Abstract
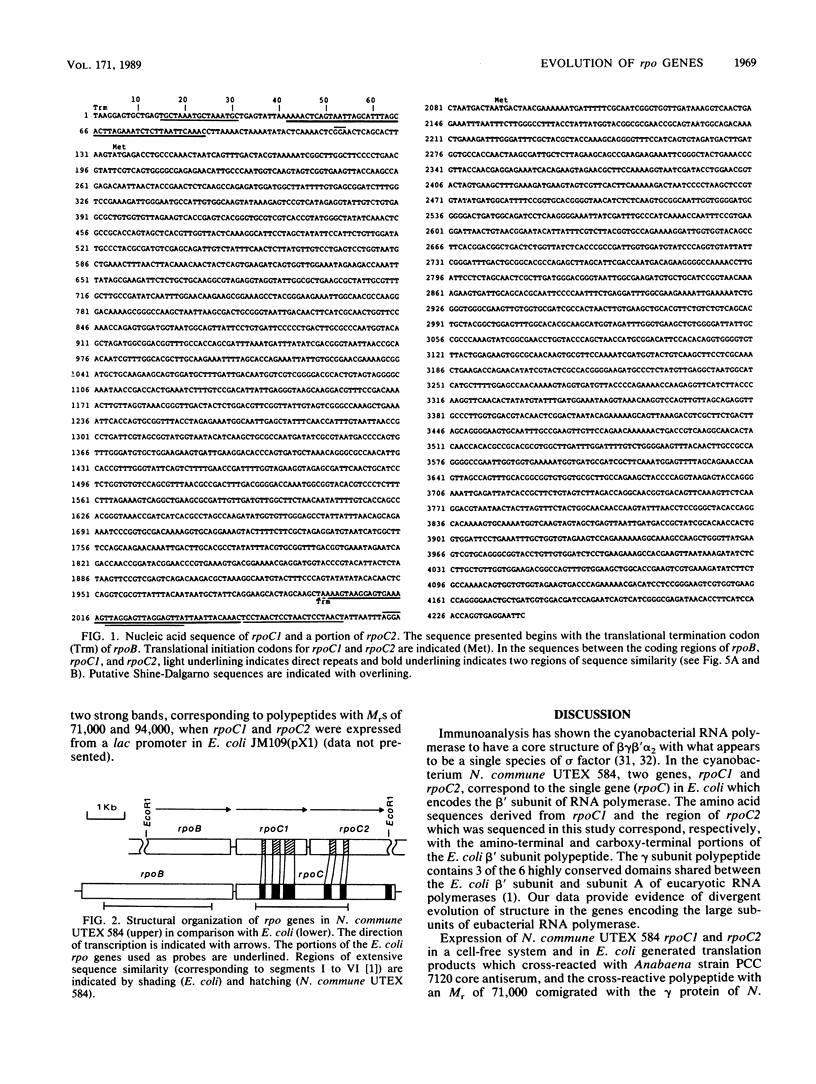
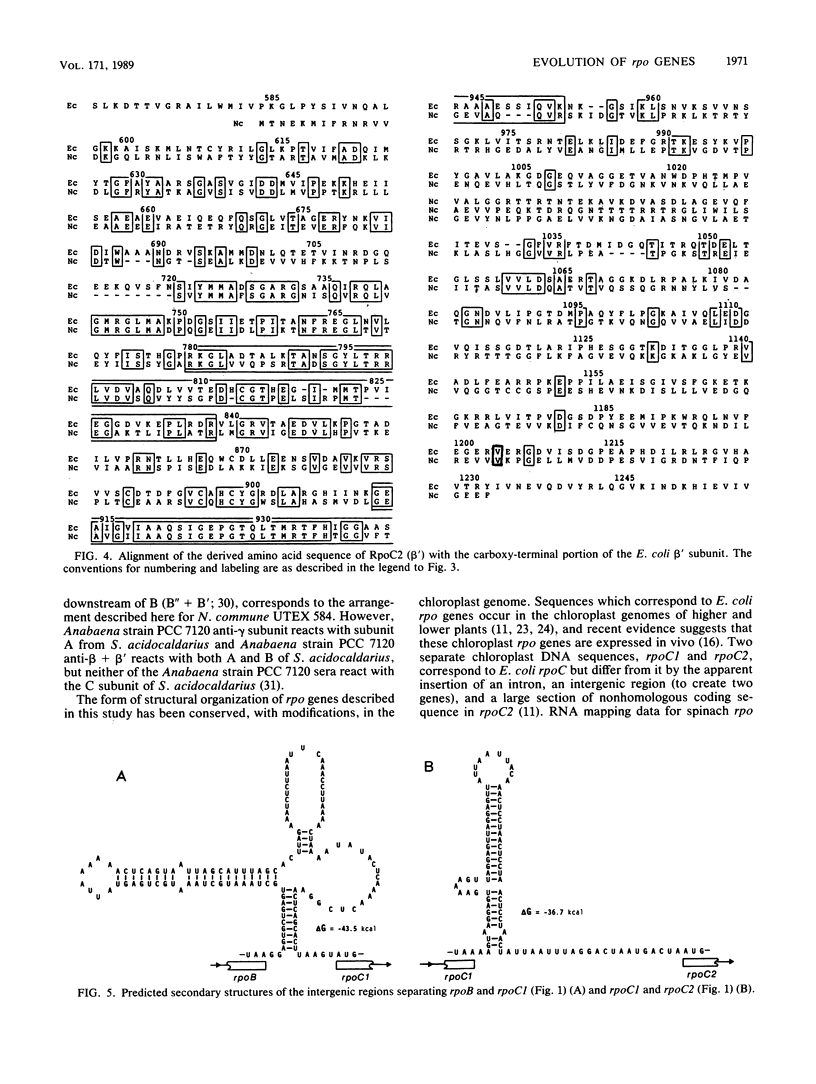
The DNA-dependent RNA polymerase (ribonucleoside triphosphate:RNA nucleotidyltransferase, EC 2.7.7.6) of cyanobacteria contains a unique core component, gamma, which is absent from the RNA polymerases of other eubacteria (G. J. Schneider, N. E. Tumer, C. Richaud, G. Borbely, and R. Haselkorn, J. Biol. Chem. 262:14633-14639, 1987). We present the complete nucleotide sequence of rpoC1, the gene encoding the gamma subunit, from the heterocystous cyanobacterium Nostoc commune UTEX 584. The derived amino acid sequence of gamma (621 residues) corresponds with the amino-terminal portion of the beta' polypeptide of Escherichia coli RNA polymerase. A second gene in N. commune UTEX 584, rpoC2, encodes a protein which shows correspondence with the carboxy-terminal portion of the E. coli beta' subunit. The rpoBC1C2 genes of N. commune UTEX 584 are present in single copies and are arranged in the order rpoBC1C2, and the coding regions are separated by short AT-rich spacer regions which have the potential to form very stable secondary structures. Our data indicate the occurrence of divergent evolution of structure in the eubacterial DNA-dependent RNA polymerase.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Berghöfer B., Kröckel L., Körtner C., Truss M., Schallenberg J., Klein A. Relatedness of archaebacterial RNA polymerase core subunits to their eubacterial and eukaryotic equivalents. Nucleic Acids Res. 1988 Aug 25;16(16):8113–8128. doi: 10.1093/nar/16.16.8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhler J. M., Huet J., Davies K. E., Sentenac A., Fromageot P. Immunological studies of yeast nuclear RNA polymerases at the subunit level. J Biol Chem. 1980 Oct 25;255(20):9949–9954. [PubMed] [Google Scholar]
- Burgess R. R. Separation and characterization of the subunits of ribonucleic acid polymerase. J Biol Chem. 1969 Nov 25;244(22):6168–6176. [PubMed] [Google Scholar]
- Dennis P. P., Nene V., Glass R. E. Autogenous posttranscriptional regulation of RNA polymerase beta and beta' subunit synthesis in Escherichia coli. J Bacteriol. 1985 Feb;161(2):803–806. doi: 10.1128/jb.161.2.803-806.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkenburg D., Dworniczak B., Faust D. M., Bautz E. K. RNA polymerase II of Drosophila. Relation of its 140,000 Mr subunit to the beta subunit of Escherichia coli RNA polymerase. J Mol Biol. 1987 Jun 20;195(4):929–937. doi: 10.1016/0022-2836(87)90496-7. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Herzfeld F., Kiper M. The reconstitution of Anacystis nidulans DNA-dependent RNA polymerase from its isolated subunits. Eur J Biochem. 1976 Feb 2;62(1):189–192. doi: 10.1111/j.1432-1033.1976.tb10112.x. [DOI] [PubMed] [Google Scholar]
- Hudson G. S., Holton T. A., Whitfield P. R., Bottomley W. Spinach chloroplast rpoBC genes encode three subunits of the chloroplast RNA polymerase. J Mol Biol. 1988 Apr 20;200(4):639–654. doi: 10.1016/0022-2836(88)90477-9. [DOI] [PubMed] [Google Scholar]
- Huet J., Sentenac A., Fromageot P. Spot-immunodetection of conserved determinants in eukaryotic RNA polymerases. Study with antibodies to yeast RNA polymerases subunits. J Biol Chem. 1982 Mar 10;257(5):2613–2618. [PubMed] [Google Scholar]
- Little M. C., Hallick R. B. Chloroplast rpoA, rpoB, and rpoC genes specify at least three components of a chloroplast DNA-dependent RNA polymerase active in tRNA and mRNA transcription. J Biol Chem. 1988 Oct 5;263(28):14302–14307. [PubMed] [Google Scholar]
- Miller S. S., Bogorad L. Purification and Characterization of RNA Polymerase from Fremyella diplosiphon. Plant Physiol. 1978 Dec;62(6):995–999. doi: 10.1104/pp.62.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Ishihama A., Kajitani M., Takahashi T., Nakada N., Yoshinaga K. Promoter selectivity of Escherichia coli RNA polymerase. II: Altered promoter selection by mutant holoenzymes. Mol Gen Genet. 1984;193(1):8–16. doi: 10.1007/BF00327407. [DOI] [PubMed] [Google Scholar]
- Ohme M., Tanaka M., Chunwongse J., Shinozaki K., Sugiura M. A tobacco chloroplast DNA sequence possibly coding for a polypeptide similar to E. coli RNA polymerase beta-subunit. FEBS Lett. 1986 May 5;200(1):87–90. doi: 10.1016/0014-5793(86)80516-6. [DOI] [PubMed] [Google Scholar]
- Ohyama K., Kohchi T., Sano T., Yamada Y. Newly identified groups of genes in chloroplasts. Trends Biochem Sci. 1988 Jan;13(1):19–22. doi: 10.1016/0968-0004(88)90013-8. [DOI] [PubMed] [Google Scholar]
- Potts M. Protein synthesis and proteolysis in immobilized cells of the cyanobacterium Nostoc commune UTEX 584 exposed to matric water stress. J Bacteriol. 1985 Dec;164(3):1025–1031. doi: 10.1128/jb.164.3.1025-1031.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schallenberg J., Moes M., Truss M., Reiser W., Thomm M., Stetter K. O., Klein A. Cloning and physical mapping of RNA polymerase genes from Methanobacterium thermoautotrophicum and comparison of homologies and gene orders with those of RNA polymerase genes from other methanogenic archaebacteria. J Bacteriol. 1988 May;170(5):2247–2253. doi: 10.1128/jb.170.5.2247-2253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G. J., Hasekorn R. RNA polymerase subunit homology among cyanobacteria, other eubacteria and archaebacteria. J Bacteriol. 1988 Sep;170(9):4136–4140. doi: 10.1128/jb.170.9.4136-4140.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G. J., Tumer N. E., Richaud C., Borbely G., Haselkorn R. Purification and characterization of RNA polymerase from the cyanobacterium Anabaena 7120. J Biol Chem. 1987 Oct 25;262(30):14633–14639. [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Xie W. Q., Potts M. Quick screening of plasmid deletion clones carrying inserts of desired sizes for DNA sequencing. Gene Anal Tech. 1989 Jan-Feb;6(1):17–20. doi: 10.1016/0735-0651(89)90008-3. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zillig W., Palm P., Reiter W. D., Gropp F., Pühler G., Klenk H. P. Comparative evaluation of gene expression in archaebacteria. Eur J Biochem. 1988 May 2;173(3):473–482. doi: 10.1111/j.1432-1033.1988.tb14023.x. [DOI] [PubMed] [Google Scholar]
- Zillig W., Stetter K. O., Tobien M. DNA-dependent RNA polymerase from Halobacterium halobium. Eur J Biochem. 1978 Nov 2;91(1):193–199. doi: 10.1111/j.1432-1033.1978.tb20951.x. [DOI] [PubMed] [Google Scholar]



