Abstract
Hibernation is a physiological adaptation characterized by dramatic decreases in heart rate, body temperature, and metabolism, resulting in long-term dormancy. Hibernating mammals survive for periods up to 6 mo in the absence of food by minimizing carbohydrate catabolism and using triglyceride stores as their primary source of fuel. The cellular and molecular mechanisms underlying the changes from a state of activity to the hibernating state are poorly understood; however, the selective expression of genes offers one level of control. To address this problem, we used a differential gene expression screen to identify genes that are responsible for the physiological characteristics of hibernation in the heart of the thirteen-lined ground squirrel (Spermophilus tridecemlineatus). Here, we report that genes for pancreatic lipase and pyruvate dehydrogenase kinase isozyme 4 are up-regulated in the heart during hibernation. Pancreatic lipase is normally expressed exclusively in the pancreas, but when expressed in the hibernating heart it liberates fatty acids from triglycerides at temperatures as low as 0°C. Pyruvate dehydrogenase kinase isozyme 4 inhibits carbohydrate oxidation and depresses metabolism by preventing the conversion of pyruvate to Ac-CoA. The resulting anaerobic glycolysis and low-temperature lipid catabolism provide evidence that adaptive changes in cardiac physiology are controlled by the differential expression of genes during hibernation.
Maintenance of normal physiological function in mammals usually requires a constant body temperature of approximately 37°C. Lowering body temperature 10–15°C for an extended period of time often leads to hypothermic dysfunction of several organ systems, while lowering the temperature 30°C below the optimum usually results in death (1). An exception is seen in hibernating mammals. Certain “deep” hibernators lower their body temperature to 2–7°C, reduce their heart rate from 300 beats/min to 2–10 beats/min, and lessen their O2 consumption as much as 50-fold (2). This amazing transformation of whole-animal physiology is completely reversible and serves as an adaptation to conserve energy reserves during extended periods of harsh climate and little or no food. Energy reserves in the form of fat sustain vital functions during long bouts of torpor and support periodic rewarming to euthermic temperatures during interbout arousals. Despite years of work on the ecology and physiology of hibernation in mammals, the underlying molecular genetic basis of this adaptation has received little attention.
The reduction in metabolism that accompanies the hypothermia of hibernation is not only a function of thermodynamics, but also the result of precisely regulated metabolic reactions (3). Hibernating mammals survive the entire winter without feeding by limiting their carbohydrate catabolism and using fat stores as their primary source of fuel (reviewed in ref. 2). We have begun to address the genetic control of this process by using a differential gene expression screen (4) to identify genes that are responsible for the physiological characteristics of hibernation in the heart of the thirteen-lined ground squirrel (Spermophilus tridecemlineatus). Our study focused on the heart because it is a contractile organ that must continue to work despite suboptimal conditions of temperature and oxygen supply. In this paper, we report on the identification and differential expression of two genes associated with metabolic rate depression and low-temperature carbon utilization during hibernation. We have found that genes encoding pancreatic lipase (PL) and pyruvate dehydrogenase kinase isozyme 4 (PDK-4) are up-regulated in the heart when hibernation begins and that steady-state levels of both mRNAs remain high while metabolism and body temperature are greatly depressed. Up-regulation of PDK-4 and PL in the heart supports the hypothesis that the physiological characteristics of hibernation are controlled, in part, by the differential expression of genes common to all mammals (5).
MATERIALS AND METHODS
Animals.
Animal care and use was in accordance with institutional animal care and use committee guidelines. Thirteen-lined ground squirrels were raised in captivity on a diet of rodent chow supplemented with sunflower seeds and water ad libitum. Squirrels were observed daily and maintained at 23°C from mid-March through August, 17°C in September, 11°C in October, and 5°C from November through mid-March. Animals were housed with a daily light cycle of 12-hr dark:12-hr light from mid-March through the end of October. From November to mid-March the animals were housed in total darkness with only water ad libitum; under these conditions all of the animals hibernated. Length of individual hibernation bouts was confirmed by the sawdust technique (6), and body temperatures (rectal) were measured at the time of sacrifice. Animals in interbout arousal were collected after at least three previous hibernation bouts of 8 days or more.
cDNA Cloning, Subtraction, and Sequencing.
Total RNA was isolated from ground squirrel hearts according to the method described below in RNA Isolation and Northern Blot Analysis. Poly(A)+ mRNA was purified using mini oligo(dT)- cellulose spin columns (5 Prime → 3 Prime). Subtractive hybridization was done according to the PCR-based gene expression screen developed by Wang and Brown (4). Modifications were made in the linkers ligated to the ends of cDNA fragments such that distinct sets of linkers were used for active-state (driver) cDNAs and for hibernating-state (tracer) cDNAs. To maintain identical melting and annealing temperatures, the majority of the sequence of the driver cDNA linker (5′-GAGAACGAAGAATTCCCTGAT-3′) was the reverse of the tracer cDNA linker (5′-TAGTCCGAATTCAAGCAAGAG-3′), the exception being that both contained EcoRI sites for purposes of cloning. Because the linkers were used as PCR primers, the sequence difference allowed for more thorough subtraction due to state-specific priming sites for amplifications performed throughout the screen. After eight rounds of subtraction, double-stranded cDNA fragments of up-regulated genes were cleaved with EcoRI and ligated to pBluescript II KS(+) (Stratagene) for transformation into electrocompetent DH10B cells. Colonies containing up-regulated sequences were screened by hybridization with probes prepared from up-regulated (+8) and down-regulated (−8) populations of cDNA fragments generated by the subtraction. Colonies that uniquely hybridized to the +8 probes were selected for further analysis. We did not select for down-regulated sequences in this study.
Up-regulation was confirmed by using the selected clones to probe Northern blots of heart RNA prepared from animals at various states of activity. cDNA fragments that proved to be up-regulated on Northern blots were used to screen a hibernating heart cDNA library to obtain full-length cDNA inserts. This directional library was created by ligating double-stranded cDNA into NotI/SalI-cleaved pSPORT1 vector (GIBCO/BRL). Positive clones selected from the cDNA library were sequenced by automated cycle sequencing using fluorescently tagged terminator base analogs (Applied Biosystems). Sequences were compared with the National Center for Biotechnology Information database using BLAST.
RNA Isolation and Northern Blot Analysis.
Total RNA was isolated from ground squirrel organs using a modification of the Chomczynski and Sacchi method (7) where tissues were homogenized in 4 M guanidinium isothiocyanate, followed by addition of sodium acetate to a final concentration of 0.2 M. Samples were extracted by water-saturated phenol and chloroform:isoamyl alcohol (49:1), followed by two extractions with phenol:chloroform:isoamyl alcohol (50:49:1). All extractions were centrifuged through Phase Lock Gel (5 Prime → 3 Prime). RNA was precipitated with isopropanol and quantified by absorption spectrophotometry.
For Northern blots of heart RNA, 15 μg of total RNA from hearts of active and hibernating 13-lined ground squirrels was separated on an agarose gel containing formaldehyde and blotted to a nylon membrane. Ten micrograms of total RNA from various ground squirrel organs was separated in each lane to show the distribution of PDK-4 and PL mRNA from a single hibernating animal sacrificed in January during hypothermic torpor (Tb = 5°C). Both sets of Northern blots were hybridized to 32P-labeled full-length ground squirrel PDK-4 (GenBank accession no. AF020845) and PL cDNAs (GenBank accession no. AF027293) isolated from heart tissue. A human β-actin probe (8) was hybridized to the same blots as a control for gel loading and RNA integrity. PDK-4 and PL mRNA levels were quantified by a phosphorimager (Molecular Dynamics) and normalized to actin mRNA levels from the same gel lane to compute the relative value above that of an active August animal on each individual blot. One-way ANOVA was used to determine whether there was a significant difference in PDK-4 and PL mean mRNA levels with respect to their August counterparts. P values were calculated using Version 6.12 of the SAS system (SAS Institute, Cary, NC).
PL Assays.
Ground squirrel hearts were processed into acetone powder (9) and solubilized in 0.2 M Tris⋅HCl (pH 9.2) and 1 M ethylene glycol. Soluble heart protein was assayed for PL activity by a modification of the method of Lowe (10) using 14C-labeled triolein (NEN). Assays were performed in the presence of the bile salt sodium deoxycholate (19.8 mM) with or without 2.8 μg/ml of the PL-specific activator protein colipase (Sigma). The reaction mixture (60 μl total) was incubated at temperatures of 0, 7, 17, 27, and 37°C for 30 min and stopped with 975 μl of methanol:chloroform:heptane (14:12.5:10 vol/vol/vol) and 315 μl of 0.1 M boric acid and 0.1 M K2CO3 (pH 10.5). After organic extraction, 100 μl of the aqueous layer (containing free fatty acids) was removed and counted in 5 ml of scintillation fluid. PL activity was calculated based on nM free fatty acids liberated/min/mg of total soluble heart protein. As a control for possible PL contamination in the colipase preparation, colipase was incubated alone and with soluble heart extract that had been heat treated at 100°C for 5 min. One-way ANOVA was used to determine whether there were significant differences in PL activity at 7 and 37°C and in percentage of PL activity at 0, 7, and 17°C, with respect to their August counterparts. P values were calculated using Version 6.12 of the SAS system (SAS Institute).
RESULTS
A differential gene expression screen (4) was used to identify genes controlling metabolic changes in the hearts of 13-lined ground squirrels during hibernation. After an initial screen of up-regulated sequences, we confirmed a substantial increase in mRNA levels for two genes that appear to regulate metabolic flux in the hibernating heart, PDK-4 and PL. PDK-4 catalyzes phosphorylation of the mitochondrial enzyme pyruvate dehydrogenase, leading to its inactivation and thereby preventing the oxidation of pyruvate to Ac-CoA and CO2 (11). Inactivation of pyruvate dehydrogenase is a key step in reducing carbohydrate catabolism since glucose cannot be oxidized to produce ATP via the tricarboxylic acid cycle and oxidative phosphorylation. PDK-4 was originally identified in humans (11) and is the newest member of the pyruvate dehydrogenase kinase family (12). Ground squirrel PDK-4 (GenBank accession no. AF020845) shares 91% amino acid identity with the human enzyme and has a predicted molecular mass of 46,833 Da.
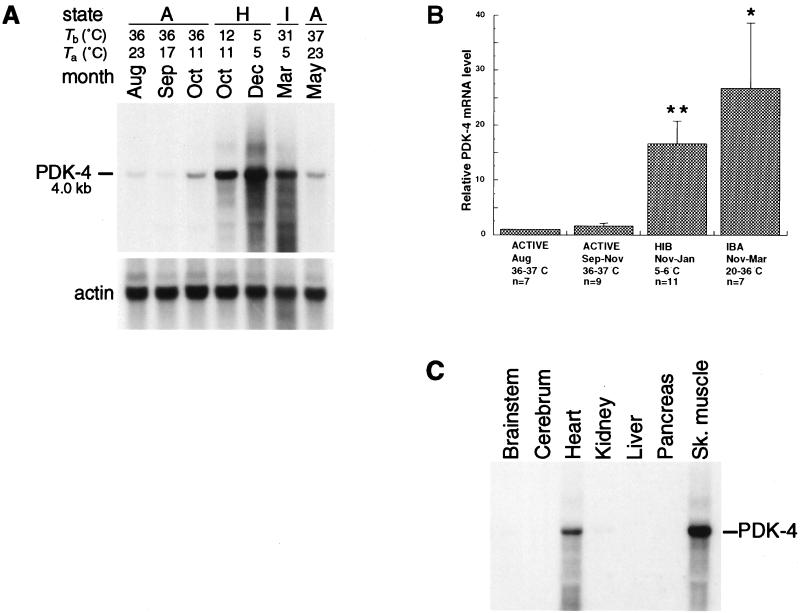
PDK-4 mRNA is present at low levels in hearts of active animals during August and September, but shows a significant increase (P ≤ 0.01) during hibernation. This pattern of expression can be seen in Fig. 1A where the amount of PDK-4 mRNA from a hibernating October animal is greater than that from a nonhibernating sibling on the same day. Levels of PDK-4 mRNA remain high during the hibernation season (Fig. 1B), including regular interbout arousals (0.05 > P > 0.01) where body temperatures can approach 37°C for periods of up to 24 hr (13). Steady-state levels of PDK-4 mRNA are eventually reduced in the spring after the animal resumes full activity. Tissue distribution of PDK-4 mRNA during hibernation (Fig. 1C) is primarily in heart and skeletal muscle; however, longer exposures show some PDK-4 mRNA in kidney and brain (data not shown).
Figure 1.
Expression of PDK-4 during the hibernation season. (A) Northern blot of total RNA from the hearts of active and hibernating 13-lined ground squirrels. The position and size of PDK-4 mRNA are indicated. Body temperature (Tb), ambient temperature (Ta), and month of sacrifice are indicated above each lane. The state of each animal is indicated as active (A), hibernating (H), or in interbout arousal (I). The actin profile is shown directly below. August through October animals and December through May animals represent two separate sibling groups from mothers captured in Michigan. (B) Bar graph summarizing PDK-4 mRNA levels in the hearts of sibling and nonsibling ground squirrels captured in Michigan and Illinois (TLS Research, Bartlett, IL). PDK-4 mRNA levels were quantified by a PhosphorImager (Molecular Dynamics). For each experiment the PDK-4 value for the August animal was normalized to 1.00. Each bar represents mean values derived from RNA blot experiments (±SEM). Body temperature, state, month(s) of sacrifice, and the number of animals (n) are indicated below each group. HIB, hibernating; IBA, interbout arousal. Statistically significant comparisons to August mRNA levels were determined by one-way ANOVA: ∗, 0.01 < P < 0.05; ∗∗, P ≤ 0.01. (C) Tissue distribution of PDK-4 mRNA from a single hibernating animal sacrificed in January during hypothermic torpor (Tb = 5°C). Tissues are as indicated above each lane and the actin profile for this blot is shown in Fig. 2C. Sk. muscle, skeletal muscle.
The increased concentration of PDK-4 mRNA in the heart closely parallels the hibernating state and appears to explain an observed 96% reduction in heart pyruvate dehydrogenase activity during hibernation (14). Inhibition of pyruvate dehydrogenase by PDK-4 offers a mechanism for the well-established findings of Lyman and colleagues (15), who showed that glycolytic intermediates were blocked from entering the tricarboxylic acid cycle during hibernation. Arrest of carbohydrate oxidation by phosphorylation of pyruvate dehydrogenase is reversed by a phosphatase which restores the conversion of pyruvate to Ac-CoA (reviewed in ref. 16). The reversibility of this inhibition resembles the reversibility of hibernation itself, because it allows for the resumption of glucose oxidation seen shortly after arousal (15).
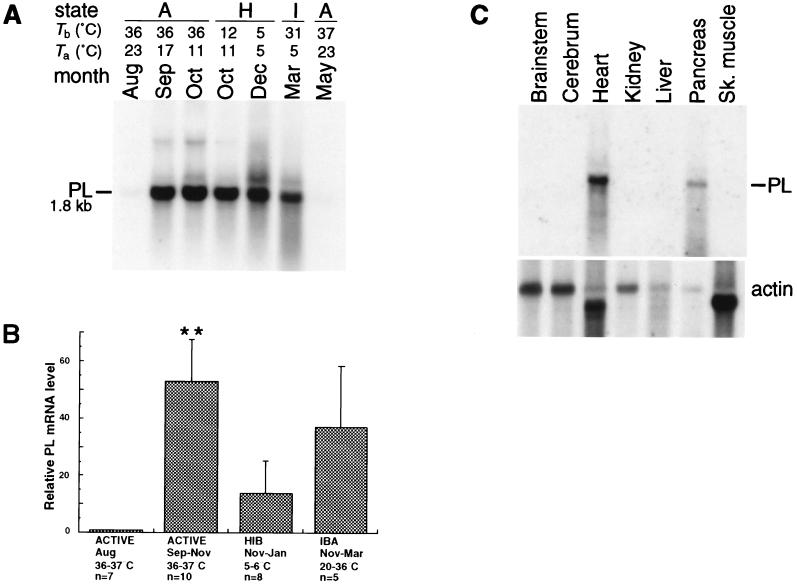
Although PDK-4 is up-regulated at the onset of hibernation, elevated levels of PL mRNA are seen in ground squirrel hearts before hibernation begins. cDNA sequencing indicates that PL from heart tissue (GenBank accession no. AF027293) has an unprocessed molecular mass of 51,243 Da and shares 85% amino acid identity with the unprocessed form of human PL (17). Expression of PL is normally confined to the pancreas, where the enzyme is secreted into the small intestine and hydrolyzes dietary fat in the form of triglycerides (18). However, in 13-lined ground squirrels, PL is strongly up-regulated in the heart as the hibernation season approaches (Fig. 2 A and B). PL expression is barely detectable in August, but a significant up-regulation (P ≤ 0.01) of the PL gene is seen in active animals as early as September. Elevated levels of heart PL mRNA continue throughout the hibernation season until they decrease after activity resumes in the spring. Tissue distribution of PL mRNA during hibernation (Fig. 2C) is restricted to the heart and, as expected, the pancreas.
Figure 2.
Expression of PL during the hibernation season. (A) The position and size of heart PL mRNA are indicated. The RNA blot and its actin profile are the same as the one shown in Fig. 1A. (B) Bar graph summarizing PL mRNA levels in the hearts of sibling and nonsibling squirrels captured in Michigan and Illinois. PL mRNA levels were quantified by a PhosphorImager. For each experiment the PL value for the August animal was normalized to 1.00. Each bar represents mean values derived from RNA blot experiments (±SEM). Body temperature, state, month(s) of sacrifice, and the number of animals (n) are indicated below each group. HIB, hibernating; IBA, interbout arousal. Statistically significant comparisons to August mRNA levels were determined by one-way ANOVA: ∗∗, P ≤ 0.01. (C) Tissue distribution of PL mRNA using same blot as shown in Fig. 1C. Tissues are as indicated above each lane and the actin profile for this blot is shown directly below. Sk. muscle, skeletal muscle.
The results shown in Fig. 2 raise the question: Why is an enzyme thought to hydrolyze dietary fat exclusively in the gut expressed in ground squirrel hearts? Catabolism of lipid is the major source of energy during hibernation (2); therefore, it is critical that the working heart has the ability to utilize this fuel at low body temperatures. Since PL from the pancreas of pigs shows catalytic activity at temperatures as low as 13°C (19), we reasoned that differential expression of PL in the heart could be an adaptation that provides low-temperature lipolysis during hibernation. To test this hypothesis, we used an in vitro assay (10) to determine PL activity in hearts of summer-active (August), fall-active (September–November), and hibernating (December–January) ground squirrels at temperatures associated with states of activity (37°C) and hibernation (7°C). Specificity for PL was attained by performing assays in the presence of the inhibitory bile salt sodium deoxycholate. Inhibition of lipolysis is overcome by addition of the PL-specific activator protein colipase which selectively stimulates PL activity in the presence of bile salts (reviewed in ref. 20). As a control for possible PL contamination in the colipase preparation, colipase was incubated with soluble heart extract that had been previously heat treated at 100°C for 5 min. Neither the heat-treated sample plus colipase, nor colipase alone, showed any PL activity when incubated under assay conditions in the presence of 14C-labeled triolein (data not shown).
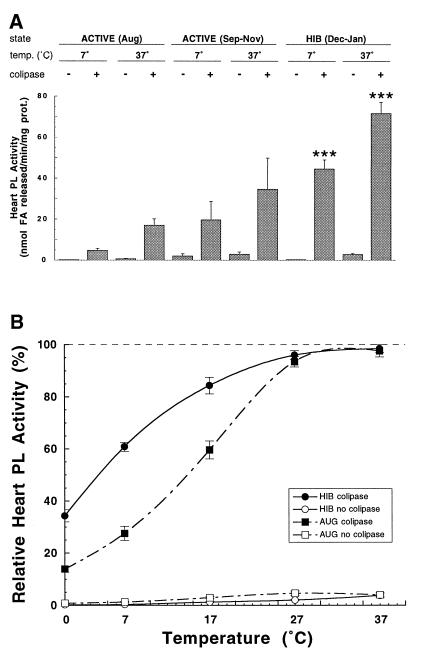
Lipase assays of soluble heart protein from hibernating ground squirrels in the months of December and January showed approximately four times more PL activity in the presence of colipase at 37°C (P = 0.0001) and nine times more activity at 7°C (P = 0.0001) when compared with August animals at the same temperatures (Fig. 3A). This increase in PL-mediated lipolysis correlates with the observed increase in PL mRNA concentration during hibernation (Fig. 2). However, a similar correlation was not seen in hearts of euthermic (Tb = 36–37°C) animals from September through November (Fig. 3A). These active animals showed an approximate 50-fold increase (P ≤ 0.01) in PL mRNA levels above their August counterparts (Fig. 2B), but only a 2-fold increase in PL activity at 37°C (P, nonsignificant) and a 4-fold increase at 7°C (P, nonsignificant).
Figure 3.
Heart PL activity determinations in vitro at various temperatures. (A) Mean heart PL activity (±SEM) at the indicated assay temperatures from five summer-active (ACTIVE, August, Tb = 36–37°C), five fall-active (ACTIVE, September–November, Tb = 36–37°C), or five hibernating (HIB, December–January; Tb = 5–7°C) ground squirrels with (+) or without (−) PL-specific activator colipase. Statistically significant comparisons to respective August PL activity were determined by one-way ANOVA: ∗∗∗, P = 0.0001. (B) Mean relative percentage of heart PL activity of five summer-active (AUG, August,; Tb = 36–37°C) or five hibernating (HIB, December–January, Tb = 5–7°C) animals at assay temperatures of 0, 7, 17, 27, and 37°C. For each individual animal the assay temperature showing the highest activity was designated as 100%. Open symbols represent mean percentage of activity in the absence of colipase and closed symbols represent mean percentage of activity with colipase. Bars flanking each point represent ±SEM. Mean relative percentage of HIB PL activity in the presence of colipase is significantly different from that of percentage of AUG PL activity as determined by one-way ANOVA at P = 0.0001 for 0 and 7°C, and P = 0.0007 for 17°C.
Our observation that PL activity in hibernators at 7°C was more than twice that of August animals at 37°C prompted us to analyze the percentage of heart PL activity at a range of temperatures from 37°C to 0°C (Fig. 3B). When PL from the heart of hibernators is assayed at a typical hibernating body temperature of 7°C, it retains 61% of maximal activity; even at 0°C the enzyme retains 34% maximal activity. At temperatures of 17°C and below, the percentage of PL activity from hibernators (in the presence of colipase) is significantly greater than that seen in active August animals (P = 0.0001 for 0 and 7°C; P = 0.0007 for 17°C). These differences are indicative of a more cold-tolerant form of PL in the hearts of hibernating squirrels. We conclude that ground squirrel PL hydrolyzes triglycerides at low body temperatures associated with the hibernating state, thus providing a steady supply of fuel for the working heart during periods of near-freezing torpor.
DISCUSSION
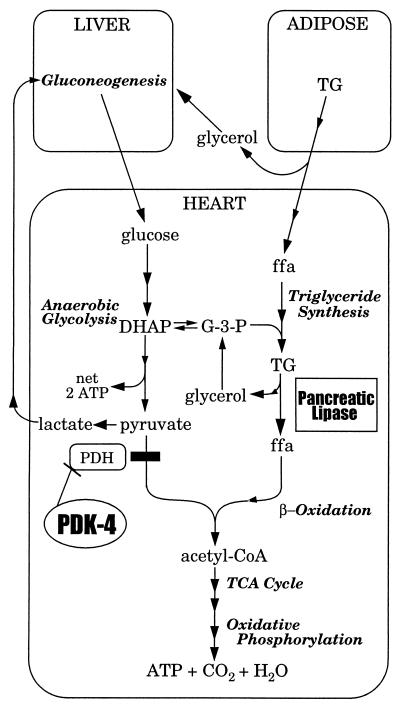
This report provides the first evidence that adaptive changes in cardiac physiology are under genetic control during hibernation. Differential expression of genes encoding PDK-4 and PL points to specific catabolic reactions that influence heart physiology during hibernation. We propose a model (Fig. 4) for the genetic control of low-temperature carbon utilization in the hibernating heart based on the results in this paper along with previous studies on the physiology of hibernation in mammals. In late summer, ground squirrels show dramatic weight gain due to an increase in the mass of their adipose tissue. Numerous studies have confirmed that this stored lipid, not carbohydrate, serves as the primary source of energy during hibernation (for review see ref. 2). We propose that the inhibition of glucose oxidation by PDK-4 impedes carbohydrate catabolism and contributes to the observed metabolic rate depression. The resulting anaerobic glycolysis (21) generates three-carbon molecules such as lactate and dihydroxyacetone phosphate that can participate in gluconeogenesis (15, 22) and cardiac triglyceride synthesis, respectively. Gluconeogenic activity is enhanced during hibernation (23, 24) and is additionally fueled by the release of glycerol from the hydrolysis of triglycerides in adipose tissue (22, 25). Fatty acids mobilized from adipose are transported to the heart where they are stored as triglycerides based on an observed increase in mitochondria-associated lipid droplets (26). Lipolysis by PL provides a steady release of fatty acids at low temperatures, thereby supplying substrate for mitochondrial β-oxidation and the generation of ATP by oxidative phosphorylation.
Figure 4.
Model showing the metabolic involvement of PDK-4 and PL in the heart of a hibernating 13-lined ground squirrel. Names of metabolic pathways are shown in italics. Arrows with a single arrowhead indicate a single reaction. Continuous arrows with two or more arrowheads indicate multistep pathways. DHAP, dihydroxyacetone phosphate; ffa, free fatty acid; G-3-P, l-glycerol 3-phosphate; PDH, pyruvate dehydrogenase; TCA cycle, tricarboxylic acid cycle; TG, triglyceride.
The majority of land-dwelling lineages of mammals contain species that hibernate (2). These include the primates, carnivores, marsupials, monotremes, bats, insectivores, and rodents. The latter three lineages are known to contain the so-called deep hibernators. A distinguishing characteristic of a deep hibernator is its ability to maintain and survive extremely low body temperatures for periods of weeks. Body temperatures as low as −2.9°C have been recorded in the Arctic ground squirrel (Spermophilus parryii), which routinely maintains a hibernating core body temperature below 0°C (27). In the absence of antifreeze proteins or abnormal solute concentrations in the plasma, the Arctic ground squirrel survives ambient temperatures as low as −18°C during hibernation and is able to rewarm itself using endogenously generated heat. A lipolytic enzyme such as PL, that retains one-third its maximal activity at 0°C, would clearly provide an advantage for this species and related hibernators.
We have observed a massive increase in the level of PL mRNA in the hearts of euthermic squirrels several weeks before the onset of hibernation (Fig. 2). However, these fall-active animals do not show significantly higher levels of PL-mediated lipolysis (Fig. 3A). It is possible that the PL mRNA produced prior to hibernation is not translated until a physiological signal, such as reduced food intake, lower body temperature, or some combination of related factors, stimulates PL protein synthesis. Alternatively, it has been shown that PL in the pancreas undergoes a pair of posttranslational modifications: removal of a 16-amino acid signal sequence at the amino terminus and N-glycosylation of a specific asparagine residue (18). Posttranslational modification of PL in the ground squirrel heart may also be required for optimization of enzymatic activity and for the generation of a more cold-tolerant form of PL seen in hibernating animals (Fig. 3B).
Involvement of PDK-4 and PL in the intermediary metabolism of the ground squirrel heart contributes to the observed respiratory quotient of 0.7 during hibernation (2). Respiratory quotient is a unit-less value representing the moles of CO2 respired per moles of O2 consumed. A value of 1.0 indicates combustion of carbohydrate; however, a respiratory quotient of 0.7 indicates that fat is the major substrate for energy production. This fuel selection of fat over carbohydrate seen during hibernation also occurs in starvation, when conservation of glucose for utilization by the brain is essential for survival and is controlled by the phosphorylation of pyruvate dehydrogenase (28, 29). Since hibernating ground squirrels can survive for 5–6 mo without feeding, mechanistic insight into the activation of genes such as PDK-4 and PL in hibernators could provide a better understanding of how nutrient–gene interactions influence an animal’s capacity to cope with the effects of starvation.
Acknowledgments
We thank V. W. Bauer and J. P. Hawley for technical assistance, R. F. Burlington for help with the capture and care of 13-lined ground squirrels, and M. E. Lowe for helpful discussions on PL. We thank C. S. Levings, E. S. Miller, and J. C. Swaffield for critical review of the manuscript. This work was supported by the U.S. Army Research Office (Grant DAAH04–96-1–0072 and Augmentation Awards for Science and Engineering Research Training DAAG55–97-1–0175).
ABBREVIATIONS
- PL
pancreatic lipase
- PDK-4
pyruvate dehydrogenase kinase isozyme 4
Footnotes
References
- 1.Adolph E F. Am J Physiol. 1951;166:75–91. doi: 10.1152/ajplegacy.1951.166.1.75. [DOI] [PubMed] [Google Scholar]
- 2.Lyman C P, Willis J S, Malan A, Wang L C H. Hibernation and Torpor in Mammals and Birds. New York: Academic; 1982. [Google Scholar]
- 3.Hochachka P W. Science. 1986;231:234–241. doi: 10.1126/science.2417316. [DOI] [PubMed] [Google Scholar]
- 4.Wang Z, Brown D D. Proc Natl Acad Sci USA. 1991;88:11505–11509. doi: 10.1073/pnas.88.24.11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srere H K, Wang L C, Martin S L. Proc Natl Acad Sci USA. 1992;89:7119–7123. doi: 10.1073/pnas.89.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pengelley E T, Fisher K C. Can J Zool. 1961;39:105–120. [Google Scholar]
- 7.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Gunning P, Ponte P, Okayama H, Engel J, Blau H, Kedes L. Mol Cell Biol. 1983;3:787–795. doi: 10.1128/mcb.3.5.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garfinkel A S, Schotz M C. J Lipid Res. 1972;13:63–68. [PubMed] [Google Scholar]
- 10.Lowe M E. J Biol Chem. 1992;267:17069–17073. [PubMed] [Google Scholar]
- 11.Rowles J, Scherer S W, Xi T, Majer M, Nickle D C, Rommens J M, Popov K M, Harris R A, Riebow N L, Xia J, Tsui L C, Bogardus C, Prochazka M. J Biol Chem. 1996;271:22376–22382. doi: 10.1074/jbc.271.37.22376. [DOI] [PubMed] [Google Scholar]
- 12.Bowker-Kinley M M, Davis W I, Wu P, Harris R A, Popov K M. Biochem J. 1998;329:191–196. doi: 10.1042/bj3290191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L C H. Can J Zool. 1979;57:149–155. [Google Scholar]
- 14.Brooks S P J, Storey K B. J Comp Physiol B. 1992;162:23–28. [Google Scholar]
- 15.Tashima L S, Adelstein S J, Lyman C P. Am J Physiol. 1970;218:303–309. doi: 10.1152/ajplegacy.1970.218.1.303. [DOI] [PubMed] [Google Scholar]
- 16.Patel M S, Roche T E. FASEB J. 1990;4:3224–3233. doi: 10.1096/fasebj.4.14.2227213. [DOI] [PubMed] [Google Scholar]
- 17.Lowe M E, Rosenblum J L, Strauss A W. J Biol Chem. 1989;264:20042–20048. [PubMed] [Google Scholar]
- 18.Lowe M E. Gastroenterology. 1994;107:1524–1536. doi: 10.1016/0016-5085(94)90559-2. [DOI] [PubMed] [Google Scholar]
- 19.Granon S, Semeriva M. Eur J Biochem. 1980;111:117–124. doi: 10.1111/j.1432-1033.1980.tb06082.x. [DOI] [PubMed] [Google Scholar]
- 20.Lowe M E. Annu Rev Nutr. 1997;17:141–158. doi: 10.1146/annurev.nutr.17.1.141. [DOI] [PubMed] [Google Scholar]
- 21.Burlington R F, Wiebers J E. Comp Biochem Physiol. 1966;17:183–189. doi: 10.1016/0010-406x(66)90019-3. [DOI] [PubMed] [Google Scholar]
- 22.Galster W, Morrison P R. Am J Physiol. 1975;228:325–330. doi: 10.1152/ajplegacy.1975.228.1.325. [DOI] [PubMed] [Google Scholar]
- 23.Burlington R F, Klain G J. Comp Biochem Physiol. 1967;22:701–708. doi: 10.1016/0010-406x(67)90763-3. [DOI] [PubMed] [Google Scholar]
- 24.Green C J, Brosnan J T, Fuller B J, Lowry M, Stubbs M, Ross B D. Comp Biochem Physiol B Biochem Mol Biol. 1984;79:167–171. doi: 10.1016/0305-0491(84)90009-9. [DOI] [PubMed] [Google Scholar]
- 25.Yeh I, Tam C F, Catuira E, Le T T, Papa V, Pena L, Vasquez M, Vu C, Wang S, Lopez G A. Comp Biochem Physiol B Biochem Mol Biol. 1995;111:651–663. doi: 10.1016/0305-0491(95)00020-9. [DOI] [PubMed] [Google Scholar]
- 26.Burlington R F, Bowers W D, Jr, Daum R C, Ashbaugh P. Cryobiology. 1972;9:224–228. doi: 10.1016/0011-2240(72)90037-5. [DOI] [PubMed] [Google Scholar]
- 27.Barnes B M. Science. 1989;244:1593–1595. doi: 10.1126/science.2740905. [DOI] [PubMed] [Google Scholar]
- 28.Randle P J. Biochem Soc Trans. 1986;14:799–806. doi: 10.1042/bst0140799. [DOI] [PubMed] [Google Scholar]
- 29.Wu P, Sato J, Zhao Y, Jaskiewicz J, Popov K M, Harris R A. Biochem J. 1998;329:197–201. doi: 10.1042/bj3290197. [DOI] [PMC free article] [PubMed] [Google Scholar]