described (4).
Analysis of PrP-res on Western Blots.
Analysis of PrP-res on Western blots was performed as described (4). Quantitative analysis of PrP-res glycoform ratio was performed on immunoblots stained with 3F4. Enhanced chemiluminescence films (Amersham) were scanned at 42-μm resolution and were analyzed quantitatively by using quantity one software (PDI Imageware Systems, Huntington Station, NY).
Purification of PrP-res Fragments.
The PrP-res fragments were purified according to a published procedure (17) as modified by Chen et al. (18). Purified samples were loaded onto the micropreparative SDS/PAGE (14% gel) (Mini Prep cell, Bio-Rad). The protein bands were eluted with 0.05% SDS in 5 mM ammonium bicarbonate (pH 8.0) at the flow rate of 70 μl/min and were collected into 400-μl fractions. The fractions containing purified PrP-res fragments were pooled. An aliquot of the pooled fraction was saved for mass spectrometric analysis whereas the rest of the fractions were concentrated by lyophilization for protein sequence analysis.
Protein Sequence Analysis.
Proteins were separated on 16% SDS/PAGE minigels (Bio-Rad) and were transferred onto a Problott membrane (Applied Biosystems). The membrane strips containing protein bands (2 mm × 6 mm) were subjected to automated Edman chemistry on an Applied Biosystems Model 494 Procise instrument carried out at the ARIAD Protein Microsequencing Facility.
Endoproteinase Digestion.
An aliquot of the pooled fraction containing purified PrP-res fragments was adjusted to 1% octyl glucoside, 1 mM EDTA, and 5 mM tricine (pH 8.0) in a total volume of 10 μl. The sample was digested with endoproteinase Lys-C (Boeringer Mannheim) at 37°C for 8 h with an enzyme:substrate ratio of 1:10–20. The digestion mixture was subjected to mass spectrometric measurements.
Mass Spectrometric Analysis.
The polypeptides were analyzed by matrix-assisted laser desorption/ionization time-of flight mass spectrometry. The spectrometer was equipped with a pulsed nitrogen laser source (λ = 337 nm) and an acceleration voltage of 28 kV operated in the linear mode (Voyager Biospectrometry Workstation, PerSeptive Biosystems, Framingham, MA). A mass accuracy ranging from 0.01 to 0.1% was obtained. Samples were mixed with an equal volume of α-cyano-4-hydroxycinnamic acid matrix solution (10 mg/ml in 60% acetonitrile, 0.3% trifluoroacetic acid). Typically, 1 μl of the mixture was applied onto the laser target probe and was air-dried before being introduced into the mass spectrometer.
RESULTS
Immunoblot Analysis of PrP-res.
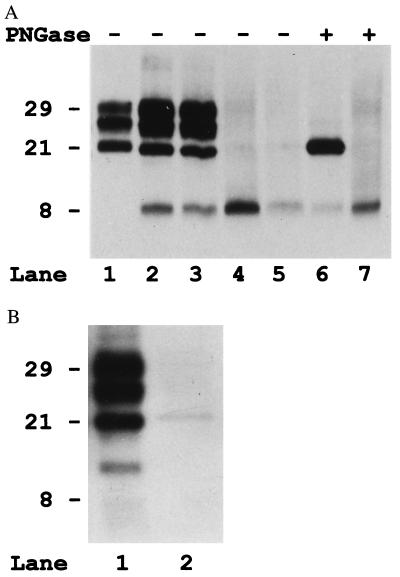
Immunoblot analysis of proteinase K (PK)-treated brain tissue showed, after 3F4 staining, two patterns of PrP-res that included either one or two major peptides, which could be distinguished on the basis of Mr and glycosylation (Fig. 1A). Two subjects showed a single major immunoreactive band migrating at ≈8 kDa in all brain regions analyzed. In the other five subjects, in addition to the 8-kDa peptide, there were three additional bands migrating at ≈29, ≈27, and ≈21 kDa (Fig. 1A). Deglycosylation of these samples resulted in two immunoreactive bands of 21 and 8 kDa, indicating that the 29- and 27-kDa bands represent the glycosylated forms of the 21-kDa peptide and that the 8-kDa fragment is unglycosylated (Fig. 1A).
Figure 1.
(A) Immunoblot analysis with the 3F4 antibody of the PrP-res extracted from the cerebral cortex of four subjects with GSS P102L (lanes 2–7) and one subjects with sporadic CJD (lane 1). Lanes 6 and 7 include samples that also were deglycosylated with PNGase F (PNGase, lanes +). Lanes: 1, sporadic CJD, codon 129 M/M, and PrP-res type1; 2 and 6, subject 2; 3, subject 5; 4 and 7, subject 7; 5, subject 6. (B) Immunoblot analysis of PrP-res from subjects 2 and 7 probed with the anti-C antiserum.
Immunoblotting with the Anti-C antiserum revealed the 21-kDa fragment but not the 8-kDa peptide (Fig. 1B) whereas the Anti-N antiserum failed to recognize both fragments, indicating that the 21-kDa fragment is N-terminally truncated whereas the 8-kDa fragment is ragged at both the N and C termini. In addition, the Anti-C antiserum revealed an immunoreactive peptide migrating at ≈13 kDa in the five subjects showing the 21-kDa band (Fig. 1B). This fragment was not recognized by the antibody 3F4, indicating that it is N-terminally truncated beyond residue 112.
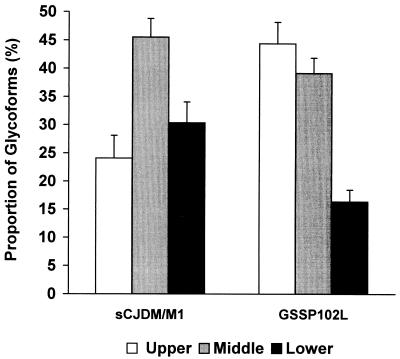
Comparison with samples from CJD-affected subjects revealed that the 21-kDa fragment from GSS subjects corresponds in electrophoretic mobility to the type 1 PrP-res identified in sporadic CJD (Fig. 1A) (4, 5). However, the ratio of the three major glycoforms of PrP-res was significantly different between the GSSP102L samples and sporadic CJD (Fig. 2).
Figure 2.
Relative proportion of the three PrP-res glycoforms of the 21-kDa fragment (PrP-res type 1) in GSS P102L compared with that of sporadic CJD (sCJD). Mean ± SD. Upper, high-molecular mass glycoform; middle, low-molecular-mass glycoform; low, unglycosylated form.
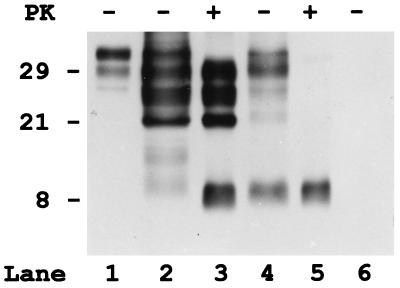
To determine whether the two fragments already were truncated in vivo, we analyzed partially purified (P3) PrP preparation without PK treatment. In the untreated state, significant amounts of truncated fragments with similar or identical Mr to the PrP-res peptides were present in the GSS (Fig. 3) but not normal samples. In addition, either full-length or truncated detergent-insoluble PrP peptides were detected in the GSS subjects whereas, in controls, only traces of full-length PrP were present (Fig. 3).
Figure 3.
Immunoblot analysis of purified detergent insoluble fractions (P3) of PrP from the cerebral cortex of one control subject (lane 6) and two subjects (2 and 7) with GSS P102L (lanes 2–5), before (lanes 1, 2, 4, and 6) and after PK digestion (lanes 3 and 5). Although in the control (lane 6), no PrP is detectable in the insoluble fraction not treated with PK, in both GSS samples, significant amount of truncated PrP fragments, in addition to some full-length PrP, are present. However, pattern and properties of the truncated PrP peptides are different in the two GSS cases. In subject 2 (lane 2), there are large amount of 29-, 27-, and 21-kDa fragments that comigrate with the PrP-res peptides visible after PK treatment. In addition, two bands migrating at 11- and 9-kDa are present. Patient 7 (lane 4) shows similar bands at 29, 27, and 21 kDa, but only before PK treatment, indicating that these peptides are insoluble but not protease resistant. In addition, a band at 8 kDa that has similar intensity and comigrates with the PrP-res fragment is visible. Full-length PrP from a crude homogenate of the control subject is represented in lane 1.
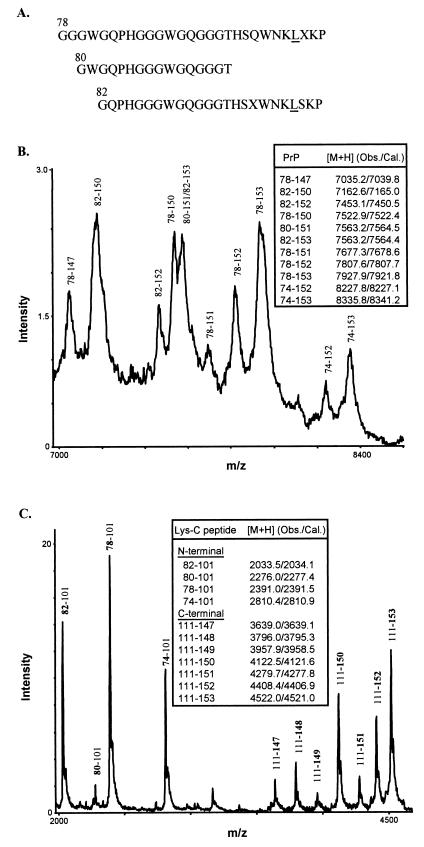
N-terminal Sequencing and Mass Spectrometry.
To further characterize the primary structure of the 8- and 21-kDa PrP-res fragments identified by a standard mini-gel electrophoresis, we performed N-terminal sequencing and mass spectrometry on purified fractions of PrP-res. Protein sequencing in three cases (3, 5, and 7) showed almost identical N termini in the two PrP-res fragments. The 8-kDa peptide demonstrated N-terminal cleavage sites at residues 78, 80, and 82 (Fig. 4A) whereas sequencing of the 21-kDa fragment showed major N-terminal starting points at residues 78 and 82 (data not shown). Thus, the 8- and the 21-kDa fragments mainly differ at the C terminus because the 8-kDa peptide is truncated at both N- and C-terminal ends.
Figure 4.
Characterization of 8-kDa PrP-res by protein sequencing and mass spectrometry. (A) N-terminal sequences of purified 8-kDa PrP-res. The experimentally determined amino acid signals at each Edman degradation cycle were aligned with the known human PrP sequence to derive three major N-terminal species, with the residue positions shown above the first amino acids and the mutant residue at position 102 underlined. Unassigned residues were indicated as X. (B) Spectrum of 8-kDa PrP-res obtained by matrix-assisted laser desorption/ionization mass spectrometry. Major PrP species having Leu at position 102 and inclusive of N and C termini were assigned according to the observed and calculated protonated mass signals ([M+H] values shown in Inset). (C) Matrix-assisted laser desorption/ionization mass spectrum of the Lys-C digest of 8-kDa PrP-res. The N- and C-terminal PrP peptides generated by Lys-C digestion were assigned as in B. The N-terminal peptides corresponded to fragments that begin (bold) at positions 74, 78, 80, and 82, respectively, and end at the cleavage site at position 101 (the presence of Leu at position 102 introduces a specific cleavage at position 101). No wild-type N-terminal peptides were observed because, if present, they would correspond to PrP peptides having Pro at position 102 and spanning residues 74–106, 78–106, 80–106, and 82–106 (calculated [M+H] values 3348.6, 2929.2, 2815.1, 2571.8, respectively). The C-terminal peptides corresponded to fragments that begin at the cleavage site at positions 111 and end (bold) at positions 147, 148, 149, 150, 151, 152, and 153, respectively.
Using mass spectrometry, we confirmed the N termini of the 8-kDa fragment as determined by protein sequencing and demonstrated an additional N-terminal residue at position 74. Furthermore, we established the C-terminal cleavage site of the 8-kDa peptide by the mass spectrometry measurements (Fig. 4 B and C). In addition, because the mutation at position 102 creates a cleavage site for Lys-C, we were able to determine the allelic origin (mutated vs. normal allele) for both the 8- and 21-kDa fragments. The mass spectrometry data for the 8-kDa PrP-res were consistent with the major signals to derive only from the mutated protein and to have multiple C terminus ending points spanning positions 147–153 (Fig. 4 B and C). Similarly, the pattern of fragments generated by the Lys-C indicated that the 21-kDa fragment also originated exclusively from the mutated allele (data not shown).
Correlation Between the Biochemical and the Histopathological Findings.
The presence of either the 21- or 8-kDa fragment clearly correlated with distinct pathologic features of the disease (Table 2). The 8-kDa PrP-res peptide correlated with the presence of the amyloid plaques and/or of the PrP plaque-like deposits, which were seen in all of the samples examined. In contrast, significant amounts of the 21-kDa fragment were found only in tissue from brain regions with spongiform degeneration (Table 2). Similarly, two patterns of PrP-res deposition, not invariably associated, were identified by immunohistochemistry by using the antibody 3F4. The presence of spongiform degeneration correlated with a punctate, “synaptic” type of immunoreactivity, as in the most common CJD phenotype (Table 2 and Fig. 5A) (4). In addition, PrP deposits colocalizing with the multicentric amyloid plaques were seen in all areas analyzed (Table 2 and Fig. 5A). These deposits, in addition to a more diffuse staining resembling the diffuse plaques or preamyloid deposits described in Alzheimer’s disease, were the only findings in patient 7 (Fig. 4B).
Table 2.
Correlation between pathologic and immunochemical findings
| Patient | Cerebral cortex
|
Cerebellum
|
||||
|---|---|---|---|---|---|---|
| Pathology (H&E) | PrP IHC (3F4) | Mr of PrP-res | Pathology (H&E) | PrP IHC (3F4) | Mr of PrP-res | |
| 1 | SP + PL | SYN + PL | NA | PL | PL | 21 (traces) and 8 |
| 2 | SP + PL | SYN + PL | 21 and 8 | SP + PL | SYN + PL | 21 and 8 |
| 3 | SP + PL | NA | 21 and 8 | SP + PL | NA | NA |
| 4 | SP + PL | NA | 21 and 8 | NA | NA | NA |
| 5 | SP + PL | SYN + PL | 21 and 8 | SP + PL | SYN + PL | 21 and 8 |
| 6 | PL (F) | PL (F), DPL | 8 | PL | PL | 8 |
| 7 | PL | NA | 8 | PL | NA | NA |
IHC, immunohistochemistry; H&F, hematoxylin and eosin; Mr, molecular weight; SP, spongiosis; PL, amyloid plaques (pathology) or plaque-like deposits (IHC); SYN, “synaptic” PrP deposition; NA, not available; DPL, diffuse plaque deposits (preamyloid); (F), Focal.
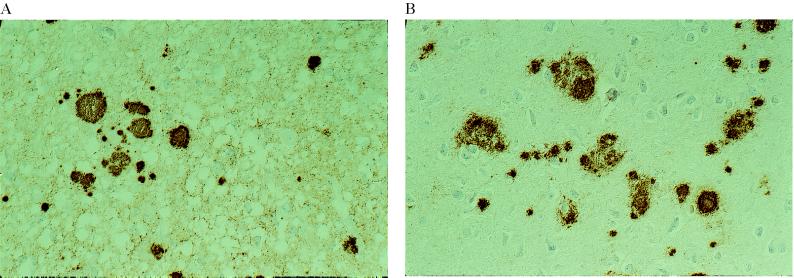
Figure 5.
PrP immunoreactivity in two GSS P102L subjects showing distinct patterns of PrP-res on immunoblot. (A) Cerebral cortex of patient 5. A diffuse, synaptic staining is seen in association with multiple PrP immunopositive plaques. Immunolabeling with 3F4, ×250. (B) Cerebral cortex of patient 6. There are multiple PrP immunopositive plaques. No synaptic staining is visible. Immunolabeling with 3F4, ×250.
The synaptic staining was also visible with the Anti-C but not with the Anti-N antisera, suggesting that full-length PrP is not a major component of these deposits. In contrast, amyloid plaques were stained by both the Anti-N and Anti-C antisera, mainly at the periphery of the cores, suggesting that they contain full-length PrP.
DISCUSSION
The present findings show that the heterogeneity of the clinicopathologic phenotype associated with the GSS P102L variant is related to the presence and relative abundance of two PrP-res fragments that have a different size and degree of glycosylation. The 8-kDa fragment, which is unglycosylated, is found in all subjects and, as similar fragments found in other GSS subtypes (2, 6, 7), correlates with the presence of multicentric amyloid plaques. In contrast, the 21-kDa peptide, which has the same size as the type 1 PrP-res found in CJD (4, 5), is present only in subjects who had significant spongiform degeneration. This observation provides strong support to the notion that the neuropathology of prion diseases largely depends on the type of PrP-res fragment(s) that form in vivo.
It commonly is thought that PrP-res results from the conversion of full-length PrP and is a full-length molecule in vivo whose treatment with PK in vitro results in the generation of a PK resistant C-terminal fragment, which comprises residues ≈90–231 and has been designated PrP 27–30 (19). Increasing evidence, however, indicates that this is not always the case. We have shown (16) that, in CJD, a significant amount of truncated 21- or 19-kDa PrP-res is formed in vivo. In vivo formation of N-terminal truncated PrP-res fragments also occurs in scrapie-infected mouse brains and scrapie-infected neuroblastoma cells (20, 21). Similarly, these as well as previous data indicate that the formation in vivo of PrP-res peptides with ragged N and C termini characterize most, if not all, GSS variants (2, 6, 7). Thus, the in vivo formation of aberrant truncated PrP-res peptides represents a common, possibly critical, pathogenetic event in prion disease.
Different PrP fragments may have different neurotoxicity and may cause distinct lesions as a consequence of their different properties, such as aggregability (22). In view of the fact of the comparatively long duration of illness in most GSS subtypes and the typically shorter duration of most CJD variants, it is possible that the formation of small fragments, such as the 8-kDa peptide, with a high tendency toward aggregation and plaque formation may provide a relative protection with less neuronal dysfunction than the 21- and 19-kDa PrP-res glycosylated fragments associated with CJD that form more diffuse and smaller deposits. Consistent with this hypothesis is the observation that, among the GSS P102L patients, those showing a CJD-like phenotype have, on average, a significantly shorter course (8, 9, 11).
Our data also have implications for the classification, diagnosis, and transmissibility of human prion diseases. In GSS P102L (as in the other GSS variants previously studied), the 8-kDa PrP fragment with ragged N and C termini is consistently found to colocalize with the amyloid plaques and provides a marker for the distinction between CJD and GSS at the molecular level. PrP-res peptides truncated at both N and C termini appear to be highly specific for GSS and were not found by us in a large series of CJD cases (5).
Diagnosis of prion diseases increasingly relies on immunoblot detection of PrP-res (23). In at least one GSS subtype, however, PrP-res has been reported to be undetectable, despite the demonstration of numerous PrP positive amyloid deposit (24). Our data, which show that, in GSS P102L subjects without significant spongiform degeneration, the 8-kDa fragment is the only detectable PrP-res fragment, underline the importance of the search of smaller PrP-res peptides in addition to PrP 27–30. A systematic search for small PrP-res peptides in all subtypes of GSS will be of paramount importance and should answer the question of whether PrP-res fragments (that resist a harsh PK digestion) are indeed present in all forms of naturally occurring prion diseases.
It also would be important to determine whether the 21- and 19-kDa PrP-res fragments, commonly associated with CJD, can be detected in other GSS subtypes. Spongiform degeneration, although inconspicuous, has been observed in other GSS variants in addition to GSS P102L. Previous studies on GSS (2, 6, 7) have focused mainly on the characterization of PrP fragments extracted from purified amyloid preparations and may have missed longer, nonamyloidogenic peptides. Of interest, a 19-kDa PrP-res fragment, although supposedly not glycosylated, has been detected in GSS F198S (25). This PrP-res fragment might represent a type 2 CJD fragment. The presence of a type 1 PrP-res in GSS P102L and of a type 2 PrP-res in GSS F198S would be consistent with the fact that, as shown for sporadic CJD, valine at codon 129, which is invariably linked to the F198S mutation, favors the formation of PrP-res type 2 whereas methionine, which cosegregates with the P102L mutation, usually is associated with PrP-res type 1 (5).
Transmission to experimental animals has yet to be accomplished for all human prion diseases. Although all CJD variants and fatal familial insomnia have been propagated successfully in experimental animals, the GSS P102L, as well as an individual GSS case carrying an insertion mutation in PRNP, are the only subtypes of GSS that have been transmitted to date (13, 26–30). Among these subjects, all but one showed significant spongiform degeneration, and, in three of four tested, including the transmitted case carrying the insertion mutation (P.P., P.B., and P.G., unpublished observation), we were able to demonstrate the presence of significant amount of PrP-res type 1. Thus, all subtypes of human prion diseases associated with the formation of either type 1 or type 2 PrP-res have been shown to be transmissible. In contrast, we are aware of only one case of GSS (case 7 in this series) that transmitted the disease and yet lacked significant spongiform degeneration and detectable PrP-res type 1 (in two brain samples). The disease could be transmitted to rodents but not to primates (26, 29, 30). Taken together, these data indicate that the formation in vivo of distinct truncated PrP-res fragments not only correlated with heterogeneous phenotypes, but also with other fundamental properties of prions such as infectivity.
The reason why, in a subset of subjects with the P102L mutation, PrP-res type 1 is not detectable is puzzling. It may depend on yet-unidentified genetic factors of the host or may represent a strain-specific phenomenon. Transmission studies, aimed to determine whether brain homogenates from subjects with GSS P102L will induce heterogeneous phenotypes in the recipient syngenic animals, may provide significant clues to answer the question. Similarly, neuropatholopic examination and PrP-res typing of animals inoculated with GSS P102L that developed disease will be important to unravel the critical factors that lead to the formation of the 8-kDa amyloidogenic fragment in GSS P102L. Because all 10 cases of GSS P102L transmitted to date have induced in three different animal species a spongiform encephalopathy indistinguishable from that caused by the inoculation of homogenates from sporadic CJD subject with a typical phenotype (28–30), it seems likely that the formation of amyloid plaques and the 8-kDa PrP-res fragment are specifically linked to the presence of the mutation. The determination of whether the PrP-res that forms in the infected animals includes the 8-kDa peptide will give the definitive answer and will further elucidate the causative role of this and similar peptides in amyloid formation.
Acknowledgments
Supported by National Institutes of Health grants AG-08155 and AG08992 and the Britton Fund. Part of this work was made within the European Union Concerted Action on Human TSEs.
ABBREVIATIONS
- GSS
Gerstmann—Sträussler–Scheinker disease
- CJD
Creutzfeldt–Jakob disease
- PrP
Prion protein
- PrP-res
protease resistant prion protein
- PRNP
Human prion protein gene
- PK
proteinase K
References
- 1.Parchi P, Piccardo P, Gambetti P, Ghetti B. In: Progress in Pathology 4. Kirkham N, Lemoine N R, editors. Edimburgh: Churchill Livingstone; 1998. pp. 39–77. [Google Scholar]
- 2.Ghetti B, Piccardo P, Frangione B, Bugiani O, Giaccone G, Young K, Prelli F, Farlow M R, Dlouhy S R, Tagliavini F. Brain Pathol. 1996;6:127–145. doi: 10.1111/j.1750-3639.1996.tb00796.x. [DOI] [PubMed] [Google Scholar]
- 3.Monari L, Chen S G, Brown P, Parchi P, Petersen R B, Mikol J, Gray F, Cortelli P, Montagna P, Ghetti B, et al. Proc Natl Acad Sci USA. 1994;91:2839–2842. doi: 10.1073/pnas.91.7.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parchi P, Castellani R, Capellari S, Ghetti B, Young K, Chen S G, Farlow M, Dickson D W, Sima A A F, Trojanowski J Q, et al. Ann Neurol. 1996;39:767–778. doi: 10.1002/ana.410390613. [DOI] [PubMed] [Google Scholar]
- 5.Parchi P, Capellari S, Chen S G, Petersen R B, Gambetti P, Kopp N, Brown P, Kitamoto T, Tateishi J, Giese A, et al. Nature (London) 1997;386:232–234. doi: 10.1038/386232a0. [DOI] [PubMed] [Google Scholar]
- 6.Tagliavini F, Prelli F, Ghiso J, Bugiani O, Serban D, Prusiner S B, Farlow M R, Ghetti B, Frangione B. EMBO J. 1991;10:513–519. doi: 10.1002/j.1460-2075.1991.tb07977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ghetti B, Piccardo P, Spillantini M G, Ichimiya Y, Porro M, Perini F, Kitamoto T, Tateishi J, Seiler C, Frangione B, et al. Proc Natl Acad Sci USA. 1996;93:744–748. doi: 10.1073/pnas.93.2.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hainfellner J A, Brantner-Inthaler S, Cervenakova L, Brown P, Kitamoto T, Tateishi J, Diringer H, Liberski P P, Regele H, Feucht M, et al. Brain Pathol. 1995;5:201–211. doi: 10.1111/j.1750-3639.1995.tb00596.x. [DOI] [PubMed] [Google Scholar]
- 9.Barbanti P, Fabbrini G, Salvatore M, Petraroli R, Cardone F, Maras B, Equestre M, Macchi G, Lenzi G L, Pocchiari M. Neurology. 1996;47:734–741. doi: 10.1212/wnl.47.3.734. [DOI] [PubMed] [Google Scholar]
- 10.Collard M, Bakchine S, Duyckaerts C. Rev Neurol (Paris) 1991;147:323–328. [PubMed] [Google Scholar]
- 11.Adam J, Crow T J, Duchen L W, Scaravilli F, Spokes E. J Neurol Neurosurg Psychiatry. 1982;45:37–45. doi: 10.1136/jnnp.45.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown P, Goldfarb L G, Brown W T, Goldgaber D, Rubenstein R, Kascsak R J, Guiroy D C, Piccardo P, Boellaard J W, Gajdusek D C. Neurology. 1991;41:375–379. doi: 10.1212/wnl.41.3.375. [DOI] [PubMed] [Google Scholar]
- 13.Masters C L, Gajdusek D C, Gibbs C J., Jr Brain. 1981;104:559–588. doi: 10.1093/brain/104.3.559. [DOI] [PubMed] [Google Scholar]
- 14.Furukawa H, Kitamoto T, Tanaka Y, Tateishi J. Mol Brain Res. 1995;30:385–388. doi: 10.1016/0169-328x(95)00034-p. [DOI] [PubMed] [Google Scholar]
- 15.Kascsak R J, Rubenstein R, Merz P A, Tonna-DeMasi M, Fersko R, Carp R J, Wisniewski H M, Diringer H. J Virol. 1987;61:3688–3693. doi: 10.1128/jvi.61.12.3688-3693.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen S G, Teplow D B, Parchi P, Teller J K, Gambetti P, Autilio-Gambetti L. J Biol Chem. 1995;270:19173–19180. doi: 10.1074/jbc.270.32.19173. [DOI] [PubMed] [Google Scholar]
- 17.Bolton, D. C., Bendheim, P. E., Marmorstein, A. D. & Potemska, A. Arch. Biochem. Biophys.258, 579–590. [DOI] [PubMed]
- 18.Chen S G, Parchi P, Capellari S, Brown P, Capellari S, Zou W, Cochran E J, Vnencak-Jones C L, Julien J, Vital C, Mikol J, Lugaresi E, et al. Nat Med. 1997;3:1009–1015. doi: 10.1038/nm0997-1009. [DOI] [PubMed] [Google Scholar]
- 19.Prusiner S B. Science. 1991;252:1515–1522. doi: 10.1126/science.1675487. [DOI] [PubMed] [Google Scholar]
- 20.Caughey B, Raymond G J, Ernst D, Race R E. J Virol. 1991;65:6597–6603. doi: 10.1128/jvi.65.12.6597-6603.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borchelt D R, Scott M, Taraboulos A, Stahl N, Prusiner S B. J Cell Biol. 1990;110:743–752. doi: 10.1083/jcb.110.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tagliavini F, Prelli F, Verga L, Giaccone G, Sarma R, Gorevic P, Ghetti B, Passerini F, Ghibaudi E, Forloni G, et al. Proc Natl Acad Sci USA. 1993;90:9678–9682. doi: 10.1073/pnas.90.20.9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Castellani R, Parchi P, Stahl J, Capellari S, Cohen M, Gambetti P. Neurology. 1996;46:1690–1693. doi: 10.1212/wnl.46.6.1690. [DOI] [PubMed] [Google Scholar]
- 24.Hegde R S, Mastrianni J A, Scott M R, DeFea K A, Trembley P, Torchia M, DeArmond S J, Prusiner S B, Lingappa V R. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- 25.Piccardo P, Seiler C, Dlouhy S R, Young K, Farlow M R, Prelli F, Frangione B, Bugiani O, Tagliavini F, Ghetti B. J Neuropath Exp Neurol. 1996;55:1157–1163. doi: 10.1097/00005072-199611000-00007. [DOI] [PubMed] [Google Scholar]
- 26.Brown P, Gibbs C J, Jr, Rodgers-Johnson P, Asher D M, Sulima M P, Bacote A, Goldfarb L G, Gajdusek D C. Ann Neurol. 1994;35:513–529. doi: 10.1002/ana.410350504. [DOI] [PubMed] [Google Scholar]
- 27.Tateishi J, Brown P, Kitamoto T, Hoque Z M, Roos R, Wollman R, Cervenakova L, Gajdusek D C. Nature (London) 1995;376:434–435. doi: 10.1038/376434a0. [DOI] [PubMed] [Google Scholar]
- 28.Baker H F, Duchen L W, Jacobs J M, Ridley R M. Brain. 1990;113:1891–1909. doi: 10.1093/brain/113.6.1891. [DOI] [PubMed] [Google Scholar]
- 29.Tateishi J, Kitamoto T, Doh-ura K, Sakaki Y, Steinmetz G, Tranchant C, Warter J M, Heldt N. Neurology. 1990;40:1578–1581. doi: 10.1212/wnl.40.10.1578. [DOI] [PubMed] [Google Scholar]
- 30.Tateishi J. Semin Virol. 1996;7:175–180. [Google Scholar]