Abstract
Lipid peroxidation plays an important role in cell membrane damage. We investigated the effect of lipid peroxidation on the properties of 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphatidylcholine (PLPC) lipid bilayers using molecular dynamics simulations. We focused on four main oxidation products of linoleic acid with either a hydroperoxide or an aldehyde group: 9-trans, cis-hydroperoxide linoleic acid, 13-trans, cis-hydroperoxide linoleic acid, 9-oxo-nonanoic acid, and 12-oxo-9-dodecenoic acid. These oxidized chains replaced the sn-2 linoleate chain. The properties of PLPC lipid bilayers were characterized as a function of the concentration of oxidized lipids, with concentrations from 2.8% to 50% for each oxidation product. The introduction of oxidized functional groups in the lipid tail leads to an important conformational change in the lipids: the oxidized tails bend toward the water phase and the oxygen atoms form hydrogen bonds with water and the polar lipid headgroup. This conformational change leads to an increase in the average area per lipid and, correspondingly, to a decrease of the bilayer thickness and the deuterium order parameters for the lipid tails, especially evident at high concentrations of oxidized lipid. Water defects are observed in the bilayers more frequently as the concentration of the oxidized lipids is increased. The changes in the structural properties of the bilayer and the water permeability are associated with the tendency of the oxidized lipid tails to bend toward the water interface. Our results suggest that one mechanism of cell membrane damage is the increase in membrane permeability due to the presence of oxidized lipids.
INTRODUCTION
Lipid peroxidation alters the physiological functions of cell membranes and plays an important role in cellular membrane damage. Peroxidation is believed to be involved in cellular aging and in various diseases, such as Parkinson's and Alzheimer's disease (1–9) as well as schizophrenia (10), atherosclerosis (11,12), inflammatory diseases (13), and cardiac ischemia reperfusion injury (14,15). Unsaturated lipids are easily susceptible to peroxidation (16). The effect of both unsaturation and peroxidation on the properties of lipid bilayers has been well characterized experimentally (17–22). Still, the exact mechanism of membrane damage by oxidized lipids is unclear. Oxidized lipid tails are more polar and can be shorter in length, due to the presence of aldehyde or hydroperoxide groups (23,24). Lipid peroxidation has been shown to perturb the bilayer structure and modify membrane properties such as membrane fluidity, permeability to different substances, and bilayer thickness. The existence of a direct relationship between lipid peroxidation and membrane leakiness has been suggested (25–28). Increased membrane permeability caused by oxidation of lipids and membrane proteins can disrupt ion gradients, therefore altering metabolic processes. Lipid peroxidation can influence the permeability of lipid membranes by increasing the dielectric constant of the membrane interior and by increasing the microviscosity, possibly through cross-linking of lipid radicals (23). Focusing on structural and dynamic properties, a decrease in membrane thickness upon oxidation has been observed using x-ray diffraction analysis, along with interdigitation of the terminal methyl segments (22). The effect of peroxidation on lipid dynamics and membrane order is less clear. According to some researchers, peroxidation does not affect the fluidity of the membrane (29) nor the reorientational dynamics of the lipids (18). According to others, membrane fluidity is decreased (30–33) and the decrease is higher near the double bonds of the bilayer, whereas other regions are less affected (30). Some have reported an increase in the lipid tails order parameter (30,33–36), others no change (37) or a decrease (18,38). Several reasons can explain the differences in experimental results, including the use of different methodologies to generate peroxides, leading to different (and usually not well-defined) lipid compositions of the membrane (22). Despite the numerous studies on the effect of oxidation on the structure and dynamics of lipid membranes, the relationship between increased membrane permeability, and modifications in the structure and dynamics of lipid bilayers is not clear.
In recent years, computational studies of model membranes proved to be particularly useful in the description of the structure and dynamics of lipid bilayers (39,40) and in the interpretation of experimental results (41). Pure lipid bilayers, including either a single type of lipid (42–44) and mixtures of two or more lipids (43,45–50), have been investigated using computational methods, as well as mixtures of lipids with proteins (51). Unsaturated lipid bilayers also have been studied using computer simulations (52–56), but the structural consequences of the presence of oxidized lipids have never been investigated using computational methods, to the best of our knowledge.
In this work, we use molecular dynamics simulations to characterize the effect of lipid oxidation on the properties of 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphatidylcholine (PLPC) lipid bilayers. In particular, we describe the effect of four different products of PLPC peroxidation (57) at five concentrations, ranging from 2.8% to 50%. We focused on four main oxidation products of linoleic acid, including either a hydroperoxide or an aldehyde group: 9-trans, cis-hydroperoxide linoleic acid (9-tc), 13-trans, cis-hydroperoxide linoleic acid (13-tc), 9-oxo-nonanoic acid (9-al), and 12-oxo-9-dodecenoic acid (12-al) (Fig. 1). These oxidized chains replaced the sn-2 linoleate chain in PLPC. The goal of this work is to understand how oxidized lipids change the membrane properties. In particular, we seek to characterize the relationship between the changes in membrane permeability and the modifications of structural and dynamic properties of the lipid bilayer, and to provide a detailed description at the atomistic level of the chemical interactions responsible for the changes in the properties of the membrane. The article is organized as follows. First, we describe the methods used to derive the simulation parameters for the lipids and the simulation methods. Then we describe a number of properties of a PLPC bilayer and how they change upon increasing the concentration of oxidized lipids, and compare simulation results to experimental ones reported in the literature. Finally, we discuss the relationship between permeability, structure, and dynamics of oxidized bilayers.
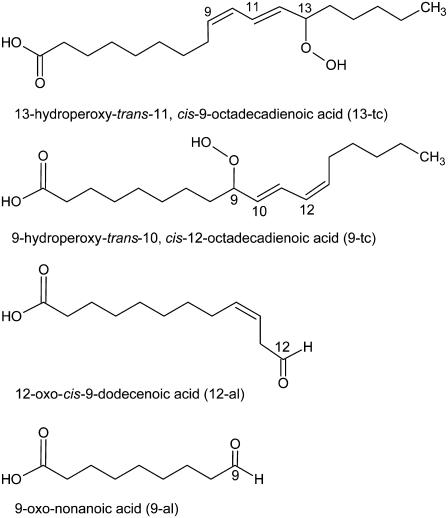
FIGURE 1.
Products of the oxidation of linoleic acid considered for this work.
METHODS
Force-field parameters
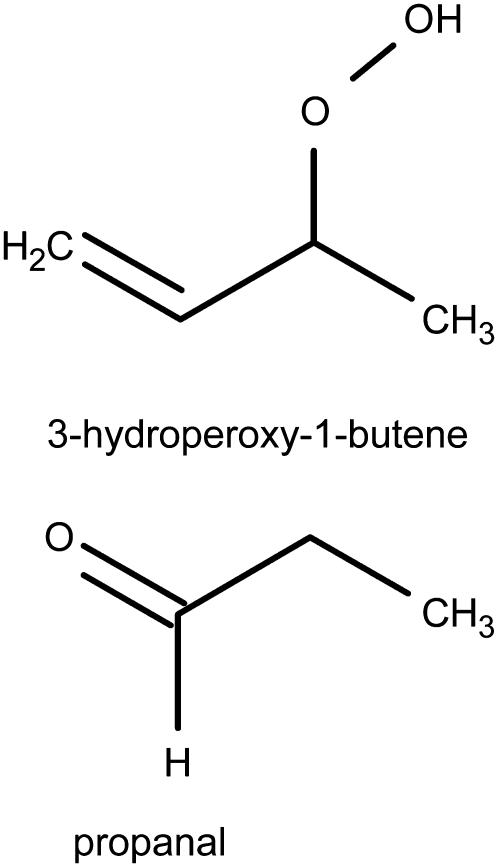
A united-atom force field was used for the lipids in all simulations. The parameters for the phosphatidylcholine (PC) headgroup and the lipid tails were taken from previous works on PLPC and DPPC lipids (42,54). The hydroperoxide lipid tails were created by addition of a hydroperoxide group at position C9 or C13 of the linoleate tail and shifting the double bonds, as shown in Fig. 1, a and b. The aldehyde lipid tails were also built starting from linoleic acid (Fig. 1, c and d). The bonded parameters for the O-O and O-H bonds and for the O-O-H angle were taken from previous calculations on hydrogen peroxide (58). The dihedral angle parameters and the partial charges for the peroxide and aldehyde groups were derived using quantum chemistry calculations on 3-hydroperoxy-1-butene and propanal (Fig. 2, a and b), while Lennard-Jones parameters were taken from hydroxide and carbonyl groups already present in the force field. We used the Jaguar software package (59) for all quantum calculations, with the B3LYP method of density functional theory (60,61) and the LACV3P**++ basis set. Partial atomic charges were estimated using natural population analysis (62) and the electrostatic surface potential fitting method with Merz-Kollman atomic radii (63) after the geometry optimization. The results for partial charges are reported in Table 1. For the calculation of bond and angle force constants, we restrained the bond lengths and angles at seven different values, then fitted a harmonic potential functions to the energy profile. For dihedral parameters, dihedral angles were restrained at 36 different values from 0 to 360°, and the standard proper dihedral function 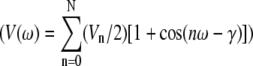 was fitted to the potential energy. For all bonded parameters, the Lennard-Jones and electrostatic energy were calculated for different geometries and subtracted from the total energy before fitting. Results for all bonded parameters are reported in Tables 2 and 3.
was fitted to the potential energy. For all bonded parameters, the Lennard-Jones and electrostatic energy were calculated for different geometries and subtracted from the total energy before fitting. Results for all bonded parameters are reported in Tables 2 and 3.
FIGURE 2.
Lipid fragments used for the calculation of bonded parameters and partial charges for the oxidized lipid tails.
TABLE 1.
Partial atomic charges in the oxidized lipid tails
| Functional group | CH | O | O | H | Reference molecule |
|---|---|---|---|---|---|
| Hydroperoxide tails | 0.30 | −0.30 | −0.45 | 0.45 | 3-hydroperoxy-1-butene |
| Aldehyde tails | 0.53 | −0.53 | — | — | propanal |
TABLE 2.
Force constant FC for optimized bonds and angles in the peroxide group
| Bond | r0 (nm) | FC [kJ/(mol × nm2)] | Reference molecule |
|---|---|---|---|
| C-O | 0.14180 | 225670 | 3-hydroperoxy-1-butene |
| O-O | 0.14430 | 269580 | H2O2 (58) |
| O-H | 0.09810 | 444130 | H2O2 (58) |
| Angle | α0 (degree) | FC [kJ/(mol × rad2)] | Reference molecule |
| =C-C-O | 104.00 | 418.40 | 3-hydroperoxy-1-butene |
| C-C-O | 109.50 | 418.40 | 3-hydroperoxy-1-butene |
| C-O-O | 105.90 | 598.37 | 3-hydroperoxy-1-butene |
| O-O-H | 100.00 | 506.92 | H2O2 (58) |
TABLE 3.
Force constants for dihedral angles in the peroxide and aldehyde groups
| Dihedral angles | a* | b† | c* | d† | e* | f† | Reference molecule |
|---|---|---|---|---|---|---|---|
| C=C-C-O | 2.12 | 223.90 | 0 | 0 | 3.62 | 180.50 | 3-hydroperoxy-1-butene |
| C-C-O-O | 2.13 | 334.25 | 0 | 0 | 7.04 | 8.10 | 3-hydroperoxy-1-butene |
| C-O-O-H | 8.46 | 23.30 | 6.51 | 18.40 | 0 | 0 | 3-hydroperoxy-1-butene |
| C-C-C=O | 0.47 | 180.00 | 1.58 | 180.00 | 2.67 | 180.00 | propanal |
The functional form is the following: 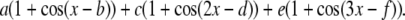
kJ/(mol × rad2).
Degrees.
Bilayer setup and simulation details
The systems were generated starting from the equilibrium structure of a PLPC bilayer containing 72 lipids. We replaced 2, 4, 8, 18, and 36 PLPC lipid molecules with each oxidized lipid, obtaining 20 different bilayers with oxidized lipid concentrations of 2.8%, 5.6%, 11.1%, 25%, and 50%, respectively. The two bilayer leaflets always contained the same number of oxidized lipids. All 21 simulation systems contained 72 lipid (including PLPC and oxidized lipids) and 2880 water molecules. All simulations were carried out with version 3.3.1 of the GROMACS package (64). After energy minimization, molecular dynamics simulations were run for 180 ns, and the initial 80 ns were considered as an equilibrium period. The integration time step was 2 fs. Periodic boundary conditions were applied in all dimensions. A 1.0 nm cutoff was employed for the electrostatic and Lennard-Jones interactions and the neighbor list was updated at every time step. The long-range electrostatics was calculated using particle-mesh Ewald (65,66); the real-space interactions were evaluated using a 1.0 nm cutoff, and the reciprocal-space interactions were evaluated on a 0.12 nm grid with fourth-order B-spline interpolation. The relative error for the Ewald sum in the direct and reciprocal space, controlled in GROMACS by the parameter ewald_rtol, was set to 10−5. The LINCS algorithm was used to constrain all bond lengths (67). The weak temperature coupling scheme was applied separately to the lipids and water (68), with a temperature of 298 K and a time constant of 0.1 ps. The semiisotropic pressure was applied (68), with an equilibrium pressure of 1 bar both in the x-y plane and in the z direction (bilayer normal) with a time constant of 4.0 ps and a compressibility of 4.5 × 10−5 bar−1. Molecular graphics were made using VMD (69).
Potential of mean force and permeability of water
Constraint simulations were used to calculate the potential of mean force (PMF) of water as a function of the distance from the center of the bilayer, the local diffusion coefficient at different depths, and water permeability through the lipid bilayer (70,71). A series of 31 simulations was run with the distance between water and the center of bilayer constrained between 0 and 3.0 nm, with 0.1 nm spacing. Only the component of the distance along the bilayer normal (z axis) was constrained, while water was completely free to move in the x and y directions. The SHAKE algorithm was used, with a relative tolerance of 10−5. Two water molecules were constrained at the chosen depths inside the bilayer, at a distance of 3.0 nm (along the z axis) from each other. In the first simulation, one water molecule was restrained at 0 nm (corresponding to the center of the bilayer) and the second at 3 nm (corresponding to the bulk water phase). This setup allows for increased sampling at no computational cost. Each simulation was 15 ns long and the forces were calculated as a function of the simulation time. The free energy of water transfer from the bulk phase to various depths in the membrane can be expressed as
 |
(1) |
where 〈F(z′)〉t indicates the average force over the simulation time; the limits of the integration range from bulk water to depth z. To estimate the error on the force, we used a block averaging procedure on five intervals, each 3 ns long. We used the force autocorrelation method for the calculation of the local time-dependent friction coefficient, ξ(t), as described by Marrink (70,71). The static friction coefficient, ξ, is inversely proportional to the local diffusion coefficient (D(z′)) (71–73) and the permeability coefficient (P) of the solute is defined as the inverse of the resistance (R). The resistance R to permeate through the bilayer is obtained by integrating over the local resistances 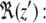
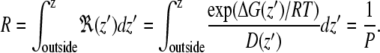 |
(2) |
RESULTS AND DISCUSSION
Structural and dynamic properties of the lipid bilayer
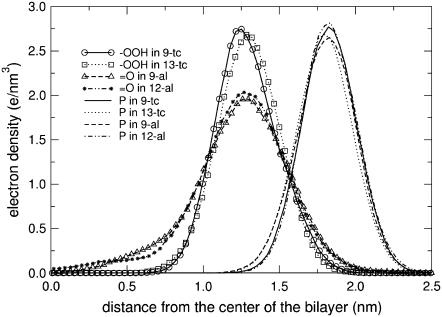
The first important change in the simulations of all the oxidized lipids is in the conformation of the lipid tails. Snapshots showing the conformation of oxidized and nonoxidized lipids are shown in Fig. 3. The portion of the lipid tail containing oxygen atoms is found, on average, close to the interface region. This is confirmed by the distribution of aldehyde and peroxide oxygen atoms in the bilayer, shown in Fig. 4. For both the aldehyde-containing and the peroxide-containing lipids, the maximum density of oxygen atoms is around the carbonyl group, and the distribution is broader for aldehyde-containing lipids. Together with the conformational change, hydrogen bonding is observed between the oxidized lipid tail and water, carbonyl groups, and phosphate groups. Table 4 shows the average number of hydrogen bonds formed by the hydroperoxide and aldehyde groups with other groups, in each simulation. In all cases, oxidized lipid tails form hydrogen bonds mostly with water molecules. For hydroperoxide-containing lipids, hydrogen bonds with phosphate groups are more probable than with carbonyl groups. The average total number of hydrogen bonds per lipid does not change significantly with the concentration of oxidized lipids. Its average value is 1.00 ± 0.13 for hydroperoxide lipids and 0.48 ± 0.05 for aldehyde lipids. This highlights a correlation between oxygen density distribution and hydrogen bonding: hydroperoxide lipids have higher propensity to form hydrogen bonds with water and narrower density distributions. These findings corroborate the model initially proposed by van Kuijk et al. (74), suggesting that the hydroperoxide moieties reside in the proximity of the lipid headgroup region, because of their hydrophilic character.
FIGURE 3.
Snapshots of a single PLPC and 13-tc taken at 5 ns intervals. Lipids are colored by atom type: nitrogen is blue, carbon is cyan, oxygen is red, and phosphorus is tan. Molecules are oriented along the z axis and superimposition was done on the phosphorus and oxygen atoms.
FIGURE 4.
Electron density profiles for the oxygen atoms of the oxidized acyl chains and the phosphorus atoms of the headgroup in PLPC bilayers containing 11.1% of oxidized lipids.
TABLE 4.
Average of the number of hydrogen bonds per oxidized lipid molecule
| Lipid bilayer | Concentration of oxidized lipids (%) | Carbonyl group | Phosphate group | Water |
|---|---|---|---|---|
| PLPC with 9-tc | 2.8 | 0.35 | 0.41 | 1.06 |
| 5.6 | 0.50 | 0.20 | 0.93 | |
| 11.1 | 0.21 | 0.50 | 1.15 | |
| 25.0 | 0.22 | 0.53 | 1.04 | |
| 50.0 | 0.26 | 0.51 | 0.96 | |
| PLPC with 13-tc | 2.8 | 0.34 | 0.41 | 0.77 |
| 5.6 | 0.36 | 0.44 | 0.85 | |
| 11.1 | 0.14 | 0.62 | 1.04 | |
| 25.0 | 0.30 | 0.45 | 0.94 | |
| 50.0 | 0.20 | 0.38 | 0.89 | |
| PLPC with 9-al | 2.8 | — | — | 0.45 |
| 5.6 | — | — | 0.45 | |
| 11.1 | — | — | 0.52 | |
| 25.0 | — | — | 0.54 | |
| 50.0 | — | — | 0.46 | |
| PLPC with 12-al | 2.8 | — | — | 0.54 |
| 5.6 | — | — | 0.45 | |
| 11.1 | — | — | 0.54 | |
| 25.0 | — | — | 0.45 | |
| 50.0 | — | — | 0.41 |
Hydrogen bonds are between hydroperoxide or aldehyde groups and the lipid headgroup or water.
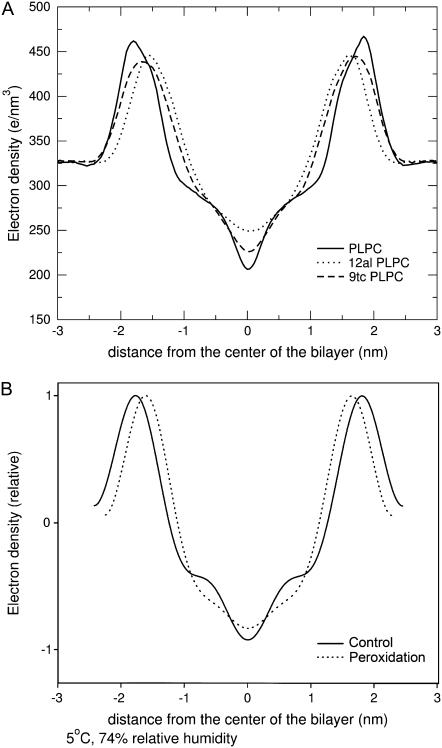
The presence of hydrogen-bonding interactions involving the lipid tails affects most of the properties of the lipid bilayer. Fig. 5 A shows the electron density profile calculated from our simulations of pure PLPC and for bilayers containing 50% concentration of each oxidized lipid. The total density at the center of the bilayer is increased in the presence of oxidation, and the maxima are shifted toward the center. The increase of the density at the center of the bilayer corresponds to partial interdigitation of the phospholipids acyl-chain terminal methyl segment when the thickness of bilayer decreases. While experimental data on PLPC are not available, the change in the electron density profile upon peroxidation has been characterized experimentally for dilinoleoyl phosphatidylcholine (DLPC) bilayers, as shown in Fig. 5 B (reproduced from (22)). Simulation results on PLPC compare favorably with the experimental ones on DLPC, showing a decrease in the bilayer thickness and a higher density in the center.
FIGURE 5.
(A) Total electron density in simulations of PLPC bilayers including two oxidation products, 12-al and 9-tc, at 50% concentration. (B) Relative electron density in x-ray diffraction experiments on DLPC and peroxidized products (reproduced from (22)).
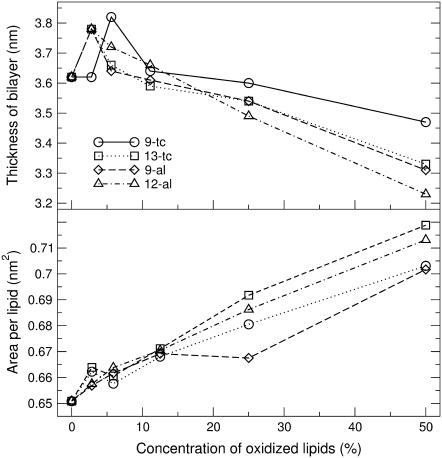
We calculated the average area per lipid and bilayer thickness in the 21 simulated systems (Table 5), and compared the results to previous simulations and experimental data. The thickness of the bilayer was calculated from the simulations as the average distance between phosphate groups in the two leaflets, computed from the total electron density profile. Errors were estimated using a block averaging procedure described by Hess (75). Fig. 6 shows the area per lipid molecule and the bilayer thickness at different concentrations of each oxidized lipid. For the pure PLPC bilayer, we found an average area per lipid of 0.651 ± 0.015 nm2 and an average thickness of 3.62 ± 0.01 nm. The difference with previous calculations (54,76) and experimental findings (53,77) is within 6% for the area and 3% for the thickness. For all the bilayers containing oxidized lipids, the area increases with increasing concentrations of the oxidized lipids, and the thickness decreases. Visual inspection of the trajectories suggests that the increase in area per lipid and the corresponding decrease in the thickness are related to the preference of the more polar oxidized tails for the interface and the headgroup region. The relationship between area and thickness is not straightforward, since both the length of the oxidized lipid tail and the position of the oxygen in tail have a specific effect on the structural properties of the bilayer. The thickness is generally less when the bilayer contains aldehyde lipids, for which one of the acyl tails is shorter. On the other hand, lipids with the peroxide or aldehyde groups farther away from the carbonyl ester tend to give larger areas. Bilayers containing 13-tc generally have the largest area per lipid, but not the smallest thickness.
TABLE 5.
Average area per lipid and bilayer thickness in all the simulations
| Concentration of oxidized lipids (%) | Avg. area (nm2) | e.e.* (nm2) | Avg. thickness (nm) | e.e.* (nm) | |
|---|---|---|---|---|---|
| PLPC | 0 | 0.651 | 0.003 | 3.620 | 0.003 |
| 2.8 | 0.662 | 0.017 | 3.620 | 0.001 | |
| 5.6 | 0.658 | 0.003 | 3.820 | 0.001 | |
| 9-tc | 11.1 | 0.668 | 0.002 | 3.640 | 0.001 |
| 25 | 0.681 | 0.005 | 3.600 | 0.000 | |
| 50 | 0.703 | 0.002 | 3.470 | 0.000 | |
| 2.8 | 0.664 | 0.006 | 3.780 | 0.001 | |
| 5.6 | 0.661 | 0.003 | 3.660 | 0.001 | |
| 13-tc | 11.1 | 0.671 | 0.006 | 3.590 | 0.001 |
| 25 | 0.692 | 0.004 | 3.540 | 0.000 | |
| 50 | 0.719 | 0.002 | 3.330 | 0.001 | |
| 2.8 | 0.657 | 0.007 | 3.780 | 0.001 | |
| 5.6 | 0.662 | 0.002 | 3.640 | 0.001 | |
| 9-al | 11.1 | 0.669 | 0.002 | 3.610 | 0.000 |
| 25 | 0.668 | 0.003 | 3.540 | 0.000 | |
| 50 | 0.702 | 0.003 | 3.310 | 0.001 | |
| 2.8 | 0.658 | 0.002 | 3.780 | 0.001 | |
| 5.6 | 0.664 | 0.005 | 3.720 | 0.000 | |
| 12-al | 11.1 | 0.670 | 0.002 | 3.660 | 0.000 |
| 25 | 0.686 | 0.003 | 3.490 | 0.000 | |
| 50 | 0.713 | 0.004 | 3.230 | 0.001 |
Error estimate, evaluated using a block averaging procedure described by Hess (75).
FIGURE 6.
The thickness and area per lipid of lipid bilayer, containing various concentrations of each oxidized lipid. Statistical error estimates are less than the size of the symbols for all the simulations (see Table 5), and are therefore omitted for the sake of clarity.
The increase in the area per lipid observed in our simulations is consistent with experimental results by Pradhan et al. (78) showing that peroxidized lipids increase the phospholipids spacing in erythrocyte membranes. Sabatini et al. (79) characterized DPPC monolayers containing oxidized lipids, in particular 9-al (referred to as PoxnoPC, in their study) and the carboxylic acid analog. They found that both oxidized lipids expanded the monolayers, similarly to our results. Interestingly, film expansion was larger with the carboxylic terminal group, the more polar group. They also proposed a model for the arrangement of the sn-2 acyl chains in monolayers that is similar to the model of van Kuijk (74), and consistent with our results.
The degree of ordering of the tail is also influenced by the presence of the oxygen atoms, as shown by the deuterium order parameter. The deuterium order parameter can be measured by NMR, and is defined as
 |
(3) |
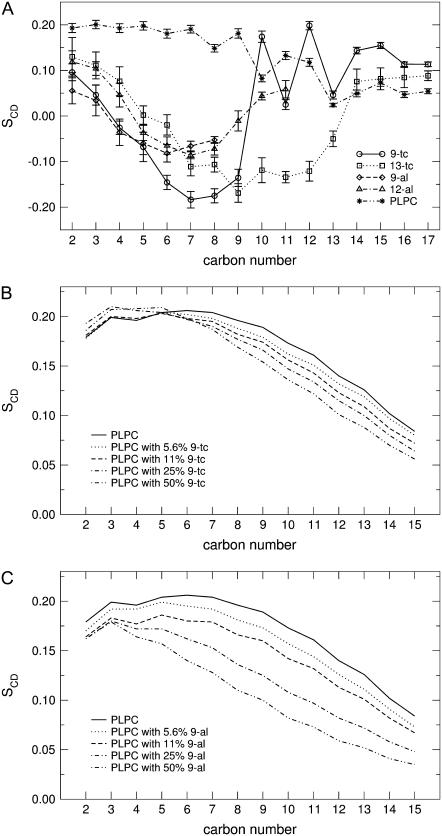
where θj is the angle between a C-D bond and the membrane normal. The brackets indicate averaging over the two bonds in a certain CD2 group, over all the lipids and over time. Since we used a united-atoms representation in our simulations, the positions of the deuterium atoms were reconstructed assuming ideal tetrahedral geometry of the methylene groups and trigonal planar geometry of the C=C double bonds. The order parameter was calculated for all the CD bonds in the sn-1 and sn-2 chains of both PLPC and the oxidized lipids in all the simulations. Fig. 7 A shows the deuterium order parameter calculated on the sn-2 chain in the simulation of pure PLPC and in the presence of 11% oxidized lipids. Standard errors were calculated using a block averaging procedure. Table 6 reports the average order parameter for the sn-1 and the sn-2 chains of PLPC and each oxidized lipids in all the simulations. As expected, both in PLPC and in the oxidized lipids the unsaturated tail has lower order parameters compared to the saturated one. In general, the order parameter of the acyl chains for all the lipids decreases with increasing concentration of oxidized lipids. Fig. 7, B and C, show the deuterium order parameter of the sn-1 chain of PLPC in the presence of 9-tc and 9-al at different concentrations. The disordering effect induced by the presence of the oxidized lipid tail is stronger for aldehyde lipids than for hydroperoxide ones (see also Table 6). This effect can be related to the lower hydrogen-bonding propensity of the aldehyde tails, which makes them more mobile, and to the larger free volume available to the acyl chains when the sn-2 chain is shorter.
FIGURE 7.
(A) Deuterium order parameter in the sn-2 lipid chains of PLPC and each oxidized lipid, in the simulations with 11% oxidized lipids. (B) Deuterium order parameter of the sn-1 chain of PLPC in the presence of 9-tc and (C) 9-al at different concentrations of oxidized lipid. Error bars were omitted in panels B and C, for the sake of clarity.
TABLE 6.
Average deuterium order parameter of the sn-1 and sn-2 chain of PLPC and the oxidized lipids in all the simulations
| Concentration of oxidized lipids (%) | Avg SCDsn-1 PLPC | Avg SCDsn-2 PLPC | Avg SCDsn-1 ox | Avg SCDsn-2 ox | |
|---|---|---|---|---|---|
| Pure PLPC | 0.168 | 0.129 | |||
| 2.8 | 0.161 | 0.126 | 0.176 | 0.023 | |
| 5.6 | 0.163 | 0.129 | 0.164 | 0.020 | |
| 9-tc | 11.1 | 0.159 | 0.126 | 0.161 | 0.023 |
| 25.0 | 0.156 | 0.119 | 0.152 | 0.030 | |
| 50.0 | 0.149 | 0.111 | 0.151 | 0.035 | |
| 2.8 | 0.160 | 0.123 | 0.154 | −0.016 | |
| 5.6 | 0.162 | 0.126 | 0.164 | 0.017 | |
| 13-tc | 11.1 | 0.161 | 0.117 | 0.168 | −0.011 |
| 25.0 | 0.157 | 0.117 | 0.161 | 0.006 | |
| 50.0 | 0.145 | 0.107 | 0.116 | 0.032 | |
| 2.8 | 0.152 | 0.123 | 0.156 | −0.033 | |
| 5.6 | 0.157 | 0.118 | 0.169 | −0.034 | |
| 9-al | 11.1 | 0.145 | 0.114 | 0.149 | −0.030 |
| 25.0 | 0.124 | 0.101 | 0.139 | −0.027 | |
| 50.0 | 0.106 | 0.082 | 0.107 | −0.013 | |
| 2.8 | 0.166 | 0.120 | 0.173 | 0.015 | |
| 5.6 | 0.161 | 0.120 | 0.139 | 0.015 | |
| 12-al | 11.1 | 0.146 | 0.119 | 0.150 | 0.010 |
| 25.0 | 0.135 | 0.109 | 0.136 | 0.026 | |
| 50.0 | 0.132 | 0.091 | 0.124 | 0.037 |
Wratten et al. (18) measured the membrane ordering in PLPC and DLPC bilayers containing hydroperoxide and hydroxide groups using angle-resolved fluorescence depolarization. Their results showed that the presence of oxidized lipid molecules cause a decrease in membrane order. However, other studies showed an increase (30,34–36) or no change (37) in the order parameter. It has been suggested that this discrepancy depends on the presence of numerous oxidative products, different from our case.
Together with the changes in the structural properties of the bilayer, dynamic properties of the lipids were also modified. We calculated the lateral diffusion coefficient from the mean-square displacements (MSD) of the lipids as a function of time. We observed that the two monolayers move relative each other and relative to water during the simulations. Both types of motion are artifacts due to the finite size of the simulated systems and to periodic boundary conditions (80). We therefore subtracted the center of mass motion of each monolayer before calculating the MSD. We then utilized the model proposed by Wohlert and Edholm for the calculation of the lateral diffusion coefficient at short and long timescales (80). This model considers two different types of diffusion occurring on different timescales. The diffusion at short times (described by the D1 coefficient) takes place within a circular area of radius R and is not Brownian, while the diffusion at long times (described by the D2 coefficient) involves large displacements and is Brownian. Both diffusion coefficients can be calculated by fitting the MSD curve to the following expression (80):
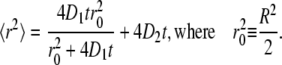 |
(4) |
To estimate the error of diffusion coefficients, we split our 100-ns trajectories in two intervals of 50-ns each and fitted the four MSD curves (two curves for each leaflet) between 0 and 10 ns. For the pure PLPC bilayer we found values of (10.4 ± 0.4) × 10−7 cm2/s for D1, 0.27 nm2 for  and (0.52 ± 0.03) × 10−7 cm2/s for D2. Previous simulations of a PLPC bilayer gave a diffusion coefficient of (1.3 ± 0.3) × 10−7 cm2/s for the diffusion at long times (56). This value is significantly higher than our result, but it was obtained at a higher temperature (310 K). Our results compare favorably to previous simulation results for a DMPC bilayer, with D1 = 13 × 10−7 cm2/s, D2 = 0.79 × 10−7 cm2/s, and
and (0.52 ± 0.03) × 10−7 cm2/s for D2. Previous simulations of a PLPC bilayer gave a diffusion coefficient of (1.3 ± 0.3) × 10−7 cm2/s for the diffusion at long times (56). This value is significantly higher than our result, but it was obtained at a higher temperature (310 K). Our results compare favorably to previous simulation results for a DMPC bilayer, with D1 = 13 × 10−7 cm2/s, D2 = 0.79 × 10−7 cm2/s, and 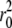 = 0.3 nm2 (80). The agreement with experimental diffusion coefficients is also very reasonable: the diffusion coefficient at short times can be compared with results from neutron scattering experiments, measuring D = (1 ∼ 10) × 10−7 cm2/s (81,82), while D2 can be compared with diffusion coefficients from fluorescence recovery after photobleaching experiments, which typically give values around (0.5 ∼ 1) × 10−7 cm2/s (83).
= 0.3 nm2 (80). The agreement with experimental diffusion coefficients is also very reasonable: the diffusion coefficient at short times can be compared with results from neutron scattering experiments, measuring D = (1 ∼ 10) × 10−7 cm2/s (81,82), while D2 can be compared with diffusion coefficients from fluorescence recovery after photobleaching experiments, which typically give values around (0.5 ∼ 1) × 10−7 cm2/s (83).
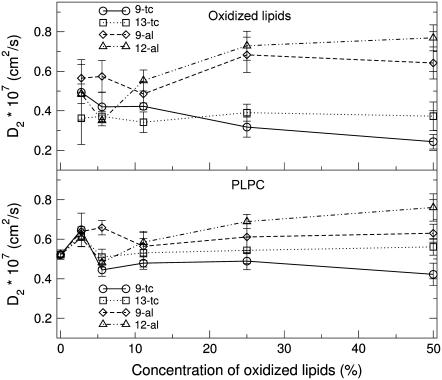
Short-time and long-time diffusion coefficients for all the simulated systems are reported in Table 7. Both the short time and the long time diffusion coefficients do not change significantly with increasing concentration of oxidized lipids. Early experimental studies suggested an increase of membrane microviscosity upon peroxidation (30–32,84), while more recent results indicated that the effect of lipid oxidation causes pronounced structural effects but minimal effects on the membrane dynamics (18,29). Our study suggests that, while the presence of oxidized lipids has a large influence on structural properties, its effect on lipid lateral diffusion is relatively small. Long time diffusion coefficients of PLPC and oxidized lipids as a function of the concentration of oxidized lipids are reported in Fig. 8. Diffusion appears to be faster for aldehyde lipids compared to hydroperoxide lipids. This is consistent with the lower degree of ordering observed in the presence of aldehyde lipids (see Fig. 7, B and C), and can be related to the stronger hydrogen-bonding interactions observed for hydroperoxide lipids (involving not only water but also polar headgroups of neighboring lipids) and the larger free volume available for the acyl chains in the presence of aldehyde lipids.
TABLE 7.
Short time diffusion coefficients, long time diffusion coefficients and r0 for PLPC and for the oxidized lipids as a function of the concentration of oxidized lipids; r0 is defined as in Eq. 4
| D1 × 107 (cm2/s) PLPC
|
D1 × 107 (cm2/s) oxidized lipids
|
D2 × 107 (cm2/s) PLPC
|
D2 × 107 (cm2/s) oxidized lipids
|
r0 (nm) PLPC
|
r0 (nm) oxidized lipids
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % ox | Avg | e.e.* | Avg | e.e.* | Avg | e.e.* | Avg | e.e.* | Avg | e.e.* | Avg | e.e.* | |
| PLPC | 0.0 | 10.4 | 0.4 | 0.52 | 0.03 | 0.52 | 0.01 | ||||||
| 2.8 | 10.9 | 0.7 | 6.0 | 1.2 | 0.65 | 0.10 | 0.49 | 0.16 | 0.51 | 0.02 | 0.51 | 0.02 | |
| 5.6 | 10.3 | 0.3 | 10.3 | 1.7 | 0.44 | 0.04 | 0.42 | 0.09 | 0.51 | 0.01 | 0.42 | 0.01 | |
| 9-tc | 11.1 | 10.2 | 0.2 | 7.9 | 0.5 | 0.48 | 0.04 | 0.42 | 0.02 | 0.54 | 0.00 | 0.44 | 0.01 |
| 25.0 | 9.7 | 0.3 | 7.6 | 1.1 | 0.49 | 0.05 | 0.32 | 0.06 | 0.53 | 0.01 | 0.45 | 0.01 | |
| 50.0 | 9.8 | 0.4 | 7.5 | 0.6 | 0.42 | 0.07 | 0.24 | 0.04 | 0.52 | 0.01 | 0.44 | 0.01 | |
| 2.8 | 10.6 | 0.3 | 7.1 | 1.5 | 0.62 | 0.03 | 0.36 | 0.15 | 0.51 | 0.01 | 0.47 | 0.04 | |
| 5.6 | 10.6 | 0.6 | 6.5 | 0.7 | 0.51 | 0.05 | 0.37 | 0.06 | 0.51 | 0.01 | 0.42 | 0.00 | |
| 13-tc | 11.1 | 10.3 | 0.5 | 7.6 | 1.1 | 0.53 | 0.08 | 0.34 | 0.06 | 0.54 | 0.02 | 0.41 | 0.02 |
| 25.0 | 10.3 | 0.7 | 7.5 | 0.4 | 0.54 | 0.08 | 0.39 | 0.05 | 0.52 | 0.02 | 0.41 | 0.01 | |
| 50.0 | 8.9 | 0.4 | 6.2 | 0.6 | 0.56 | 0.05 | 0.37 | 0.08 | 0.48 | 0.01 | 0.41 | 0.01 | |
| 2.8 | 10.4 | 0.7 | 7.2 | 0.5 | 0.64 | 0.03 | 0.57 | 0.11 | 0.53 | 0.02 | 0.49 | 0.01 | |
| 5.6 | 10.2 | 0.3 | 10.7 | 1.6 | 0.66 | 0.04 | 0.57 | 0.09 | 0.53 | 0.01 | 0.44 | 0.01 | |
| 9-al | 11.1 | 9.7 | 0.5 | 6.9 | 0.3 | 0.56 | 0.08 | 0.49 | 0.07 | 0.55 | 0.02 | 0.52 | 0.01 |
| 25.0 | 9.2 | 0.2 | 8.2 | 0.8 | 0.61 | 0.08 | 0.68 | 0.10 | 0.56 | 0.01 | 0.48 | 0.01 | |
| 50.0 | 8.2 | 0.1 | 6.8 | 0.1 | 0.63 | 0.05 | 0.64 | 0.09 | 0.61 | 0.02 | 0.54 | 0.01 | |
| 2.8 | 10.8 | 0.7 | 10.7 | 2.2 | 0.61 | 0.05 | 0.49 | 0.06 | 0.52 | 0.02 | 0.44 | 0.02 | |
| 5.6 | 9.9 | 0.2 | 6.9 | 0.8 | 0.48 | 0.06 | 0.35 | 0.06 | 0.53 | 0.01 | 0.52 | 0.03 | |
| 12-al | 11.1 | 10.6 | 0.3 | 8.6 | 0.8 | 0.58 | 0.06 | 0.55 | 0.06 | 0.54 | 0.01 | 0.47 | 0.01 |
| 25.0 | 10.3 | 0.3 | 9.3 | 0.9 | 0.69 | 0.04 | 0.73 | 0.08 | 0.54 | 0.01 | 0.45 | 0.01 | |
| 50.0 | 9.7 | 0.5 | 7.6 | 0.3 | 0.76 | 0.08 | 0.77 | 0.08 | 0.55 | 0.02 | 0.49 | 0.00 | |
Error estimate.
FIGURE 8.
Average long time (D2) lateral diffusion coefficients of PLPC and oxidized lipids as a function of the concentration of oxidized lipids.
Water permeation through PLPC bilayers
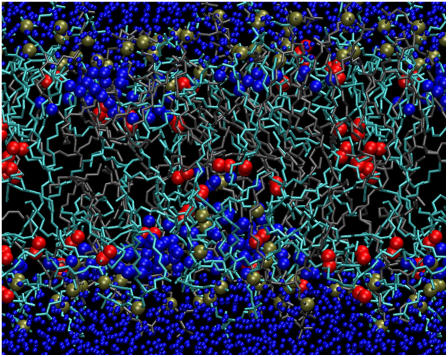
The presence of oxidized lipids has a profound influence on the permeability of water through PLPC bilayers. Water pores are observed in all the simulations containing 5% or more oxidized lipids, and they are relatively stable at higher oxidation levels. Fig. 9 shows a water defect in a bilayer containing 13-tc lipids at 50% concentration. Based on visual inspection of the trajectories, water defects can form independently in both leaflets and they are larger in the presence of 12-al and 13-tc. This difference correlates well with the higher area per lipid in bilayers containing 12-al and 13-tc lipids, which have polar oxygen atoms closer to the terminal methyl group. It is possible that water defects form more easily upon clustering of oxidized lipids, but the simulations do not provide sufficient statistics in this respect. Demixing of oxidized lipids is not observed on the simulation timescale, and the clusters appear to have a limited stability.
FIGURE 9.
Snapshot showing a stable water defect in a PLPC bilayer containing 50% 13-tc lipids. PLPC is shown as sticks in gray, 13-tc in cyan; all phosphate atoms are shown as tan spheres, oxygen atoms in the hydroperoxide group as red spheres, and water oxygen atoms are shown as blue spheres (with bigger size inside the bilayer).
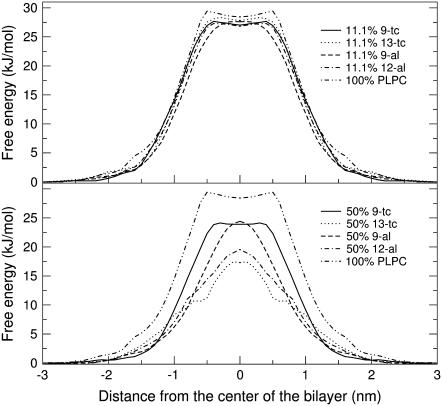
To understand the energetics of water penetration inside different bilayers, we calculated the potential mean force (PMF) of water as a function of the distance from the center of the bilayer using constraint simulations. Due to the high computational cost, we limited the calculations to nine systems, including pure PLPC and bilayers containing 11.1% and 50% of each oxidized lipid. For the same systems we also calculated the local diffusion coefficient and the permeability of water across the bilayer. PMF profiles are shown in Fig. 10. The shape of free energy profile for pure PLPC is very similar to previously published profiles for DPPC (70,71,85). The free energy increases moving from the water interface to the hydrophobic tail region, reaching a maximum of 29.4 ± 2.3 kJ/mol at 0.5 nm from the center of bilayer. A dip of ∼1 kJ/mol is observed at the center of bilayer, which can be interpreted as the effect of the slightly lower local density (70,71). Free energy values in our simulation are higher than those obtained by Marrink and Bemporad simulating DPPC bilayers (70,85). The difference might be explained by the different nature of the lipids and the lower temperature used in our simulations (300 K instead of 350 K or higher). For the pure PLPC bilayer, we compared PMF results obtained from constraint and umbrella sampling simulations (data not shown), and did not find significant differences. For the mixtures containing oxidized lipids, the shape of the PMF becomes Gaussian-like when the concentrations of oxidized lipids increases, and the dip at the center of the bilayer disappears. Since the density at the center of the bilayer increases with increasing level of oxidation, this result is consistent with the idea that the dip in the PMF depends on the local density. The free energy barrier to translocation across the bilayer decreases as the concentration of oxidized lipids increases, with all four oxidized lipids. The free energy at the center of bilayer is decreased by 0.6 to 1.6 kJ/mol in the presence of 11.1% oxidized lipids, and by 4.0 to 11.1 kJ/mol in the presence of 50% oxidized lipids. Like for the area per lipid, 12-al and 13-tc lipids have a stronger effect on the free energy barrier compared to 9-tc and 9-al.
FIGURE 10.
Potential of mean force for water as a function of the distance from the center of the bilayer, in a bilayer containing pure PLPC or 11.1% oxidized lipids or 50% oxidized lipids. Error bars are omitted for clarity.
Local diffusion coefficients and the local resistance were calculated for water at different depths in the membrane. In the case of the pure PLPC bilayer, we observe a decrease in the local diffusion coefficient from bulk water to lipid tail interior and an increase in the central portion of the bilayer. Although this behavior is qualitatively similar to that observed by Marrink and Bemporad on DPPC bilayers (70,85), the discrepancy in the actual values is significant. In the central region, the local diffusion coefficient of water is lower in oxidized bilayers than in the nonoxidized bilayer. This is related to the increase in the local density at the center of oxidized bilayers. As expected, the free energy barrier dominates the local resistance profiles for water in the bilayers, and therefore water permeability. To our knowledge, water permeability through PLPC bilayer has never been reported in the literature. However, several articles reported experimental results for water permeability through different saturated and unsaturated lipid bilayers. A value of (4.9 ± 0.6) × 10−3 cm/s has been reported for 1-stearoyl-2-linoleoyl phosphatidylcholine, and (9.1 ± 2.4) × 10−3 cm/s for 1,2-dilinoleoyl phosphatidylcholine (DLPC) (86). Although comparable, these values are higher than our findings for pure PLPC, (1.6 ± 0.5) × 10−3 cm/s at 25°C.
Table 8 shows the permeability of water through PLPC bilayers at two different concentrations of oxidized lipids. As all oxidized lipids decrease the free energy barrier for water penetration, all of them increase water permeability. This effect is not very large at the concentration of 11.1%, but at 50% the increase in permeability is between one and two orders of magnitude. The increase in water permeability with increasing concentration of oxidized lipids is consistent with experimental results (25–28) and correlates with looser packing of lipids. In our simulations, 12-al and 13-tc lipids have a larger effect on water permeability compared to 9-al and 9-tc lipids. This result suggests a relationship between permeability and the bilayer structural features. Water permeability appears to be related to the position of the oxygen in the lipid tail more than to the length of the tail and the hydrogen-bonding capability of the oxidized moiety. Based on our results, water permeability correlates better with the area per lipid than with the bilayer thickness and hydrogen-bonding capability.
TABLE 8.
Water permeability through PLPC bilayers containing four different oxidized lipids (concentrations of 11.1% and 50%)
| Lipid bilayer | Concentration of oxidized lipids (%) | Permeability of water (× 10−3 cm/s) |
|---|---|---|
| PLPC | 0 | 1.6 ± 0.5 |
| PLPC with 9-tc | 11.1 | 3.3 ± 0.6 |
| 50 | 11.6 ± 4.5 | |
| PLPC with 13-tc | 11.1 | 2.1 ± 0.8 |
| 50 | 92 ± 40 | |
| PLPC with 9-al | 11.1 | 3.2 ± 1.9 |
| 50 | 16.1 ± 4.7 | |
| PLPC with 12-al | 11.1 | 4.0 ± 2.2 |
| 50 | 66 ± 32 |
Although our calculations are limited to the permeability of water through a lipid membrane, we expect similar trends to be valid for any polar solute (27). An increase in the permeability of all polar substances would lead to an imbalance for numerous substances in a cell and therefore to cell death.
CONCLUSIONS
In this work we studied the effect of lipid peroxidation on the properties of 1-palmitoyl-2-linoleoyl-sn-glycero-3-phosphatidylcholine (PLPC) lipid bilayers using molecular dynamics simulations. We simulated PLPC lipid bilayers containing different concentrations of four oxidation products of linoleic acid, containing either a hydroperoxide or an aldehyde group, in two different positions in the lipid tails. The aldehyde lipids have different chain length compared to the hydroperoxide ones, and the hydroperoxide group is more hydrophilic than the aldehyde group. These simple chemical features of the individual lipid molecules lead to well-defined changes in the properties of the bilayers. Structural and dynamic properties of the lipid membrane are affected by the presence of oxidized lipids to different extents. All the oxidized tails show a significant conformational difference compared to nonoxidized tails: the oxidized moiety is generally close to the lipid headgroup region and forms hydrogen bonds mainly with water. This tendency is more pronounced for hydroperoxide lipids compared to aldehyde lipids, due to the higher hydrophilicity of the hydroperoxide group. Structural properties of the lipid bilayer are found to depend on the different chemical features of the oxidized lipids: nature and position of the functional group, length of the lipid tail. The area per lipid has a stronger dependence on the position of the oxidized moiety, while the bilayer thickness depends strongly also on the length of the tail. The changes in area per lipid and bilayer thickness are reflected by a decrease in the order parameter of the lipid tails, which is stronger in the case of aldehyde lipids. Despite the large changes in the structural properties of the lipid bilayer, lipid translational dynamics does not appear to be significantly affected by the presence of oxidized lipids. Water permeability through the bilayer is increased in the presence of oxidized lipids, and water defects are observed frequently at high concentrations of oxidized lipids. Our results suggest that one mechanism of cell membrane damage is the increase in membrane permeability due to the presence of oxidized lipids.
Acknowledgments
Calculations were performed in part on WestGrid facilities.
This research has been supported by the Natural Science and Engineering Research Council. J.W.-e. and I.-M.T. are supported by the Royal Golden Jubilee PhD Program (grant No. PHD/0240/2545). W.T. is supported by the National Center for Genetic Engineering and Biotechnology and Thailand Research Fund. D.P.T. is an Alberta Heritage Foundation for Medical Research Senior Scholar and Canadian Institutes of Health Research New Investigator, and L.M. is an Alberta Heritage Foundation for Medical Research postdoctoral fellow.
Zhitao Xu's present address is Materials and Process Simulation Center, California Institute of Technology, Pasadena, CA.
Editor: Helmut Grubmüller.
References
- 1.Markesbery, W. R. 1997. Oxidative stress hypothesis in Alzheimer's disease. Free Radic. Biol. Med. 23:134–147. [DOI] [PubMed] [Google Scholar]
- 2.Sohal, R. S., and R. Weindruch. 1996. Oxidative stress, caloric restriction, and aging. Science. 273:59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dexter, D. T., C. J. Carter, F. R. Wells, F. Javoyagid, Y. Agid, A. Lees, P. Jenner, and C. D. Marsden. 1989. Basal lipid peroxidation in Substantia nigra is increased in Parkinson's disease. J. Neurochem. 52:381–389. [DOI] [PubMed] [Google Scholar]
- 4.Mark, R. J., Z. Pang, J. W. Geddes, K. Uchida, and M. P. Mattson. 1997. Amyloid β-peptide impairs glucose transport in hippocampal and cortical neurons: Involvement of membrane lipid peroxidation. J. Neurosci. 17:1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mark, R. J., M. A. Lovell, W. R. Markesbery, K. Uchida, and M. P. Mattson. 1997. A role for 4-hydroxynonenal, an aldehydic product of lipid peroxidation, in disruption of ion homeostasis and neuronal death induced by amyloid beta-peptide. J. Neurochem. 68:255–264. [DOI] [PubMed] [Google Scholar]
- 6.Halliwell, B., and J. M. Gutteridge. 1990. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 186:1–85. [DOI] [PubMed] [Google Scholar]
- 7.Halliwell, B. 2000. The antioxidant paradox. Lancet. 355:1179–1180. [DOI] [PubMed] [Google Scholar]
- 8.Butterfield, D. A., J. Drake, C. Pocernich, and A. Castegna. 2001. Evidence of oxidative damage in Alzheimer's disease brain: central role for amyloid β-peptide. Trends Mol. Med. 7:548–554. [DOI] [PubMed] [Google Scholar]
- 9.Everse, J., and P. W. Coates. 2005. Role of peroxidases in Parkinson disease: a hypothesis. Free Radic. Biol. Med. 38:1296–1310. [DOI] [PubMed] [Google Scholar]
- 10.Zhang, X. Y., Y. L. Tan, L. Y. Cao, G. Y. Wu, O. Xu, Y. Shen, and D. F. Zhou. 2006. Antioxidant enzymes and lipid peroxidation in different forms of schizophrenia treated with typical and atypical antipsychotics. Schizophr. Res. 81:291–300. [DOI] [PubMed] [Google Scholar]
- 11.Berliner, J. A., and J. W. Heinecke. 1996. The role of oxidized lipoproteins in atherogenesis. Free Radic. Biol. Med. 20:707–727. [DOI] [PubMed] [Google Scholar]
- 12.Diaz, M. N., B. Frei, J. A. Vita, and J. F. Keaney. 1997. Mechanisms of disease—antioxidants and atherosclerotic heart disease. N. Engl. J. Med. 337:408–416. [DOI] [PubMed] [Google Scholar]
- 13.Wood, L. G., P. G. Gibson, and M. L. Garg. 2003. Biomarkers of lipid peroxidation, airway inflammation and asthma. Eur. Respir. J. 21:177–186. [DOI] [PubMed] [Google Scholar]
- 14.Downey, J. M. 1990. Free radicals and their involvement during long-term myocardial ischemia and reperfusion. Annu. Rev. Physiol. 52:487–504. [DOI] [PubMed] [Google Scholar]
- 15.McCord, J. M. 1985. Oxygen-derived free radicals in post ischemic tissue injury. N. Engl. J. Med. 312:159–163. [DOI] [PubMed] [Google Scholar]
- 16.Brodnitz, M. H. 1968. Autooxidation of saturated fatty acids. A review. J. Agric. Food Chem. 16:994–999. [Google Scholar]
- 17.Binder, H., and K. Gawrisch. 2001. Effect of unsaturated lipid chains on dimensions, molecular order and hydration of membranes. J. Phys. Chem. B. 105:12378–12390. [Google Scholar]
- 18.Wratten, M. L., G. Vanginkel, A. A. Vantveld, A. Bekker, E. E. Vanfaassen, and A. Sevanian. 1992. Structural and dynamic effects of oxidatively modified phospholipids in unsaturated lipid membranes. Biochemistry. 31:10901–10907. [DOI] [PubMed] [Google Scholar]
- 19.Baenziger, J. E., H. C. Jarrell, R. J. Hill, and I. C. P. Smith. 1991. Average structural and motional properties of a diunsaturated acyl chain in a lipid bilayer—effects of 2 cis-unsaturated double bonds. Biochemistry. 30:894–903. [DOI] [PubMed] [Google Scholar]
- 20.Chamulitrat, W., and R. P. Mason. 1989. Lipid peroxyl radical intermediates in the peroxidation of poly-unsaturated fatty acids by lipoxygenase—direct electron spin resonance investigations. J. Biol. Chem. 264:20968–20973. [PubMed] [Google Scholar]
- 21.Tamm, L. K., and J. Seelig. 1983. Lipid solvation of cytochrome-C oxidase—deuterium, N-14, and P-31 nuclear magnetic resonance studies on the phosphocholine headgroup and on cis-unsaturated fatty acyl chains. Biochemistry. 22:1474–1483. [DOI] [PubMed] [Google Scholar]
- 22.Mason, R. P., M. F. Walter, and P. E. Mason. 1997. Effect of oxidative stress on membrane structure: small-angle x-ray diffraction analysis. Free Radic. Biol. Med. 23:419–425. [DOI] [PubMed] [Google Scholar]
- 23.Stark, G. 1991. The effect of ionizing radiation on lipid membranes. Biochim. Biophys. Acta. 1071:103–122. [DOI] [PubMed] [Google Scholar]
- 24.Porter, N. A., S. E. Caldwell, and K. A. Mills. 1995. Mechanisms of free radical oxidation of unsaturated lipids. Lipids. 30:277–290. [DOI] [PubMed] [Google Scholar]
- 25.Mandal, T. K., and S. N. Chatterjee. 1980. Ultraviolet- and sunlight-induced lipid peroxidation in liposomal membrane. Radiat. Res. 83:290–302. [PubMed] [Google Scholar]
- 26.Chatterjee, S. N., and S. Agarwal. 1988. Liposomes as membrane model for study of lipid peroxidation. Free Radic. Biol. Med. 4:51–72. [DOI] [PubMed] [Google Scholar]
- 27.Kunimoto, M., K. Inoue, and S. Nojima. 1981. Effect of ferrous ion and ascorbate-induced lipid peroxidation on liposomal membranes. Biochim. Biophys. Acta Biomembr. 646:169–178. [DOI] [PubMed] [Google Scholar]
- 28.Nakazawa, T., S. Nagatsuka, and O. Yukawa. 1986. Effects of membrane stabilizing agents and radiation on liposomal membranes. Drugs Exp. Clin. Res. 12:831–835. [PubMed] [Google Scholar]
- 29.Leyko, W., D. Ertel, and G. Bartosz. 1991. Effect of hyperthermia and lipid peroxidation on the erythrocyte membrane structure. Int. J. Radiat. Biol. 59:1185–1193. [DOI] [PubMed] [Google Scholar]
- 30.Bruch, R. C., and W. S. Thayer. 1983. Differential effect of lipid peroxidation on membrane fluidity as determined by electron spin resonance probes. Biochim. Biophys. Acta. 733:216–222. [DOI] [PubMed] [Google Scholar]
- 31.Dobretsov, G. E., T. A. Borschevskaya, V. A. Petrov, and Y. U. Vladimirov. 1977. The increase of phospholipid bilayer rigidity after lipid peroxidation. FEBS Lett. 84:125–128. [DOI] [PubMed] [Google Scholar]
- 32.Sevanian, A., M. L. Wratten, L. L. McLeod, and E. Kim. 1988. Lipid peroxidation and phospholipase A2 activity in liposomes composed of unsaturated phospholipids: a structural basis for enzyme activation. Biochim. Biophys. Acta Lipids Lipid Metab. 961:316–327. [DOI] [PubMed] [Google Scholar]
- 33.Richter, C. 1987. Biophysical consequences of lipid peroxidation in membranes. Chem. Phys. Lipids. 44:175–189. [DOI] [PubMed] [Google Scholar]
- 34.Cavatorta, P., L. Masotti, G. Sartor, M. B. Ferrari, E. Casali, S. Borello, G. Minotti, and T. Galeotti. 1985. Cell Membranes and Cancer. Elsevier, New York.
- 35.Gut, J., S. Kawato, R. J. Cherry, K. Winterhalter, and C. Richter. 1985. Lipid peroxidation decreases the rotational mobility of cytochrome P-450 in rat liver microsomes. Biochim. Biophys. Acta. 817:217–228. [DOI] [PubMed] [Google Scholar]
- 36.Eichenberger, K., P. Bohni, K. Winterhalter, and C. Richter. 1982. Microsomal lipid peroxidation causes an increase in the order of the membrane lipid domain. FEBS Lett. 142:59–62. [DOI] [PubMed] [Google Scholar]
- 37.Galeotti, T., S. Borello, G. Palombini, L. Masotti, M. B. Ferrari, P. Cavatorta, A. Arcioni, C. Stremmena, and C. Zanonni. 1984. Lipid peroxidation and fluidity of plasma membranes from rat liver and Morris hepatoma 3924A. FEBS Lett. 169:169–173. [DOI] [PubMed] [Google Scholar]
- 38.Grzelinska, E., G. Bartosz, K. Gwozdzinski, and W. Leyko. 1979. A spin-label study of the effect of gamma radiation on erythrocyte membrane. Influence of lipid peroxidation on membrane structure. Int. J. Radiat. Biol. 36:325–334. [DOI] [PubMed] [Google Scholar]
- 39.Feller, S. E. 2000. Molecular dynamics simulations of lipid bilayers. Curr. Op. Colloid Inter. 5:217–223. [Google Scholar]
- 40.Tieleman, D. P., S. J. Marrink, and H. J. C. Berendsen. 1997. A computer perspective of membranes: molecular dynamics studies of lipid bilayer systems. Biochim. Biophys. Acta Rev. Biomembr. 1331:235–270. [DOI] [PubMed] [Google Scholar]
- 41.Nagle, J. F., and S. Tristram-Nagle. 2000. Structure of lipid bilayers. Biochim. Biophys. Acta. 1469:159–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berger, O., O. Edholm, and F. Jahnig. 1997. Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure, and constant temperature. Biophys. J. 72:2002–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindahl, E., and O. Edholm. 2000. Mesoscopic undulations and thickness fluctuations in lipid bilayers from molecular dynamics simulations. Biophys. J. 79:426–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mukhopadhyay, P., L. Monticelli, and D. P. Tieleman. 2004. Molecular dynamics simulation of a palmitoyl-oleoyl phosphatidylserine bilayer with Na+ counterions and NaCl. Biophys. J. 86:1601–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edholm, O., and A. M. Nyberg. 1992. Cholesterol in model membranes. A molecular dynamics simulation. Biophys. J. 63:1081–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Vries, A. H., A. E. Mark, and S. J. Marrink. 2004. The binary mixing behavior of phospholipids in a bilayer: a molecular dynamics study. J. Phys. Chem. B. 108:2454–2463. [Google Scholar]
- 47.Faller, R., and S. J. Marrink. 2004. Simulation of domain formation in DLPC-DSPC mixed bilayers. Langmuir. 20:7686–7693. [DOI] [PubMed] [Google Scholar]
- 48.Leekumjorn, S., and A. K. Sum. 2007. Molecular studies of the gel to liquid-crystalline phase transition for fully hydrated DPPC and DPPE bilayers. Biochim. Biophys. Acta Biomembr. 1768:354–365. [DOI] [PubMed] [Google Scholar]
- 49.Niemela, P. S., S. Ollila, M. T. Hyvonen, M. Karttunen, and I. Vattulainen. 2007. Assessing the nature of lipid raft membranes. PLoS Comput. Biol. 3:304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pandit, S. A., G. Khelashvili, E. Jakobsson, A. Grama, and H. L. Scott. 2007. Lateral organization in lipid-cholesterol mixed bilayers. Biophys. J. 92:440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tieleman, D. P., J. L. MacCallum, W. L. Ash, C. Kandt, Z. Xu, and L. Monticelli. 2006. Membrane protein simulations with a united-atom lipid and all-atom protein model: lipid-protein interactions, side chain transfer free energies and model proteins. J. Phys. Cond. Matter. 18:S1221–S1234. [DOI] [PubMed] [Google Scholar]
- 52.Feller, S. E., D. X. Yin, R. W. Pastor, and A. D. MacKerell. 1997. Molecular dynamics simulation of unsaturated lipid bilayers at low hydration: parameterization and comparison with diffraction studies. Biophys. J. 73:2269–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hyvonen, M. T., and P. T. Kovanen. 2005. Molecular dynamics simulations of unsaturated lipid bilayers: effects of varying the numbers of double bonds. Eur. Biophys. J. 34:294–305. [DOI] [PubMed] [Google Scholar]
- 54.Bachar, M., P. Brunelle, D. P. Tieleman, and A. Rauk. 2004. Molecular dynamics simulation of a polyunsaturated lipid bilayer susceptible to lipid peroxidation. J. Phys. Chem. B. 108:7170–7179. [Google Scholar]
- 55.Niemela, P. S., M. T. Hyvonen, and I. Vattulainen. 2006. Influence of chain length and unsaturation on sphingomyelin bilayers. Biophys. J. 90:851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ollila, S., M. T. Hyvonen, and I. Vattulainen. 2007. Polyunsaturation in lipid membranes: dynamic properties and lateral pressure profiles. J. Phys. Chem. B. 111:3139–3150. [DOI] [PubMed] [Google Scholar]
- 57.Pratt, D. A., J. H. Mills, and N. A. Porter. 2003. Theoretical calculations of carbon-oxygen bond dissociation enthalpies of peroxyl radicals formed in the autooxidation of lipids. J. Am. Chem. Soc. 125:5801–5810. [DOI] [PubMed] [Google Scholar]
- 58.Swart, M. 2002. Density Functional Theory Applied to Copper Proteins. Rijksuniversiteit Groningen, The Netherlands.
- 59.Schrodinger, I. 2000. JAGUAR 4.1. Schrodinger Inc., Portland, OR.
- 60.Becke, A. D. 1993. Density functional thermochemistry. 3. The role of exact exchange. J. Chem. Phys. 98:5648–5652. [Google Scholar]
- 61.Lee, C. T., W. T. Yang, and R. G. Parr. 1988. Development of the Colle-Salvetti correlation energy formula into a functional of the electron density. Phys. Rev. B. 37:785–789. [DOI] [PubMed] [Google Scholar]
- 62.Reed, A. E., R. B. Weinstock, and F. Weinhold. 1985. Natural population analysis. J. Chem. Phys. 83:735–746. [Google Scholar]
- 63.Singh, U. C., and P. A. Kollman. 1984. An approach to computing electrostatic charges for molecules. J. Comput. Chem. 5:129–145. [Google Scholar]
- 64.Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. (Online). 7:306–317. [Google Scholar]
- 65.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald—an N·Log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 66.Essmann, U., L. Perera, M. L. Berkowitz, T. Darden, H. Lee, and L. G. Pedersen. 1995. Smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. [Google Scholar]
- 67.Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- 68.Berendsen, H. J. C., J. P. M. Postma, W. F. van Gunsteren, A. DiNola, and J. R. Haak. 1984. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81:3684–3690. [Google Scholar]
- 69.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:27–38. [DOI] [PubMed] [Google Scholar]
- 70.Marrink, S. J., and H. J. C. Berendsen. 1994. Simulation of water transport through a lipid membrane. J. Phys. Chem. 98:4155–4168. [Google Scholar]
- 71.Marrink, S. J., and H. J. C. Berendsen. 1996. Permeation process of small molecules across lipid membranes studied by molecular dynamics simulations. J. Phys. Chem. 100:16729–16738. [Google Scholar]
- 72.Kubo, R. 1966. The fluctuation-dissipation theorem. Rep. Prog. Phys. 29:255–284. [Google Scholar]
- 73.Kirkwood, J. G. 1946. The statistical mechanical theory of transport processes. I. General theory. J. Chem. Phys. 14:180–201. [Google Scholar]
- 74.van Kuijk, F. J. G. M., A. Sevanian, G. Handelman, and E. A. Dratz. 1988. A new role for phospholipase A2: protection of membranes from lipid peroxidation damage. Trends Biochem. Sci. 12:31–34. [Google Scholar]
- 75.Hess, B. 2002. Determining the shear viscosity of model liquids from molecular dynamics simulations. J. Chem. Phys. 116:209–217. [Google Scholar]
- 76.Kupiainen, M., E. Falck, S. Ollila, P. Niemelä, A. A. Gurtovenko, M. T. Hyvönen, M. Patra, M. Karttunen, and I. Vattulainen. 2005. Free volume properties of sphingomyelin, DMPC, DPPC, and PLPC bilayers. J. Comput. Theor. Nanosci. 2:1–13. [Google Scholar]
- 77.Lewis, B. A., and D. M. Engelman. 1983. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J. Mol. Biol. 166:211–217. [DOI] [PubMed] [Google Scholar]
- 78.Pradhan, D., M. Weiser, K. Lumley-Sapanski, D. Fazier, S. Kemper, P. Williamson, and R. A. Schlegel. 1990. Peroxidation-induced perturbations of erythrocyte lipid organization. Biochim. Biophys. Acta. 1023:398–404. [DOI] [PubMed] [Google Scholar]
- 79.Sabatini, K., J. P. Mattila, F. M. Megli, and P. K. J. Kinnunen. 2006. Characterization of two oxidatively modified phospholipids in mixed monolayers with DPPC. Biophys. J. 90:4488–4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wohlert, J., and O. Edholm. 2006. Dynamics in atomistic simulations of phospholipid membranes: nuclear magnetic resonance relaxation rates and lateral diffusion. J. Chem. Phys. 125:204703–204710. [DOI] [PubMed] [Google Scholar]
- 81.Tabony, J., and B. Perly. 1990. Quasielastic neutron scattering measurements of fast local translational diffusion of lipid molecules in phospholipid bilayers. Biochim. Biophys. Acta. 1063:67–72. [DOI] [PubMed] [Google Scholar]
- 82.Konig, S., W. Pfeiffer, T. Bayerl, D. Richter, and E. Sackmann. 1992. Molecular dynamics of lipid bilayers studied by incoherent quasi-elastic neutron scattering. J. Phys. II Fr. 2:1589–1615. [Google Scholar]
- 83.Almeida, P. F., W. L. Vaz, and T. E. Thompson. 1992. Lateral diffusion in the liquid phases of dimyristoylphosphatidylcholine/cholesterol lipid bilayers: a free volume analysis. Biochemistry. 31:6739–6747. [DOI] [PubMed] [Google Scholar]
- 84.Ianzini, F., L. Guidoni, P. L. Indovina, V. Viti, G. Erriu, S. Onnis, and P. Randaccio. 1984. Gamma-irradiation effects on phosphatidylcholine multilayer liposomes: calorimetric, NMR, and spectrofluorimetric studies. Radiat. Res. 98:154–166. [PubMed] [Google Scholar]
- 85.Bemporad, D., J. W. Essex, and C. Luttmann. 2004. Permeation of small molecules through a lipid bilayer: a computer simulation study. J. Phys. Chem. B. 108:4875–4884. [Google Scholar]
- 86.Olbrich, K., W. Rawicz, D. Needham, and E. Evans. 2000. Water permeability and mechanical strength of polyunsaturated lipid bilayers. Biophys. J. 79:321–327. [DOI] [PMC free article] [PubMed] [Google Scholar]