Abstract
Sulfonylureas are a class of drugs commonly used in the management of non-insulin-dependent diabetes mellitus. Their therapeutic action results primarily from their ability to inhibit ATP-sensitive potassium (KATP) channels in the plasma membrane of pancreatic β cells and thereby stimulate insulin release. A key question is whether an endogenous ligand for the KATP channel exists that is able to mimic the inhibitory effects of sulfonylureas. We describe here the cloning of the cDNA encoding human α-endosulfine, a 13-kDa peptide that is a putative candidate for such a role. α-Endosulfine is expressed in a wide range of tissues including muscle, brain, and endocrine tissues. The recombinant protein displaces binding of the sulfonylurea [3H]glibenclamide to β cell membranes, inhibits cloned KATP channel currents, and stimulates insulin secretion. We propose that endosulfine is an endogenous regulator of the KATP channel, which has a key role in the control of insulin release and, more generally, couples cell metabolism to electrical activity.
Sulfonylureas are a class of drugs commonly used in the management of non-insulin-dependent diabetes mellitus (1). Their therapeutic action results primarily from their ability to inhibit ATP-sensitive potassium (KATP) channels in the β cell plasma membrane and thereby stimulate insulin release. The KATP channel sets the β cell resting potential, and its closure results in membrane depolarization, activation of voltage-gated Ca2+ channels, enhanced Ca2+ influx, and thus insulin secretion (2). Glucose-induced insulin release involves the same sequence of events, except that in this case KATP channel inhibition is believed to be caused by metabolically produced changes in ATP and MgADP, which inhibit and activate the channel, respectively (3).
The KATP channel is an octameric complex of two structurally unrelated types of subunit, Kir6.2 and SUR, which coassemble with a 4:4 stoichiometry (4–7). Kir6.2, a member of the inward rectifier K+ channel family, forms the channel pore and possesses intrinsic ATP sensitivity (8). The sulfonylurea receptor, SUR, belongs to a family of ATP-binding cassette transporter proteins (9). It acts as a receptor for many drugs and endows Kir6.2 with sensitivity, both to sulfonylureas and to K+ channel openers. Two types of SUR have been cloned, with different pharmacological sensitivities: the β cell KATP channel is composed of Kir6.2 and SUR1, and cardiac KATP channels comprise Kir6.2 and SUR2A subunits (10).
The existence of SUR1, which binds sulfonylureas with a high affinity (Ki glibenclamide <5 nM), and its interaction with Kir, which argues for a physiological regulatory role, suggested the presence of an endogenous regulator of KATP channel activity that interacts with the same site as these drugs. This led to the purification of α- and β-endosulfine from ovine brain (11), two peptides that inhibit binding of sulfonylureas to their receptor in vitro (12). During the search for endosulfine (11), a peptide corresponding to the high molecular mass α-form was isolated from porcine brain and partially sequenced (13). A partial cDNA sequence of α-endosulfine was determined (14) and found to show strong homology with that of a bovine brain 12- to 13-kDa cyclic AMP-regulated phosphoprotein of unknown function called ARPP-19 (15, 16). The two proteins, however, appear to be encoded by different genes (14).
In the present report, we describe the cloning of a human α-endosulfine partial cDNA and the expression of large amounts of the entire protein in a bacterial system. α-Endosulfine is expressed in a wide range of tissues including muscle, brain, and endocrine tissues. The recombinant protein inhibits binding of [3H]glibenclamide to β cell membranes as efficiently as extracted porcine α-endosulfine (13), indicating that it indeed represents human α-endosulfine. It also inhibits cloned KATP channel currents and thereby stimulates insulin secretion.
MATERIALS AND METHODS
PCR.
A 233-bp α-endosulfine cDNA fragment was first synthesized by PCR, by using a human brain λGT11 cDNA library (CLONTECH) and oligonucleotides derived from the bovine α-endosulfine sequence (14). DNA fragments were resolved by agarose gel electrophoresis, electroeluted, and purified by using the Wizard PCR-prep procedure (Promega). PCR products were subcloned into the pBluescript II/SK− (Stratagene) and sequenced on both DNA strands by using the dideoxy chain termination method (17).
Library Screening.
The α-endosulfine cDNA fragment was labeled with [α-32P]dCTP by using a random primer DNA-labeling kit (Pharmacia) and used to screen a λGT11 human brain cDNA library. The three positive clones obtained each contained a 749-bp insert, which was subcloned into pBluescript II/SK−, sequenced on both strands, and shown to correspond to human α-endosulfine cDNA (EMBL accession no. X99906).
Northern Blot.
A Northern Territory (Invitrogen) total RNA blot from normal human tissues was hybridized according to the manufacturer’s instructions in a 50% formamide hybridization solution containing 106 cpm/ml of the [α-32P]dCTP-labeled α-endosulfine cDNA probe. The blot was washed under standard conditions (18) before autoradiography. Total RNAs were extracted from MIN6 cells by using the EXTRACT-ALL reagent (Eurobio, Paris), according to the manufacturer’s instructions, and analyzed as described (14).
Expression and Purification of the Recombinant α-Endosulfine.
The human α-endosulfine cDNA-coding region was subcloned into the pCAL-n expression vector (Stratagene) in-frame with a cleavage site for thrombin and a downstream sequence encoding a calmodulin-binding peptide (CBP) tag. The hybrid protein was expressed under the control of the bacteriophage T7 gene promoter in the BL21 DE3 pLys S bacteria after a 3-h induction by isopropyl-β-d-thiogalactopyranoside (19), and its expression was checked on a 20% SDS/PAGE (20). The fusion protein was purified on a calmodulin affinity column and eluted with EGTA (21). Purified α-endosulfine was obtained after thrombin cleavage of the fusion protein and removal of the tag by adsorption on the calmodulin affinity resin. The purified protein was then extensively dialyzed against water, and 5-nmol aliquots were lyophilized.
Cell Culture.
MIN6 cells, kindly provided by H. Ishihara, (Third Department of Internal Medicine, University of Tokyo) were grown at 37°C (5% CO2 in air atmosphere) as described (22) in DMEM (GIBCO) containing 25 mM glucose and supplemented with 15% fetal calf serum (GIBCO), 100 units/ml penicillin, 100 μg/ml streptomycin, and 5 μl/ml β-mercaptoethanol.
Measurement of Insulin Release.
For insulin release experiments (23), cells culture medium was renewed 18 h before the experiment was begun. On the day of the experiment, cells were washed twice with a Krebs-Ringer bicarbonate buffer, pH 7.5 (containing 0.1% BSA) and preincubated for 1 h at 37°C in the presence of 1 mM glucose. Cells were then incubated under the same conditions for 1 h in the presence of various concentrations of glucose and/or other effectors. Insulin release was measured by radioimmunoassay (24). It must be noted that the MIN6 cells used in our set of experiments display a response to glucose in the physiological range (no effect at 1 mM, half-maximal effect at approximately 12 mM, and maximal effect at approximately 25 mM).
Binding Assay.
Membranes were prepared from MIN6 cells (25), and binding studies were conducted as described (26). In brief, membranes (20 μg/ml) were incubated for 1 h at 25°C with 0.1 nmol/liter [3H]glibenclamide (48–51 Ci/mmol, NEN) in 50 mM Tris⋅HCl, pH 7.5, with no addition of albumin or protease inhibitors, and in the presence of unlabeled glibenclamide (Pierre Fabre, Castres, France) or recombinant α-endosulfine. Specific binding was obtained by subtracting the binding in the presence of 100 μM unlabeled glibenclamide. Bound and free radiolabeled ligands were separated by filtration under vacuum on GF/B glass fiber filters (Whatman). Filters were washed twice with the same cold buffer, and membrane-bound [3H]glibenclamide was measured by directly counting filters in a liquid scintillation medium.
Electrophysiological Experiments.
Xenopus laevis oocytes were coinjected with mouse Kir 6.2 (GenBank accession no. D50581) and rat SUR1 (GenBank accession no. L40624) cRNAs (27). Macroscopic currents were recorded 1–4 d later from giant inside-out patches, by using an EPC7 patch-clamp amplifier (List Electronics, Darmstadt, Germany) at 20–24°C. The pipette solution contained 140 mM KCl, 1.2 mM MgCl2, 2.6 mM CaCl2, and 10 mM Hepes (pH 7.4 with KOH). The internal (bath) solution contained 110 mM KCl, 1.44 mM MgCl2, 30 mM KOH, 10 mM EGTA, and 10 mM Hepes (pH 7.2 with KOH). Solutions were exchanged by positioning the patch electrode in the mouth of one of a series of adjacent inflow pipes.
RESULTS
Cloning of Human α-Endosulfine cDNA.
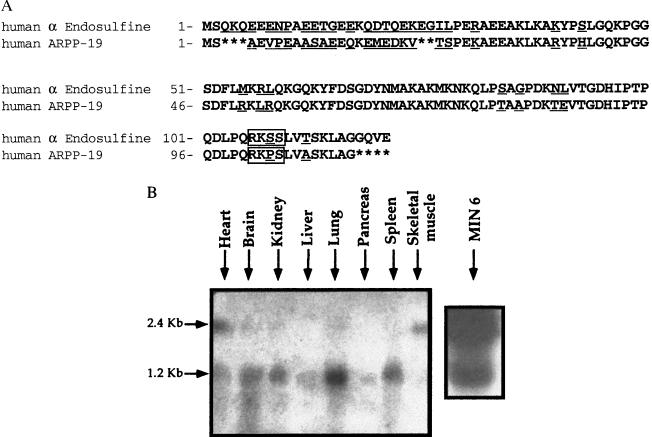
We isolated a 749-bp insert from a human brain λGT11 cDNA library. The 749-bp α-endosulfine cDNA that was cloned contains a single 363-bp ORF beginning with the first ATG. The nucleotide sequence predicts a polypeptide of 121 residues with a molecular mass of 13,517 Da (Fig. 1A). There is no obvious signal peptide sequence at the N terminus, suggesting that α-endosulfine is an intracellular protein. A partial bovine α-endosulfine sequence displayed considerable homology with that of bovine ARPP-19 (14–16), although the two proteins appeared to be encoded by different genes (14). We therefore cloned part of the human ARPP-19 cDNA (EMBL accession no. AJ223091), and compared its sequence to that of human α-endosulfine (Fig. 1A). The two human peptidic sequences display a high degree of identity, except in the N-terminal region. Both proteins contain a cAMP-dependent phosphorylation site (boxed in Fig. 1A) in the C terminus, as observed for bovine ARPP-19 (15, 16). It was verified that α-endosulfine can be phosphorylated in vitro by protein kinase A.
Figure 1.
(A) Amino acid sequence of human α-endosulfine and human ARPP-19 predicted from the respective cDNAs. Amino acids are identified by the one-letter code. Differences between the two proteins are underlined and potential cAMP-dependent phosphorylation sites are boxed. (B) Northern blot analysis of α-endosulfine mRNA expression in various human tissues. Total mRNA (20 μg/lane) from the specified tissues was hybridized with a probe consisting of the (749-bp) human α-endosulfine cDNA insert. Poly(A)+ RNAs corresponding to 500 μg of total RNA from MIN6 cells were analyzed under the same conditions.
Tissue Distribution of α-Endosulfine mRNAs.
Northern blot analysis showed that the 749-bp α-endosulfine cDNA hybridizes with two mRNA species of 2.4 and 1.2 kb, respectively (Fig. 1B). These transcripts are likely to represent different α-endosulfine mRNA forms because the probe used did not cross-hybridize with ARPP-19 cDNA (L.H., unpublished observations). Expression of the 2.4-kb transcript appears to be restricted to heart and skeletal muscle, whereas the 1.2-kb transcript is more widely expressed, with the highest levels occurring in lung and brain. All of these regions also express KATP channel subunits (4, 5). Although the large size difference observed for the two α-endosulfine transcripts raises the possibility that different proteins may be generated, depending on the tissue in which they were expressed, this does not seem to be the case, because the coding region obtained by reverse-transcription-PCR amplification of both transcripts showed complete sequence identity (data not shown). Thus, the two α-endosulfine mRNAs differ only in their 5′- and/or 3′-untranslated regions. Two transcripts with similar sizes were also detected in MIN6 cells (Fig. 1B).
Production of Recombinant α-Endosulfine in E. coli and Affinity Chromatography Purification.
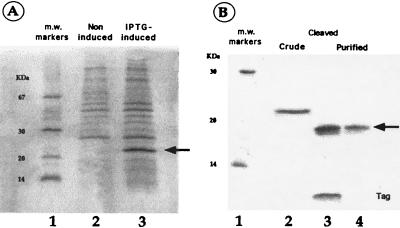
We used a bacterial system to produce recombinant human α-endosulfine for functional studies. A fusion protein was constructed by linking the cDNA encoding α-endosulfine with that of a CBP. This fusion protein is expressed in bacteria after isopropyl-β-d-thiogalactopyranoside induction (Fig. 2A, lane 3) and has an apparent Mr of 23,000. Thrombin cleavage of the purified fusion protein generated a 19-kDa protein corresponding to α-endosulfine and a low molecular mass peptide corresponding to the CBP tag (Fig. 2B, lane 3). Running thrombin cleavage products on the calmodulin affinity column enabled elimination of the tag and yielded highly purified recombinant human α-endosulfine (Fig. 2B, lane 4). About 1 mg of pure α-endosulfine was obtained from 300 ml of bacterial culture.
Figure 2.
(A) SDS/PAGE analysis of proteins extracted from bacteria transfected with a CBP/α-endosulfine construct before (lane 2) and after (lane 3) induction of the fusion protein. The arrow shows the specific fusion protein band. IPTG, isopropyl-β-d-thiogalactopyranoside. (B) SDS/PAGE of affinity chromatography-purified CBP/α-endosulfine fusion protein before (lane 2) and after (lane 3) thrombin cleavage and after elimination of the tag by affinity chromatography (lane 4). The arrow shows the purified α-endosulfine protein. m.w. markers, molecular mass markers (lanes 1).
Human α-Endosulfine Interacts with the β Cell Sulfonylurea Receptor.
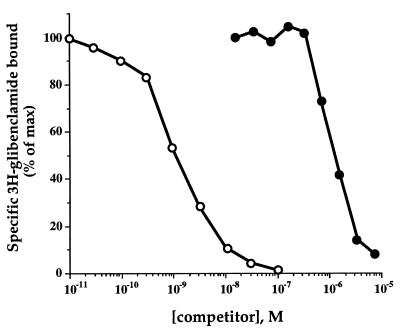
We first tested the ability of recombinant α-endosulfine to displace sulfonylurea binding to membranes from the MIN6 β cell line. The recombinant protein dose-dependently inhibited [3H]glibenclamide binding with an ED50 of ≈1 μM (Fig. 3). This binding affinity is similar to that obtained for the native protein (13) and confirms that the protein encoded by the cloned cDNA is indeed human α-endosulfine.
Figure 3.
Effects of glibenclamide (○) and α-endosulfine (•) on [3H]glibenclamide binding to MIN6 cell membranes. Half-maximal inhibition of [3H]glibenclamide binding was produced by 1nM glibenclamide and 1 μM endosulfine. Each point is the mean of duplicate determinations within a representative experiment, which has been reproduced 3-fold.
Effect of Recombinant α-Endosulfine on K+ Currents in Oocytes.
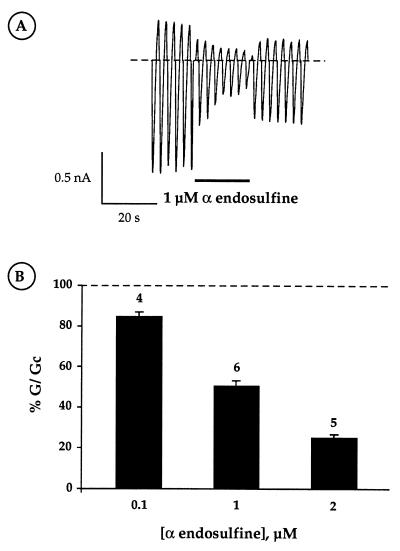
Because the sulfonylurea receptor (SUR1) forms part of the KATP channel, we next examined the functional effects of α-endosulfine on cloned β cell KATP channel currents (Kir6.2/SUR1), expressed in Xenopus oocytes (Fig. 4A). When applied to the intracellular membrane surface, 2 μM human α-endosulfine reduced the KATP conductance by 75 ± 2% (n = 5; Fig. 4B). The estimated EC50 of ≈1 μM was similar to that observed for displacement of glibenclamide binding (Figs. 3 and 4). We observed that extracellular α-endosuline also reduced KATP currents in intact oocytes, when measured with the two-electrode voltage-clamp (data not shown). α-Endosuline (2 μM) was without effect on Kir1.1a, a related inwardly rectifying K channel (data not shown), suggesting that the peptide may interact specifically with the Kir6.2/SUR1 channel.
Figure 4.
(A) Macroscopic Kir6.2/SUR1 currents recorded in a giant inside-out patch in response to a series of voltage ramps from −110 to +100 mV from a holding potential of 0 mV. α-Endosulfine (2 μM) was added to the intracellular solution as indicated. (B) Effects of intracellular α-endosulfine concentration on the macroscopic Kir6.2/SUR1 conductance recorded from inside-out patches, expressed as a percentage of its amplitude (G/Gc) in control solution (without α-endosulfine). Data are expressed as means ± SEM, and the number of experiments is indicated above each column.
Effect of Recombinant α-Endosulfine on Insulin Release.
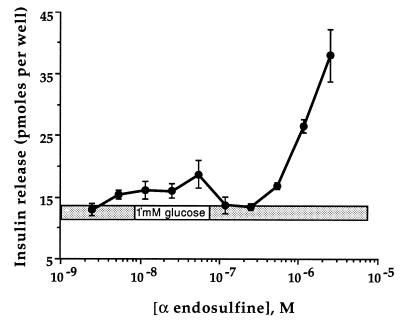
Inhibition of KATP channels plays a key role in insulin release both in response to glucose (the main physiological stimulus) and to drugs such as the sulfonylureas (2, 28). The KATP channel sets the β cell resting potential and its closure results in membrane depolarization, activation of voltage-gated Ca2+ channels, enhanced Ca2+ influx, and thus insulin secretion. The ability of α-endosulfine to block KATP channels therefore suggests that the peptide will trigger insulin release. Fig. 5 shows that this is indeed the case. When tested at a nonstimulatory glucose concentration (1 mM), α-endosulfine induced a dose-dependent stimulation of insulin release. This stimulation occurred within the same range of concentrations that caused displacement of [3H]glibenclamide binding and inhibition of KATP currents.
Figure 5.
Effect of α-endosulfine concentration on insulin release from MIN6 cells. The extracellular solution contained 1 mM glucose throughout. Data are expressed as means ± SEM of a representative experiment performed in triplicate. Similar results were obtained in three different experiments.
DISCUSSION
The results presented here demonstrate that human α-endosulfine is strongly expressed in muscle and brain, with lower levels in the pancreas. This tissue distribution resembles that of the KATP channel (4, 5) and is consistent with the ability of α-endosulfine to regulate KATP channel activity.
Sulfonylureas interact with both SUR1 and Kir6.2 subunits of the β cell KATP channel: high affinity binding is associated with SUR1 (Ki < 5 nM for glibenclamide), but there is also a low affinity site on Kir6.2 (Ki ≈ 100 μM for glibenclamide) (9, 27). The ability of α-endosulfine to displace high affinity [3H]glibenclamide binding to β cell membranes suggests that α-endosulfine may interact with SUR1. There is evidence, however, that Kir6.2 and SUR1 physically interact, because antibodies to either subunit can coimmunoprecipitate the other subunit and sulfonylureas cophotoaffinity label both SUR1 and Kir6.2 (6). We therefore cannot exclude the alternative possibility that α-endosulfine binds to Kir6.2 and thereby displaces glibenclamide binding to SUR1. The question of which subunit α-endosulfine interacts with is of importance, because Kir6.2 is more widely expressed than SUR1 (which is principally found in β cells and parts of the brain). It will be of interest to determine whether α-endosulfine also interacts with other types of KATP channels, such as those found in cardiac, skeletal, and smooth muscles and in certain neurones (2).
The relatively low binding affinity (Ki ≈ 1 μM) of α-endosulfine for the Kir6.2/SUR1 complex contrasts with the much higher affinity of the most potent sulfonylureas (Ki < 5 nM) (1). It remains possible, however, that access of α-endosulfine to its target site is restricted in microsomes and giant patches or that it requires additional cofactors not present in our solutions for full activity. It is also possible that the α-endosulfine concentrations in β cells are compatible with the observed affinity.
It is well established that all secretagogs capable of stimulating insulin secretion in the absence of glucose act by closing the β cell KATP channel. This was also the case for endosulfine, which blocked the cloned KATP current with a Ki of ≈1 μM. KATP channel closure triggers insulin release by causing membrane depolarization, activating voltage-gated Ca2+ entry and so increasing intracellular Ca2+ levels. Because endosulfine is expressed in insulin-secreting β cells, including MIN6 cells, and displays the structure of an endogenous protein with a functional protein kinase A-dependent phosphorylation site, it potentially could act as an endogenous, intracellular regulator of β cell KATP channel activity and insulin secretion in vivo. On the other hand, we observed that α-endosulfine blocked KATP currents when applied both intracellularly and extracellularly, and it stimulated insulin secretion when added to the extracellular solution. This suggests that either α-endosulfine is able to cross the plasma membrane or that it is able to act at more than one site. Binding studies do not address this issue, because these experiments were carried out on isolated membranes. Because α-endosulfine is a peptide, it is unlikely to cross the plasma membrane easily, but its uptake into the cell could be facilitated by a transport protein or by endocytosis.
We conclude that α-endosulfine may act as an endogenous regulator of β cell KATP channel activity and insulin secretion. Cloning of the cDNA encoding α-endosulfine and production of a recombinant protein will enable the mechanism of this regulation to be studied in detail. In particular it will be important to address the possibility that α-endosulfine is involved in the metabolic regulation of KATP channel activity. The functional role of the peptide in other tissues that express KATP channels, such as muscle, nerve, and endocrine cells, also requires investigation. Finally, the availability of the human α-endosulfine cDNA sequence will enable the role of this peptide in pathological states, such non-insulin-dependent diabetes, to be evaluated.
Acknowledgments
We thank the Institut de Recherches Internationales Servier, the Wellcome Trust, and the British Diabetic Association for support.
ABBREVIATIONS
- KATP
ATP-sensitive potassium
- CBP
calmodulin-binding peptide
Footnotes
References
- 1.Ashcroft F M, Ashcroft S J H. Biochim Biophys Acta. 1992;1175:45–59. doi: 10.1016/0167-4889(92)90008-y. [DOI] [PubMed] [Google Scholar]
- 2.Ashcroft S J H, Ashcroft F M. Cell Signalling. 1990;2:197–214. doi: 10.1016/0898-6568(90)90048-f. [DOI] [PubMed] [Google Scholar]
- 3.Ashcroft F M, Gribble F M. Trends Neurosci. 1998;21:288–294. doi: 10.1016/s0166-2236(98)01225-9. [DOI] [PubMed] [Google Scholar]
- 4.Inagaki N, Gonoi T, Clement J P, IV, Namba N, Inazawa J, Gonzalez G, Aguilar-Bryan L, Seino S, Bryan J. Science. 1995;270:1166–1169. doi: 10.1126/science.270.5239.1166. [DOI] [PubMed] [Google Scholar]
- 5.Sakura H, Ämmälä C, Smith P A, Gribble F M, Ashcroft F M. FEBS Lett. 1995;377:338–344. doi: 10.1016/0014-5793(95)01369-5. [DOI] [PubMed] [Google Scholar]
- 6.Clement J P, IV, Kunjilwar K, Gonzalez G, Schwanstecher M, Panten U, Aguilar Bryan L, Bryan J. Neuron. 1997;18:827–838. doi: 10.1016/s0896-6273(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 7.Shyng S L, Ferrigni T, Nichols C G. J Gen Physiol. 1997;110:655–664. doi: 10.1085/jgp.110.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tucker S J, Gribble F M, Zhao C, Trapp S, Ashcroft F M. Nature (London) 1997;387:179–183. doi: 10.1038/387179a0. [DOI] [PubMed] [Google Scholar]
- 9.Aguilar-Bryan L, Nichols C G, Wechsler S W, Clement J P, IV, Boyd A E, Gonzĺez G, Herrera-Sosa H, Nguy K, Bryan J, Nelson D A. Science. 1995;268:423–426. doi: 10.1126/science.7716547. [DOI] [PubMed] [Google Scholar]
- 10.Inagaki N, Gonoi T, Clement J P, IV, Wang C Z, Aguilar-Bryan L, Bryan J, Seino S. Neuron. 1996;16:1011–1017. doi: 10.1016/s0896-6273(00)80124-5. [DOI] [PubMed] [Google Scholar]
- 11.Virsolvy-Vergine A, Leray H, Kuroki S, Lupo B, Dufour M, Bataille D. Proc Natl Acad Sci USA. 1992;89:6629–6633. doi: 10.1073/pnas.89.14.6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Virsolvy A, Brück M, Dufour M, Cauvin A, Lupo B, Bataille D. FEBS Lett. 1988;242:65–69. doi: 10.1016/0014-5793(88)80986-4. [DOI] [PubMed] [Google Scholar]
- 13.Virsolvy-Vergine A, Salazar G, Sillard R, Denoroy L, Mutt V, Bataille D. Diabetologia. 1996;39:135–141. doi: 10.1007/BF00403955. [DOI] [PubMed] [Google Scholar]
- 14.Peyrollier K, Héron L, Virsolvy A, Le Cam A, Bataille D. Biochem Biophys Res Commun. 1996;223:583–586. doi: 10.1006/bbrc.1996.0938. [DOI] [PubMed] [Google Scholar]
- 15.Girault J A, Horiuchi A, Gustafson E, Rosen N, Greengard P. J Neurosci. 1990;10:1124–1133. doi: 10.1523/JNEUROSCI.10-04-01124.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horiuchi A, Williams K, Kurihara T, Nairn A, Greengard P. J Biol Chem. 1990;265:9476–9484. [PubMed] [Google Scholar]
- 17.Sanger F. Science. 1981;214:1205–1209. doi: 10.1126/science.7302589. [DOI] [PubMed] [Google Scholar]
- 18.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 19.Studier F W, Rosenberg H, Dunn J J, Dubendorff J W. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 20.Laemmli U K. Nature (London) 1970;227:680–686. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Vaillancourt P, Simcox T G, Zheng C F. Biotechniques. 1997;22:451–453. doi: 10.2144/97223bm17. [DOI] [PubMed] [Google Scholar]
- 22.Ishihara H, Asano T, Tsuduka K, Katagari H, Inukai K, Anai M, Kikuchi M, Yazaki Y, Miyazaki J I, Oka Y. Diabetologia. 1993;36:1139–1145. doi: 10.1007/BF00401058. [DOI] [PubMed] [Google Scholar]
- 23.Le Brigand L, Virsolvy A, Peyrollier K, Manechez D, Godfroid J J, Guardiola-Lemaître B, Bataille D. Br J Pharmacol. 1997;122:786–791. doi: 10.1038/sj.bjp.0701449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kervran A, Rieutort M, Guillaume M A. Diabete Metab. 1976;2:67–72. [Google Scholar]
- 25.Göke R, Cole T, Conlon J M. J Mol Endocrinol. 1989;2:93–98. doi: 10.1677/jme.0.0020093. [DOI] [PubMed] [Google Scholar]
- 26.Lupo B, Bataille D. Eur J Pharmacol. 1987;140:157–169. doi: 10.1016/0014-2999(87)90801-6. [DOI] [PubMed] [Google Scholar]
- 27.Gribble F M, Tucker S J, Ashcroft F M. J Physiol. 1997;504.1:35–45. doi: 10.1111/j.1469-7793.1997.00035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashcroft F M, Rorsman P. Prog Biophys Mol Biol. 1989;54:87–143. doi: 10.1016/0079-6107(89)90013-8. [DOI] [PubMed] [Google Scholar]