Abstract
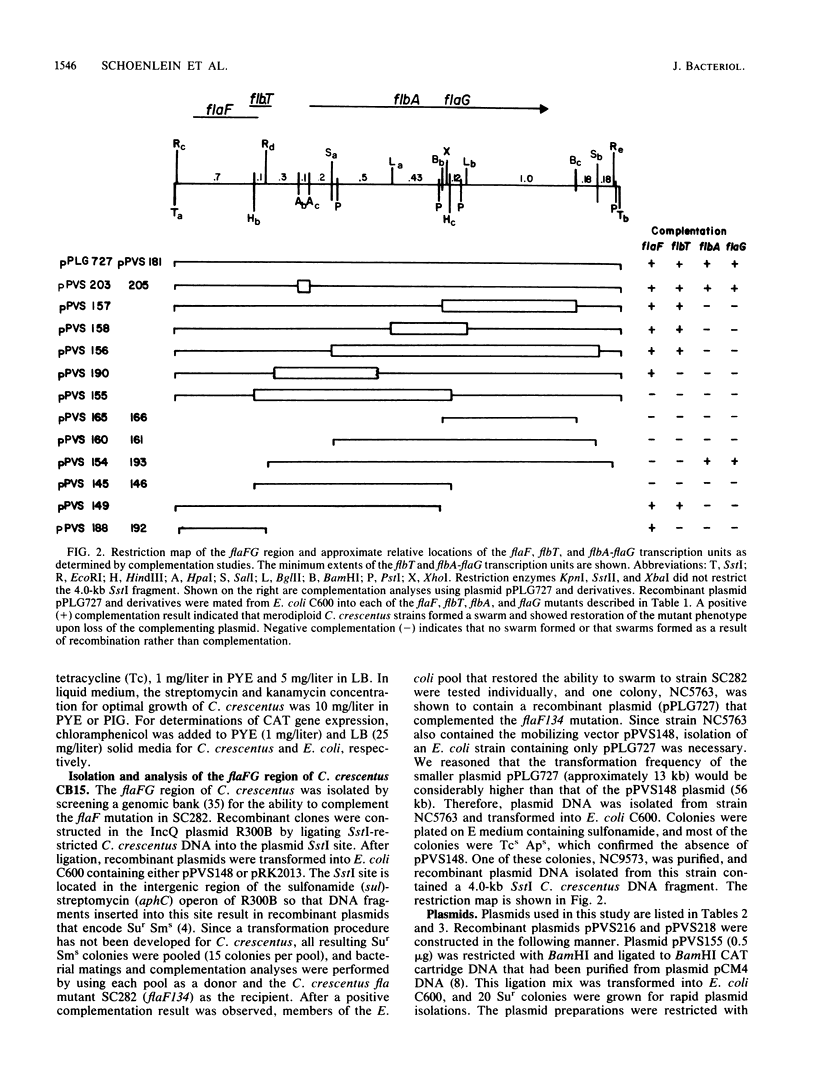
In Caulobacter crescentus, mutations have been isolated in more than 30 flagellar genes (fla, flb, and flg) which are required in the cell cycle event of flagellum biogenesis. The flaF and flaG mutations and two newly identified mutations, flbT and flbA (P.V. Schoenlein and B. Ely, J. Bacteriol. 171:000-000, 1989), have been localized to the flaFG region. In this study, the genetic and physical organization of this region was analyzed, using the cloned 4.0-kilobase flaFG region in the recombinant plasmid pPLG727. Plasmid pPLG727 complemented flaF, flaG, flbA, and flbT mutations. Further complementation studies with pPLG727 derivatives indicated that flaF and flbT are unique but overlapping transcription units, whereas flbA and flaG constitute a single transcription unit. To determine the direction of transcription of the putative flbA-flaG operon, the promoterless chloramphenicol transacetylase gene was inserted into various positions in the flbA-flaG region, and merodiploid strains containing these transcriptional fusions were assayed for gene function and expression of chloramphenicol resistance. These studies showed that transcription proceeds from flbA to flaG. To confirm the complementation analysis, Southern analyses were performed on chromosomal DNAs isolated from strains containing insertion and deletion mutations. Taken together, these studies defined the relative gene order at one end of the flaYG flagellar gene cluser as flgL-flaF-flbT-flbA-flaG.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Barrett J. T., Croft R. H., Ferber D. M., Gerardot C. J., Schoenlein P. V., Ely B. Genetic mapping with Tn5-derived auxotrophs of Caulobacter crescentus. J Bacteriol. 1982 Aug;151(2):888–898. doi: 10.1128/jb.151.2.888-898.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryan R., Champer R., Gomes S., Ely B., Shapiro L. Separation of temporal control and trans-acting modulation of flagellin and chemotaxis genes in Caulobacter. Mol Gen Genet. 1987 Feb;206(2):300–306. doi: 10.1007/BF00333588. [DOI] [PubMed] [Google Scholar]
- Champer R., Dingwall A., Shapiro L. Cascade regulation of Caulobacter flagellar and chemotaxis genes. J Mol Biol. 1987 Mar 5;194(1):71–80. doi: 10.1016/0022-2836(87)90716-9. [DOI] [PubMed] [Google Scholar]
- Chen L. S., Mullin D., Newton A. Identification, nucleotide sequence, and control of developmentally regulated promoters in the hook operon region of Caulobacter crescentus. Proc Natl Acad Sci U S A. 1986 May;83(9):2860–2864. doi: 10.1073/pnas.83.9.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Close T. J., Rodriguez R. L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982 Dec;20(2):305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Driver R. P., Lawther R. P. Physical analysis of deletion mutations in the ilvGEDA operon of Escherichia coli K-12. J Bacteriol. 1985 May;162(2):598–606. doi: 10.1128/jb.162.2.598-606.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Croft R. H., Gerardot C. J. Genetic mapping of genes required for motility in Caulobacter crescentus. Genetics. 1984 Nov;108(3):523–532. doi: 10.1093/genetics/108.3.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B., Croft R. H. Transposon mutagenesis in Caulobacter crescentus. J Bacteriol. 1982 Feb;149(2):620–625. doi: 10.1128/jb.149.2.620-625.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely B. Transfer of drug resistance factors to the dimorphic bacterium Caulobacter crescentus. Genetics. 1979 Mar;91(3):371–380. doi: 10.1093/genetics/91.3.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill P. R., Agabian N. The nucleotide sequence of the Mr = 28,500 flagellin gene of Caulobacter crescentus. J Biol Chem. 1983 Jun 25;258(12):7395–7401. [PubMed] [Google Scholar]
- Johnson R. C., Ely B. Analysis of nonmotile mutants of the dimorphic bacterium Caulobacter crescentus. J Bacteriol. 1979 Jan;137(1):627–634. doi: 10.1128/jb.137.1.627-634.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Ferber D. M., Ely B. Synthesis and assembly of flagellar components by Caulobacter crescentus motility mutants. J Bacteriol. 1983 Jun;154(3):1137–1144. doi: 10.1128/jb.154.3.1137-1144.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. C., Walsh M. P., Ely B., Shapiro L. Flagellar hook and basal complex of Caulobacter crescentus. J Bacteriol. 1979 Jun;138(3):984–989. doi: 10.1128/jb.138.3.984-989.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knauf V. C., Nester E. W. Wide host range cloning vectors: a cosmid clone bank of an Agrobacterium Ti plasmid. Plasmid. 1982 Jul;8(1):45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- Koyasu S., Asada M., Fukuda A., Okada Y. Sequential polymerization of flagellin A and flagellin B into Caulobacter flagella. J Mol Biol. 1981 Dec 5;153(2):471–475. doi: 10.1016/0022-2836(81)90292-8. [DOI] [PubMed] [Google Scholar]
- Loewy Z. G., Bryan R. A., Reuter S. H., Shapiro L. Control of synthesis and positioning of a Caulobacter crescentus flagellar protein. Genes Dev. 1987 Aug;1(6):626–635. doi: 10.1101/gad.1.6.626. [DOI] [PubMed] [Google Scholar]
- Milhausen M., Agabian N. Caulobacter flagellin mRNA segregates asymmetrically at cell division. Nature. 1983 Apr 14;302(5909):630–632. doi: 10.1038/302630a0. [DOI] [PubMed] [Google Scholar]
- Milhausen M., Gill P. R., Parker G., Agabian N. Cloning of developmentally regulated flagellin genes from Caulobacter crescentus via immunoprecipitation of polyribosomes. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6847–6851. doi: 10.1073/pnas.79.22.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minnich S. A., Newton A. Promoter mapping and cell cycle regulation of flagellin gene transcription in Caulobacter crescentus. Proc Natl Acad Sci U S A. 1987 Mar;84(5):1142–1146. doi: 10.1073/pnas.84.5.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ohta N., Swanson E., Ely B., Newton A. Physical mapping and complementation analysis of transposon Tn5 mutations in Caulobacter crescentus: organization of transcriptional units in the hook gene cluster. J Bacteriol. 1984 Jun;158(3):897–904. doi: 10.1128/jb.158.3.897-904.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osley M. A., Sheffery M., Newton A. Regulation of flagellin synthesis in the cell cycle of caulobacter: dependence on DNA replication. Cell. 1977 Oct;12(2):393–400. doi: 10.1016/0092-8674(77)90115-5. [DOI] [PubMed] [Google Scholar]
- POINDEXTER J. S. BIOLOGICAL PROPERTIES AND CLASSIFICATION OF THE CAULOBACTER GROUP. Bacteriol Rev. 1964 Sep;28:231–295. doi: 10.1128/br.28.3.231-295.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purucker M., Bryan R., Amemiya K., Ely B., Shapiro L. Isolation of a Caulobacter gene cluster specifying flagellum production by using nonmotile Tn5 insertion mutants. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6797–6801. doi: 10.1073/pnas.79.22.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer B., Henderson N. The cyclization of linear DNA in Escherichia coli by site-specific recombination. Gene. 1988 Oct 30;70(2):331–341. doi: 10.1016/0378-1119(88)90205-3. [DOI] [PubMed] [Google Scholar]
- Schoenlein P. V., Ely B. Characterization of strains containing mutations in the contiguous flaF, flbT, or flbA-flaG transcription unit and identification of a novel fla phenotype in Caulobacter crescentus. J Bacteriol. 1989 Mar;171(3):1554–1561. doi: 10.1128/jb.171.3.1554-1561.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro L. Generation of polarity during Caulobacter cell differentiation. Annu Rev Cell Biol. 1985;1:173–207. doi: 10.1146/annurev.cb.01.110185.001133. [DOI] [PubMed] [Google Scholar]
- Sheffery M., Newton A. Regulation of periodic protein synthesis in the cell cycle: control of initiation and termination of flagellar gene expression. Cell. 1981 Apr;24(1):49–57. doi: 10.1016/0092-8674(81)90500-6. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Weissborn A., Steinmann H. M., Shapiro L. Characterization of the proteins of the Caulobacter crescentus flagellar filament. Peptide analysis and filament organization. J Biol Chem. 1982 Feb 25;257(4):2066–2074. [PubMed] [Google Scholar]
- Winkler M. E., Schoenlein P. V., Ross C. M., Barrett J. T., Ely B. Genetic and physical analyses of Caulobacter crescentus trp genes. J Bacteriol. 1984 Oct;160(1):279–287. doi: 10.1128/jb.160.1.279-287.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]



