Abstract
During meiosis, self-inflicted DNA double-strand breaks (DSBs) are created by the protein Spo11 and repaired by homologous recombination leading to gene conversions and crossovers. Crossover formation is vital for the segregation of homologous chromosomes during the first meiotic division and requires the RecA orthologue, Dmc1.We analyzed repair during meiosis of site-specific DSBs created by another nuclease, VMA1-derived endonuclease (VDE), in cells lacking Dmc1 strand-exchange protein. Turnover and resection of the VDE-DSBs was assessed in two different reporter cassettes that can repair using flanking direct repeat sequences, thereby obviating the need for a Dmc1-dependent DNA strand invasion step. Access of the single-strand binding complex replication protein A, which is normally used in all modes of DSB repair, was checked in chromatin immunoprecipitation experiments, using antibody against Rfa1. Repair of the VDE-DSBs was severely inhibited in dmc1Δ cells, a defect that was associated with a reduction in the long tract resection required to initiate single-strand annealing between the flanking repeat sequences. Mutants that either reduce Spo11-DSB formation or abolish resection at Spo11-DSBs rescued the repair block. We also found that a replication protein A component, Rfa1, does not accumulate to expected levels at unrepaired single-stranded DNA (ssDNA) in dmc1Δ cells. The requirement of Dmc1 for VDE-DSB repair using flanking repeats appears to be caused by the accumulation of large quantities of ssDNA that accumulate at Spo11-DSBs when Dmc1 is absent. We propose that these resected DSBs sequester both resection machinery and ssDNA binding proteins, which in wild-type cells would normally be recycled as Spo11-DSBs repair. The implication is that repair proteins are in limited supply, and this could reflect an underlying mechanism for regulating DSB repair in wild-type cells, providing protection from potentially harmful effects of overabundant repair proteins.
Author Summary
During meiosis, DNA is deliberately damaged by formation of double-strand breaks. Programmed breaks must be repaired for cell division to be completed. Break repair enables reciprocal exchange between parental chromosomes, and this exchange acts as a link between chromosomes before anaphase separation. These links are essential to ensure that maternal and paternal chromosomes segregate into different daughter cells. Meiosis has special mechanisms to ensure the repair creates sufficient reciprocal exchanges between parental chromosomes; Dmc1 protein is essential for these mechanisms to work. When Dmc1 is absent, programmed breaks accumulate with excess single-stranded DNA nearby. Using reporter constructs integrated into yeast, we examined repair of an experimentally induced break expected not to need Dmc1. When Dmc1 is absent, programmed breaks accumulate in single-stranded form, and the experimental break is not repaired. Either preventing formation of programmed breaks, or stopping DNA near them from becoming single-stranded, relieves this repair block. We conclude that repair proteins are likely to be in limited supply during meiosis, and they run out in cells lacking Dmc1 function. Limiting protein supply may be an important regulatory mechanism, protecting DNA from potentially damaging effects of oversupply.
Introduction
In most organisms the success of meiosis is dependent on the creation of molecular joints that serve to lock homologous chromosomes together until they mediate ordered chromosome segregation at the first meiotic division. This is achieved by the creation of crossovers, which creates a covalent link between nonsister chromatids, and through the forces of sister chromatid cohesion to maintain a link between chromosome pairs until first anaphase (reviewed in [1]). Crossovers are formed during repair of programmed DNA double-strand breaks (DSBs) created by the Spo11 protein [2,3]. DSBs can also be repaired by use of the sister chromatid as template. But, because intersister repair does not create links between homologous chromosomes, meiotic cells have evolved a strong bias toward using the homologous chromosome as donor template. Much attention is currently focused on understanding how the meiotic cell enforces the preference for interhomolog repair.
Various proteins have been implicated in directing DSB repair toward the homologous chromosome and/or away from the sister chromatid. These include the meiosis-specific RecA homolog Dmc1. In the absence of Dmc1, DNA joint molecules between homologous chromosomes fail to form, causing unrepaired DSBs to accumulate [4,5]. Also implicated in enforcing interhomolog DNA repair are members of a meiosis-specific complex, Mek1-Hop1-Red1. Mek1 is a kinase with similarities to Rad53. Loss of Mek1 function bypasses the requirement for Dmc1, rendering meiotic DSB repair Rad54-dependent and increasing the frequency of intersister recombination events [6–9]. Hop1 and Red1 are phosphor-proteins that localize to meiotic chromosome axes, and mutants of both genes have been recovered in screens for increased intersister chromatid repair [6,8,10–13].
In addition to the classical homologous recombination mechanisms, DSBs can be repaired by a second homology-dependent mechanism, single-strand annealing (SSA) [14,15]. DSBs that form during mitotic or meiotic growth are resected to create short tracts of 3′ ending single-stranded DNA (ssDNA). If unrepaired by strand invasion, resection can extend for many kilobases. Extensive resection has the potential to uncover repeated sequences that flank the initial lesion, such that complementary strands anneal leaving a flap of intervening DNA that is removed by Rad1/Rad10 flap endonuclease activity [16]. Little is known about proteins that initiate resection or catalyze the formation of long tracts of ssDNA. The Mre11 complex and Exo1 certainly contribute to resection, and these proteins influence the likelihood of DSBs repairing through SSA [17–22] (R. Johnson, M. J. Neale, A. S. H. Goldman, unpublished data). Mitotic studies show SSA to be independent of RAD51, RAD54, RAD55, and RAD57, but dependent on RAD52 [17,18]. The single-stranded binding protein complex replication protein A (RPA) is also required for SSA, probably to help recruit Rad52 [19,23].
The activities of general and meiosis-specific recombination proteins are not restricted to repair of Spo11-induced DSBs during meiosis. This has been determined from studies of recombination induced by the HO-endonuclease and the meiosis-specific homing VMA1-derived endonuclease, VDE. Unlike Spo11, HO-endonuclease and VDE have strict cleavage sequence-specificity and do not become covalently bound to the DSB end, creating “clean” DSBs [2,24–27]. Despite these differences, the genetic requirements for repair of HO- and VDE-induced DSBs are similar to those of Spo11-DSBs [28–30]. For example, SAE2 is required for removal of covalently bound Spo11 from DSB ends and for single-strand resection, and VDE-DSB repair is slowed in sae2 mutants because resection at VDE-DSBs is also retarded [28,31] (A. Bishop-Bailey, A. S. H. Goldman, unpublished data). That Sae2 would be important for repair of a clean DSB is supported by more recent studies on HO-DSBs in mitotic cells [32]. DMC1 is required for gene conversion at a VDE-DSB, indicating that the commitment of meiotic cells to repair using a homologous chromosome template is not restricted to Spo11-induced DSBs [28]. Studying DSBs created by an endonuclease other than Spo11 provides insight into the regulatory significance of large numbers of Spo11-DSBs and allows study of mutations at a stage beyond the point where a phenotypic block would be observed at Spo11-DSBs [31].
Here we report that the requirement of Dmc1 for meiotic DSB repair persists even when the VDE-DSB is flanked by direct repeats, which allow repair without need for DNA strand invasion. If repair by interhomolog gene conversion is precluded, and SSA is the only pathway for VDE-DSB repair, DSB repair is still strongly Dmc1-dependent. However, SSA-mediated VDE-DSB repair becomes Dmc1-independent in the absence of active Spo11, Hop1, or Sae2, all of which influence levels of resected Spo11-DSBs. Titrating DSB-associated ssDNA using hypomorphic spo11 alleles increases SSA repair efficiency in the dmc1Δ cells. Analysis of Rfa1 binding supports the view that extensive Spo11-DSB-associated ssDNA formed in the absence of Dmc1 reduces the availability of ssDNA binding proteins necessary for SSA. The implication is that availability of proteins to perform DSB repair is limited, such that mutations or physiological conditions that alter the genomic distribution and/or timing of DSB formation may have unforseen pleiotropic consequences.
Results
Previous studies have shown that a DSB created by VDE (described as VDE-DSB1 below) can be repaired either through an interhomolog gene conversion event, or by using direct repeats that flank the DSB site, causing deletion of intervening sequences and creating “Δproduct” ([31]; Figure 1A). Both sister chromatids containing the VDE-recognition sequence are usually cleaved during meiosis, and tetrad analysis revealed both chromatids are frequently repaired to Δproduct, an outcome most compatible with repair by SSA [31]. The proportion of VDE-DSB1s repaired by interhomolog gene conversion versus repair using flanking repeated sequences, creating Δproduct, varied significantly between cells with mutations that either inhibited Spo11-DSB formation or single-strand resection at Spo11-DSBs. The former caused an increase in both the rate of VDE-DSB1 repair and the proportion of Δproduct, whereas the latter slowed repair and reduced the proportion of Δproduct. To further determine how the presence of Spo11-DSBs can influence VDE-DSB repair in trans, we have now investigated two VDE-DSB sites in dmc1Δ meiotic cells. Mutating DMC1 allows efficient Spo11-DSB formation and efficient single-strand resection, but Spo11-DSBs fail to repair due to an inability to form interhomolog joint molecules. Thus, in dmc1Δ cells the VDE-DSBs are in the context of multiple hyper-resected Spo11-DSBs.
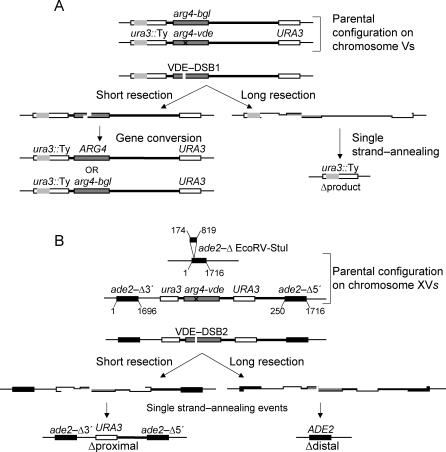
Figure 1. The arg4-vde-Containing Reporter Constructs.
(A) The ura3::arg4-vde reporter cassette containing the VDE-DSB1 site has been described previously [31]. This cassette is in a heterozygous state with a nearly identical insertion containing the arg4-bgl allele [69] on the opposite Chromosome V. Repair of the VDE-DSB1 is possible by gene conversion after short resection. Long resection of approximately 2 kb and 6.5 kb on the left and right of the VDE-DSB uncover flanking homology (URA3 sequences) that can be used for repair by SSA yielding Δproduct (the grey area within URA3::Ty is a naturally disrupting Ty element).
(B) The ade2::arg4-vde reporter cassette is hemizygous, inserted into one ADE2 locus. The opposite Chromosome XV has an internal deletion in the ADE2 locus. Resection of approximately 3.0 kb to both the left and right will uncover the proximal URA3 repeated DNA sequences. An SSA event between these yields Δproximal. Further resection to 4.5 kb and approximately 7.0 kb to the left and right will uncover the ADE2 repeated sequences; SSA between them yields Δdistal. Repair of VDE-DSB2 by gene conversion is unlikely as homology with the ade2Δ chromosome is over 3 kb and 7 kb away, on the left and right sides, respectively, leading to long nonhomologous 3′ ends.
In addition to the published reporter construct [31], we used a second VDE-DSB reporter construct, inserted on Chromosome XV. In this cassette there are two pairs of nested direct repeats flanking the VDE cleavage site (VDE-DSB2). The proximal repeats contain URA3 sequence; the distal repeats contain ADE2 sequence (Figure 1B). The second Chromosome XV has a 645-bp deletion from the ADE2 locus. No homology to repair VDE-DSB2 exists on the homologous chromosome for approximately 4 kb to the left and 7 kb to the right of VDE-DSB2, forcing DSB repair events to proceed through an intrachromosomal route, which we show is SSA.
The VDE-DSB1 Accumulates in Cells Lacking Dmc1 Function
In wild-type cells, VDE-DSB1 is repaired by both interhomolog gene conversion and SSA. We considered that knocking out DMC1 could have very different influences on VDE-DSB repair. On the one hand, RecA type strand invasion function is not required for SSA, and so in dmc1Δ cells there could have been efficient VDE-DSB repair by SSA, at the expense of gene conversion. On the other hand, in dmc1Δ cells the nucleus accumulates around 200 resecting Spo11-DSBs and hundreds of kilobases of ssDNA [5]. This might create a competition for resources (such as resection proteins or RPA) and therefore compromise VDE-DSB repair.
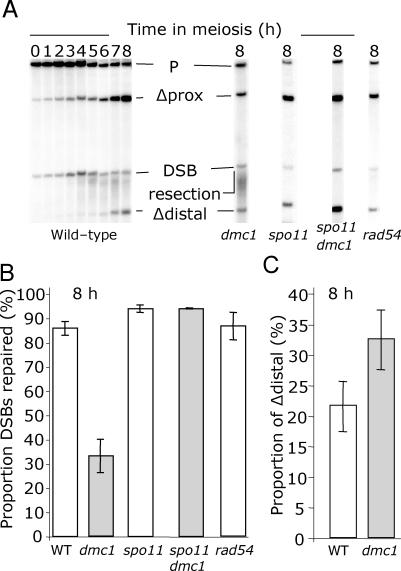
Using Southern analysis that isolates a parental arg4-vde DNA fragment from all other arg4-containing species, we determined that mutating DMC1 had no significant effect on the efficiency of forming VDE-DSB1 (Figure 2A and 2B). By contrast, mutating DMC1 causes a major delay in turnover of the VDE-DSB1 DNA (Figure 2C and 2D). In wild-type cells, the VDE-DSB1 signal accumulates up to 5 h after induction of meiosis and gradually disappears as the rate of DSB repair outpaces residual DSB formation. The VDE-DSB1 signal accumulated in dmc1Δ cells similarly to wild type up to 4 h after induction of meiosis, but did not diminish. Consistent with the observed accumulation of VDE-DSB1 molecules in dmc1Δ cells, very little Δproduct was created compared to wild-type cells (Figure 2E). As there is no significant difference in the kinetics of VDE-DSB1 formation in the different strains, the observed variation in VDE-DSB1 levels must be due to a difference in repair.
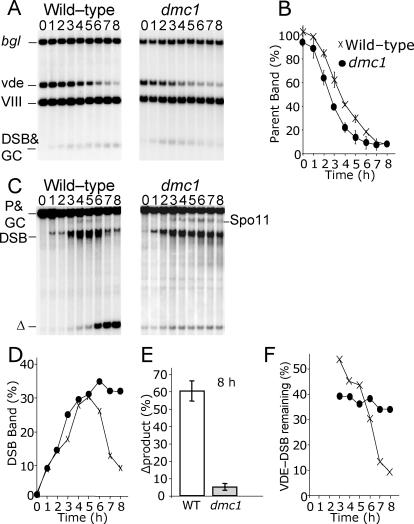
Figure 2. Repair of VDE-DSB Is Inhibited in dmc1 Cells.
(A) DNA from 0 h to 8 h of meiotic culture was digested with EcoRV and BglII and probed close to the VDE-DSB1 site to create a DNA fragment of unique size containing uncut parental arg4-vde DNA (bgl = parental arg4-bgl chromatids plus gene conversion products creating further arg4-bgl; vde = parental arg4-vde chromatids; VIII = arg4-nsp;bgl chromatids in both natural Chromosome VIII loci; DSB&GC= VDE-DSB1 molecules plus gene conversion products creating ARG4).
(B) Quantification of the parental arg4-vde band normalized to 50% of signal in band VIII. Diminution of the arg4-vde band is a consequence of VDE-DSB1 formation, which occurs at similar rates in wild-type and dmc1 cells.
(C) DNA was digested with SpeI and probed distal to the URA3 locus to isolate DNA fragments of unique size containing VDE-DSB1 molecules and Δproduct (P&GC = parental arg4-vde and arg4-bgl chromatids plus gene conversion products; DSB = chromatids with VDE-DSB1; Δ = Δproduct; Spo11 = natural Spo11-DSB site close to the arg4 insert).
(D) Quantification of VDE-DSB1 signal expressed as a proportion of arg4-vde-containing chromatids, symbols as in (B).
(E) Quantification of Δproduct expressed as a proportion of parental arg4-vde chromatids.
(F) Quantification of VDE-DSB1 signal expressed as a proportion of cumulative parental arg4-vde chromatids that have received a VDE-DSB1, symbols as in (B).
In dmc1Δ cells, the amount of VDE-DSB1 DNA detected on Southern blots was maintained from 4 h to 8 h at a steady level representing approximately 35% of chromatids (Figure 2F). However, by 8 h of meiosis, less than 10% of chromatids were detectable in the Δproduct band, even though approximately 90% of chromatids had been broken (Figure 2B). The discrepancy between these values suggested that standard Southern analysis fails to detect some VDE-DSB1 DNA. This could be due to extensive single-strand resection, which is known to cause Spo11-DSB bands to become diffuse when DNA is extracted from dmc1Δ cells [5].
Accumulated VDE-DSBs Are Resected
To test whether or not the VDE-DSB1 band was underrepresented due to diffuse smearing, we compared the amount of resected DNA that accumulates in wild-type and dmc1Δ cells using a loss-of-restriction site assay. This assay resolves DNA under denaturing conditions and quantitatively detects resected molecules as discrete bands by using a strand-specific probe (Figure 3A). In the wild-type culture after 8 h, approximately 11% of arg4-vde chromatids were detected in bands representing VDE-DSB1 resected DNA (Figure 3B). By contrast, 8 h after induction of meiosis in the dmc1Δ culture, 84% of VDE-DSB1 DNA was detected in the resection bands (Figure 3B). Since 90% of arg4-vde chromatids are cut by VDE by 8 h meiosis (Figure 2B), virtually all VDE-DSB1s in dmc1Δ cells persist in a resected, unrepaired state.
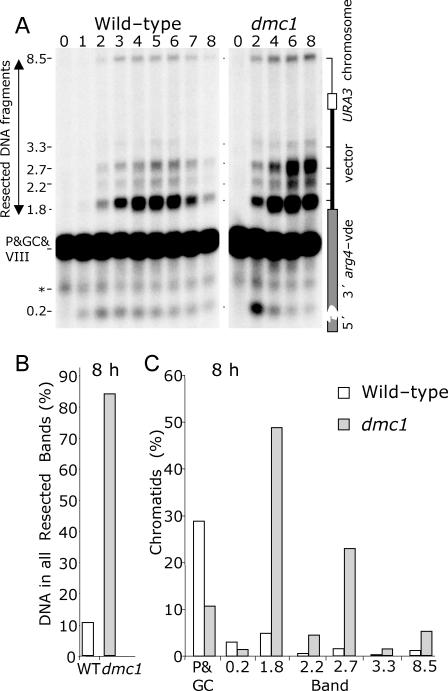
Figure 3. Most Broken arg4-vde-Containing Chromatids in dmc1 Cells Remain Unrepaired in a Resected State.
(A) DNA from 0 h to 8 h of meiotic culture DNA was digested with HaeII and fractionated by alkaline denaturing gel electrophoresis. Bands have increasing molecular weight as the restriction sites are progressively destroyed by resection. The numerals to the left indicate distance from the VDE-DSB1 site to an HaeII cut-site, and represent the maximum extent of resection for molecules in the respective bands (* = nonspecific band; other labels are as described in Figure 2; a maximum of 1/4 of the signal in the P&GC&VIII band represents the parental arg4-vde fragment; full details in Materials and Methods).
(B) Quantification of the resected bands totaled after 8 h of meiosis expressed as a proportion of parental arg4-vde chromatids. In dmc1Δ cells practically all VDE-DSB1s created remain in a resected state.
(C) Quantification of individual resected bands after 8 h of meiosis expressed as a proportion of parental arg4-vde chromatids. Accumulation of resected molecules is punctuated in dmc1Δ cells indicating likely pause sites through which resection is nonprocessive. (P&GC represents remaining parental chromatids plus gene conversion product; the numbers refer to band sizes).
Persistence of resected DNA around VDE-DSB1 is highly reminiscent of the behavior of Spo11-DSBs in dmc1Δ cells. We found most of the resected DNA appears in two apparent pause sites for resection (Figure 3A and 3C). One pause site, with resection up to 1.8 kb, is also present in earlier time points for wild-type cells, and in dmc1Δ cells contains approximately 49% of VDE-DSB1 chromatids at 8 h (Figure 3C). The second pause site, with up to 2.7 kb of resection, appears to be a major block to further processing in dmc1Δ cells with 23% of VDE-DSB1 chromatids collecting there by 8 h (Figure 3C). Repair of the VDE-DSB1 by SSA requires that resection extends as far as both flanking copies of the repeated sequence. To uncover the repeat sequence on the right side of VDE-DSB1 requires over 6 kb of resection. Thus, the difficulty in repairing the VDE-DSB1 by SSA may be caused by the inability to create sufficiently extensive resection tracts in the dmc1Δ cells.
Another important feature of this analysis is a 5-fold increase in the band representing resection between 3.3 kb and 8.5 kb in dmc1Δ cells, compared to wild-type (Figure 3A and 3C). This band is barely visible in wild-type cells, presumably due to rapid repair by SSA of DSBs with long resection tracts. Thus, even when there is sufficient resection for SSA, repair is inhibited in dmc1Δ cells.
Repair of VDE-DSB1 Is Not Dependent on Meiotic Progression
dmc1Δ cells arrest meiosis prior to exit from prophase, due to checkpoint-induced inactivation of the Ndt80 transcription factor [7,33–35]. To test the possibility that this arrest causes inhibition of resection and SSA, we repeated the assay in cells lacking NDT80 (Figure 4).
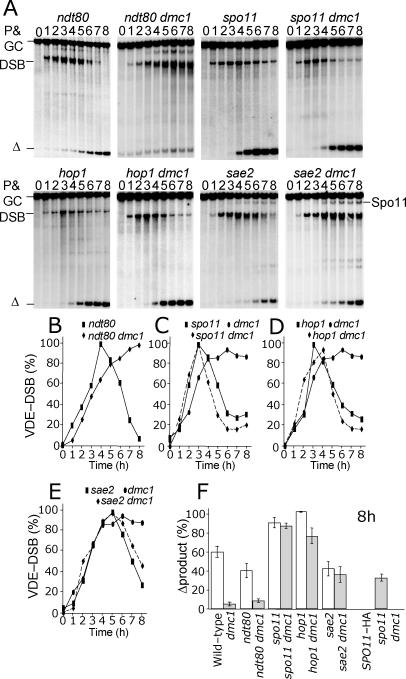
Figure 4. Meiotic Prophase Arrest Does Not Inhibit VDE-DSB1 Repair, but the Quantity and State of Spo11-DSBs Affects the Need for Dmc1.
In all cases spo11 refers to spo11-Y135F-HA3His6.
(A) DNA from 0 h to 8 h of meiotic culture was digested and displayed as described in Figure 2C.
(B) Cells do not require Ndt80 function to turnover the VDE-DSB in DMC1 cells, and dmc1Δ is epistatic to ndt80 for VDE-DSB1 repair. The repair defect imposed by loss of Dmc1 function is fully rescued by (C and D) removing all or most Spo11-DSB formation in either spo11 or hop1 mutant cells and by (E) inhibiting resection at Spo11-DSBs by mutating SAE2.
(F) The epistatic relationships between the various mutations and dmc1 are also displayed by quantification of Δproduct after 8 h of meiotic culture, expressed as a proportion of parental arg4-vde chromatids. Wild type and dmc1 are shown for comparison. Reducing the quantity of Spo11-DSBs to 50% of wild-type levels using a strain heterozygous for different SPO11 alleles leads to partial relief of Dmc1 dependence for VDE-DSB1 repair.
The kinetics of VDE-DSB1 induction and repair were similar in wild type and ndt80 meioses (Figure 4A and 4B). While ndt80 dmc1Δ mutants displayed the same defects in VDE-DSB1 repair as dmc1Δ cells, ndt80 cells exhibited amounts of Δproduct comparable to wild type (Figure 4F). Thus, the meiotic prophase arrest imposed by dmc1Δ is unlikely to be the cause of the inefficient VDE-DSB1 repair.
Mutants That Reduce the Cellular Load of Resected Spo11-DSBs Restore SSA of VDE-DSBs in dmc1Δ Cells
We next considered the possibility that VDE-DSB repair is reduced in dmc1Δ cells because of the large quantity of ssDNA produced by the resection of unrepaired Spo11-DSBs. To test this, we measured the efficiency of repairing VDE-DSB1 in dmc1Δ cells with few Spo11-DSBs (hop1Δ), completely lacking Spo11-DSBs (spo11-Y135F-HA3His6 [36], referred to as spo11, forthwith), and in sae2Δ cells with wild-type levels of Spo11-DSBs but no single-stranded resection (Figure 4A and 4C–4F).
We previously reported that mutation of HOP1, SPO11, or SAE2 alters the relative proportion of VDE-DSB1 molecules that repair using the flanking repeats versus repair by gene conversion. In that study, and here, we found that mutating HOP1 or SPO11 increased both the speed of repair and the proportion of VDE-DSB1 that repaired using flanking homology [31] (Figure 4A, 4C, 4D, and 4F). In sae2Δ cells, VDE-DSB1 repair is slower and the proportion of molecules that repair using flanking repeats is reduced, probably due to a direct role of Sae2 on resection, even at DSBs lacking covalently bound protein [32] (Figure 4A, 4E, and 4F) .
We now report that all the double mutants, spo11 dmc1Δ, hop1Δ dmc1Δ, and sae2Δ dmc1Δ behave almost the same as their DMC1 single mutant counterparts with respect to the kinetics and efficiency of VDE-DSB1 repair (Figure 4A and 4C–4F). In other words, the requirement for Dmc1 to repair a VDE-DSB using flanking homology is relieved by mutations that reduce the large quantity of resected Spo11-DSBs and ssDNA usually seen in dmc1Δ meiosis.
Lowering the Quantity of Resected Spo11-DSBs Reduces the Requirement for Dmc1 to Repair VDE-DSB1 by SSA
We next checked if intermediate levels of VDE-DSB repair could be achieved in dmc1Δ cells when intermediate levels of ssDNA accumulate in the nucleus. The number of Spo11-DSBs was reduced by using a heterozygous diploid expressing the hypomorphic SPO11-HA3His6 allele and the null allele, spo11-Y135F-HA3His6 [36]. This combination of SPO11 alleles is reported to reduce recombination and Spo11-DSB levels to approximately 50% of wild type [37]. We confirmed this to be the case in dmc1Δ cells by measuring DSB formation at the ARE1 hotspot (unpublished data). This reduction in Spo11-DSB formation increased the VDE-DSB1 repair efficiency in dmc1Δ cells approximately 6-fold (Figure 4F), supporting the contention that inefficient SSA repair of VDE-DSBs in dmc1Δ cells is related to accumulation of resecting Spo11-DSBs.
Dependency on Dmc1 for VDE-DSB1 Repair Does Not Reflect the Requirement for an Unexpected DNA Strand Invasion Event
Theoretically, VDE-DSB1 could create the Δproduct following an unequal strand exchange event with the homologous chromosome, which could be Dmc1-dependent in meiosis. It was important therefore to test repair of a VDE-DSB in a context that ruled out the possibility of Dmc1-dependent interhomolog repair. To test this, we used a second reporter cassette that contains the VDE cleavage site (VDE-DSB2) inserted on Chromosome XV between two pairs of nested direct repeats (Figure 1B). Repair of VDE-DSB2 by strand invasion with the homolog is precluded by large tracts of heterology present in the hemizygous reporter insert. Repair of VDE-DSB2 was also reduced by loss of DMC1, although not to the same extent as VDE-DSB1 (Figure 5A and 5B). By 8 h in the dmc1Δ culture, approximately 33% of VDE-DSB2 broken chromatids had repaired compared to 86% in wild type (Figure 5B). The repair level did not increase in dmc1Δ cells by 12 h (unpublished data). As for VDE-DSB1, repair of VDE-DSB2 was increased in dmc1Δ cells when the DSB forming function of Spo11 was removed (Figure 5B).
Figure 5. Southern Analysis of the VDE-DSB2 Cassette in a Hemizygous Context.
In all cases spo11 refers to spo11-Y135F-HA3His6.
(A) DNA from wild-type and mutant cells, as indicated, was digested with SpeI, which isolates unique fragments representing (P) the uncut parent arg4-vde chromatid, (Δprox) the product of SSA between URA3 proximal repeated sequences, (DSB) VDE-DSB2 molecules, and (Δdistal) the product of SSA between ADE2 distal repeated sequences. Cutting by VDE is less efficient in this assay, as indicated by the remaining parental DNA. In all but the dmc1Δ mutant cells, little VDE-DSB DNA is visible by 8 h. The amount of product accumulating is significantly reduced in dmc1Δ cells in which the VDE-DSB2 becomes smeared due to prolonged resection.
(B) The total amount of repair product (sum of Δdistal and Δproximal) visible has been measured as a proportion of arg4-vde-containing chromatids that have received a VDE-DSB by 8 h in meiosis (details in Materials and Methods).
(C) The quantity of Δdistal expressed as a proportion of sum of total repair product gives an indication of how often repaired molecules used the longer resection tract.
By mutating the RAD54 gene, which is important for strand exchange between sister chromatids, we confirmed our expectation that repair of VDE-DSB2 using flanking repeated sequences was not by unequal intersister repair [6,9,38,39]. For rad54 cells, there was a slight delay in meiosis indicated by delayed induction of VDE-DSB2 and late onset of the first division (unpublished data). Despite this delay, 87% of broken chromatids molecules were repaired by 8 h in rad54 meiosis (Figure 5B). Because VDE-DSB2 repair was as efficient in rad54 cells as in wild type, we conclude that repair using the flanking repeats does not require either a homolog or Rad54. In other words, VDE-DSB2 repair occurs by SSA, rather than via a mechanism that requires DNA strand invasion of a donor duplex.
Evidence that VDE-DSBs cannot repair using the sister chromatid in meiosis is supported by analysis of a his4-vde allele in a hybrid strain containing a Saccharomyces cerevisiae Chromosome III opposite a Saccharomyces carlsbergensis Chromosome III. Due to sequence divergence between the two chromosomes, interhomolog gene conversion is prohibited [40–42]. Since no repeats flank the his4 VDE-DSB, the only possible route to homologous repair is through invasion of the sister chromatid. However, all tetrads are two-spore viable due to loss of the his4-vde Chromosome III, indicating that intersister strand invasion and repair does not happen (K. Tittcomb and A. S. H. Goldman, unpublished data).
Resection at VDE-DSB2 Can Be Extensive in dmc1Δ Cells
Our analysis of resection intermediates at VDE-DSB1 suggested that DSB repair might be inhibited in dmc1Δ cells because of a failure to perform sufficient extensive ssDNA resection (Figure 4). Because VDE-DSB2 is flanked by nested direct repeats (Figure 1B), this site can be used to estimate how much resection had occurred in the repaired molecules. Repair by SSA using the proximal repeats (Δprox) creates a 5 kb deletion, detected on Southern blots as a 10 kb band. Repair using distal repeats (Δdistal) creates a 10.5 kb deletion and a 4.5 kb band (Figure 5A). By comparing the relative proportion of these repaired products, it is possible to estimate the proportion of molecules that repaired with longer resection tracts (Figure 5C). In wild-type cells, approximately 22% of repaired VDE-DSB2 molecules used the distal repeats. In dmc1Δ cells, the distal repeats were used more frequently, with 33% of repaired molecules being of the Δdistal type. Thus, the reduction in resection in dmc1Δ cells is not uniform across the population of cells, possibly reflecting a stochastic inhibition created by competition for resources.
Rfa1, a Component of the ssDNA Binding Complex RPA, Has Limited Access to the Repeated Sequences Flanking VDE-DSB1 in dmc1Δ Cells
The negative correlation between ability to repair the VDE-DSB by SSA and the number of DSBs undergoing extensive resection could reflect inability of ssDNA binding proteins to access homologous sequences flanking the VDE-DSBs. Failure of ssDNA binding proteins to bind DNA could result from a combination of reduced resection and ssDNA binding proteins being in limited supply at the VDE-DSBs, because they are sequestered to the long ssDNA tracts that accumulate at the many Spo11-DSBs present in the cell.
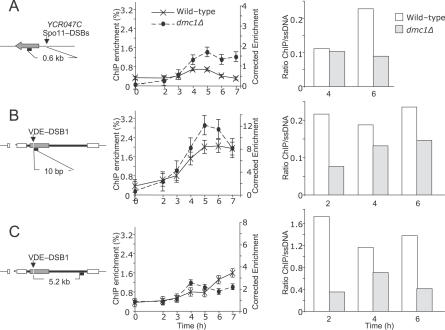
The ssDNA binding protein complex, RPA (also known as RF-A in yeast), is required to remove secondary structure in ssDNA and to recruit recombination proteins such as Rad52 during homologous recombination [43]. Using chromatin immunoprecipitation (ChIP), we compared (in wild-type and dmc1Δ cells) the association of an RPA component (Rfa1) to DNA coupled with a Spo11-DSB hotspot (YCR047C/BUD23 ORF, [44]) or with VDE-DSB1 (Figure 6A–6C). Close to the BUD23 Spo11-DSB hotspot, a small amount of DNA is enriched from wild-type cells by ChIP, the changing levels through time reflect the kinetics of appearance and disappearance of Spo11-DSBs at this site (Figure 6A). In dmc1Δ cells, the Rfa1 signal is not reduced in later time points because Spo11-DSBs are not repaired. Normally Spo11-DSBs are formed up to 6 h after induction of meiosis. Interestingly, Rfa1 does not accumulate to higher levels after 4 h as more Spo11-DSBs are created, perhaps because as Rfa1 becomes limiting it is competed away from a proportion of the sites close to the Spo11-DSB.
Figure 6. ChIP Reveals That RPA Is a Limiting Factor in dmc1Δ Cells.
(A–C) DNA associated with immunoprecipitated Rfa1, a component of RPA, was amplified and quantified by qPCR. The values are averages from duplicate experiments. Each diagram shows the distance of PCR primers from the relevant DSBs; grey arrow represents YCR047C, the cassette containing the VDE-DSB1 site is as in Figure 1A, the small black bars represent the position of the PCR products at the indicated distance from the DSB site. The left axis of the graphs show ChIP enrichment (i.e., ChIP signal relative to input signal; %); on the right axis the values have been corrected to account for overrepresentation of substrate in the input compared to the proportion of probed chromatids that can receive a DSB (see Materials and Methods). The bar charts show the ratio of corrected ChIP enrichment values to proportion of chromatids that are expected to contain ssDNA in the PCR-amplified region. In each bar graph the ratio of corrected ChIP enrichment to estimated levels of ssDNA are always higher in wild type compared to dmc1Δ, showing that the immunoprecipitation of Rfa1 is more efficient in wild-type cells. The difference in ChIP efficiencies for wild-type and dmc1Δ cells is greatest for the site far from VDE-DSB1, implying that as resection proceeds it becomes more difficult to compete for Rfa1, which may be stably bound to ssDNA created earlier, close to DSB sites.
(A) Close to a Spo11-DSB hotspot, the ChIP enrichment decreases with repair in wild-type cells but not in dmc1Δ cells.
(B) Close to VDE-DSB1, the ChIP enrichment in dmc1Δ cells is no higher than in wild-type, even though by 7 h around 80% of arg4-vde-containing chromatids are in a resected state (see Figures 2 and 3) and (C). Distant from VDE-DSB1, but close to a flanking repeated sequence used for SSA, again the ChIP enrichment in dmc1Δ cells is no higher than in wild type, even though significant amounts of DNA accumulate with resection beyond this point (see Figure 3).
We have already shown that almost all VDE-DSBs created (in close to 100% of cells; Figure 2B) in dmc1Δ remain broken and in a resected state throughout the time course (Figure 3). We reasoned that if Rfa1 coats all ssDNA in dmc1Δ cells, the Rfa1 ChIP signal should be higher in dmc1Δ cells versus wild type.
Using two different primer sets, one close to the VDE-DSB1 (Figure 6B) and one close to a flanking repeated sequence (Figure 6C), we found that the ChIP signal was not significantly enhanced in dmc1Δ cells close to the VDE-DSB and is slightly reduced far from the VDE-DSB1. Taken together, these results support the view that when Spo11-DSBs remain in a resected state, Rfa1 does not accumulate at specific ssDNA sites, even though the quantity of ssDNA is demonstrably increasing (Figure 3). Thus, for those cells, in which resection does uncover the repeated sequences flanking the VDE-DSB1, there may not be sufficient RPA to mediate repair by SSA.
We noted that the ChIP signal is not significantly reduced by 7 h in wild-type cells, when a large proportion of VDE-DSB1s are repaired. In part this is probably because 40% of the peak numbers of VDE-DSB1s are still present at this time (Figure 2D). It is also possible that RPA is not removed as rapidly from the DNA following SSA as it would be following classical homologous recombination.
Discussion
In budding yeast, repair of DSBs induced by VDE is a natural process, which propagates VDE-containing genetic elements from one chromosome to another during meiosis [45]. Previous studies have shown the timing of DSB induction by VDE and the mechanisms of repair parallel closely those of Spo11-induced recombination [28,31]. Deleting DMC1 is expected to prevent gene conversion events at VDE-DSB sites, but in addition we have found it also inhibits repair by SSA, a process that in mitosis neither requires strand invasion nor the RecA ortholog Rad51 [17].
The failure in VDE-DSB repair in dmc1Δ mutant cells can be relieved by mutations that eliminate Spo11-DSBs, reduce their frequency, or prevent their resection. Furthermore, in dmc1Δ cells resection at VDE-DSBs is reduced, and Rfa1 (a component of the ssDNA binding complex, RPA) is unable to access the repeated sequences flanking the VDE-DSB sites.
Repair Proteins Are a Limiting Factor
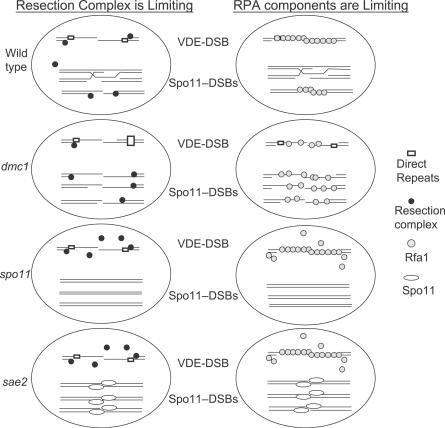
Losing Dmc1 function has an enormous impact on the normal balance of DNA transactions taking place in the meiotic nucleus. Spo11-DSBs are formed with wild-type kinetics, but breaks remain unrepaired and accumulate genome-wide, so that at later time points each cell would contain about 200 lesions [5]. Under these conditions, resection continues for many hundreds or thousands of nucleotides further than normal, and therefore the demand for both resection complex and RPA is likely to be extremely high and critical (Figure 7).
Figure 7. Limited Availability of DNA Repair Proteins Explains the Requirement of Dmc1 for SSA at VDE-DSBs.
In wild-type meiosis there is sufficient resection complex and Rfa1 to create and bind to long tracts of ssDNA at the VDE-DSB so that SSA is possible. In part, the ready supply of such proteins is likely created by the asynchronous nature of Spo11-DSB formation and repair in the nucleus, thus when some Spo11-DSBs are using these proteins others may have moved to a biochemical step that allows their release. In dmc1Δ cells, so much resection complex and Rfa1 is sequestered to multiple unprepared Spo11-DSBs that insufficient resection complex is available to create long resection tracts at the VDE-DSB; or in cases where long resection tracts appear there is not enough free Rfa1 to bind the repeated sequences. Mutating either SPO11 or SAE2 relaxes the demand on both resection proteins and ssDNA binding proteins such that resection and repair of the VDE-DSB is no longer limiting.
The data from two VDE-DSB-containing cassettes support the idea that either resection complex or ssDNA binding proteins can become limiting. Insufficient access to resection complex could reduce resection below the lengths needed to render flanking repeats single-stranded. Insufficient RPA to coat long resection tracts could prevent SSA even if resection has uncovered the repeated sequences. Whether a protein complex becomes limiting at a particular DSB may be stochastic, though genome location and immediate environment may also be important. This would explain why at VDE-DSB1 resection appears extremely limited, but less so at VDE-DSB2, which also repairs poorly but more efficiently than VDE-DSB1.
The coordinate induction and repair of DNA damage is a vital part of the meiotic developmental pathway. Ensuring that the many proteins required for DSB repair are in appropriate supply and in active form is a major task for the cell. Spo11-DSB formation is temporally linked to replication, with DSBs appearing about 1.5 h after replication has passed through [46]. Thus, like replication, the formation of Spo11-DSBs across the genome is asynchronous. In between Spo11-DSB formation and repair, Spo11-DSBs are processed by 5′ to 3′ resection. Resulting ssDNA subsequently forms joint molecules with the homologous chromosome, and DSBs are repaired 1–1.5 h after Spo11-DSB formation [47,48]. It is reasonable to assume that by this time resection would have ceased and ssDNA-binding complexes such as RPA and Dmc1/Rad51 may be liberated for reuse. Since a large number of such molecules are required at every DSB, the asynchronous induction of recombination may have evolved to spread the workload of proteins, which otherwise would have to be produced at levels that might result in pleiotropic negative consequences.
There is precedence for proteins involved in nucleic acid metabolism being in limiting supply to avoid pleiotropic effects from over abundance. Activity of ribonucleotide reductases (RNRs) is tightly controlled to ensure nucleotide pools are sustained at appropriate levels. Control is maintained at the transcriptional level and by an inhibitor protein, Sml1 ([49] and within). The Mec1 DNA damage response pathway, acting through Rad53 and Dun1, regulates both controls. DNA damage increases transcription of RNR genes and phosphorylates Sml1 causing its inactivation [50,51]. Regulation of active RNR protein supply is so tight that mutations in the Mec1 kinase cascade cause SML1 expression to become toxic, due to insufficiency of nucleotides needed for routine DNA repair [50,52,53]. Stringent control of RNRs probably reflects the fact that overabundance of nucleotides can be mutagenic due to increased risk of misincorporation.
Our data suggest that ssDNA binding proteins must also be kept in limiting supply. This suggestion fits well with the fact that the transcription regulation of RPA components is linked to the cell cycle, peaking at the G1/S boundary [54]. Furthermore, reports on in vitro DNA binding and activity of Rad52 and Rad51 indicate that RPA at low concentrations promotes strand exchange, yet at high concentrations it limits access of repair proteins to the DNA [55–57].
Limited Supply of Repair Proteins Could Have Wide-Ranging Impact on Regulating Meiotic Processes
That Rfa1 can become a limiting factor in an experimental situation raises the possibility that in wild-type cells perturbations of repair efficiency or replication could have indirect effects on other repair/replication processes. Such effects can be at sites other than the original lesion. For example, a level of DNA damage similar to that experienced by meiotic cells might perturb DNA replication, because damage repair sequesters protein factors such as RPA that are also critical for replication.
One reason for limiting the number of Spo11-DSBs in yeast meiosis could be to ensure that a safe and sustainable balance can be reached between repair protein supply and demand within a suitable time frame. Studies on mammalian and yeast vegetative cells support the view that homologous recombination is extremely sensitive to protein supply, as overexpression of Rad52 epistasis group proteins can inhibit repair of an experimentally induced DSB [58,59]. Consistent with the notion that protein supply and demand is finely balanced in meiosis, moderately hypomorphic mutants of DSB repair genes such as MRE11, NBS1, or RAD51C have significantly reduced fertility in mice due to inefficient DSB repair [60,61]. Similarly in yeast meiosis, deleting one RecA ortholog, DMC1, causes a complete block in DSB repair that can be largely rescued by either overexpressing or releasing inhibition of RAD51 [62,63].
It was recently shown that synchrony of the first meiotic division could be influenced by temporary chemical inhibition of a specific Cdc7 activity required for induction of Spo11-DSBs [64]. Nearly half of the population underwent the first meiotic division sometime between 3 h and 4 h after inhibitor wash out, with no loss of viability. This might seem to counter our argument that repair proteins are limiting. But across the population at the single Spo11-DSB analyzed, break induction was still spread over some hours. Thus, while the Wan et al. data [64] demonstrate some improved synchrony of the first division across a population of cells, it does not directly address the synchrony of DSB formation within each cell. Further analysis of highly synchronized populations, in which the time of inducing Spo11-DSBs and their life spans is well defined, will help to clarify the time limits within which Spo11-DSB formation must be limited to avoid problems of repair protein supply.
Limiting protein supply could serve a useful function other than protecting against ill effects of overproduction, such as to direct the proportion of events that take one biochemical pathway rather than another [58]. One possible example of this comes from maize, which produces more than 20-fold excess of DSBs during meiosis compared to the known crossover frequencies [65]. These DSBs are identified cytologically as Rad51 foci that often appear as opposite pairs on homologous chromosomes and may be used as pairing sites [65,66]. What prevents a much higher proportion of DSBs in maize meiosis from becoming crossovers is unknown. Perhaps a protein required for crossover-associated repair is in limited supply, so the majority of DSBs can serve a function other than forming crossovers.
The impact single mutations have on biochemistry, nucleus-wide, is rarely known. This study highlights the fact that indirect effects can easily arise from changing the balance of protein supply and substrate in the nucleus. In nature, tight control of protein supply may be essential for avoiding pleiotropic effects from oversupply, or for limiting the number of events passing down a specific pathway. In the laboratory, phenotypes ascribed to mutations may often not be caused by a direct biochemical impact of mutating a gene, but may be due to broader biochemical imbalances created across the nucleus. Genes involved in DNA metabolism may be particularly likely to cause pleiotropic phenotypes. Mutations altering the amount of damage present in a nucleus are certainly prone to pleiotropic effects that are worth serious consideration when defining protein function.
Materials and Methods
Media, genetic methods, and strains.
Diploid yeast strains of the SK1 background relevant genotypes are listed in Table S1.
The ura3::arg4-vde cassette (Figure 1A) was created as described previously in [31]. The ade2::arg4-vde reporter cassette (Figure 1B) was created by transforming parent strains with pAG408, a derivative of pBR322 based pAG137 [31]. The URA3 of pAG137 was modified by a 5′ (−17 to +129) deletion between restriction sites SdaI to XcmI. A functional URA3 (HindIII fragment) was inserted into an NruI site in the PBR322 backbone. A section of the ADE2 ORF (+250 to +1695) generated by PCR from yeast genomic DNA was inserted into the pBR322 EcoRI site and used for integration into the yeast genome following linearization at the AflII site.
Relevant mutant strains containing either the ura3::arg4-vde or ade2::arg4-vde reporter cassettes were made by mating and dissection with appropriate SK1 haploids. The source of the spo11-Y135F-HA3His6; hop1; sae2Δ; and ndt80 haploids is reported in [31].
For the ura3::arg4-vde reporter construct, dmc1 disruption was made in pAG64 using primers 5′-gccattctatgtctgatcccgg-3′ and 5′-tcgcttagttcacctctaccgc-3′ to amplify a 1,466-bp region of the dmc1 locus from haploid SK1 genomic DNA. MfeI-cleaved PCR product was ligated into EcoRI-linearized pUC19. A 2.2 kb ADE2-containing BglII fragment of pAG52 ( pMJ412 from M. Lichten) was ligated at the BglII site located 90 bp inside the DMC1 ORF. For the ade::arg4-vde reporter cassett, dmc1::ARG4 was obtained from D. Bishop in SK1 and was crossed into our experimental strains. The rad54Δ mutation was obtained from D. Bishop and transformed into relevant strains.
DNA isolation and Southern blot analysis.
40 ml samples of culture were removed at hourly intervals and processed for storage and DNA isolation according to Allers and Lichten (2001); hexamine cobalt (III) chloride was excluded from solutions.
Restriction endonuclease–digested DNA was separated under native conditions or denaturing conditions. Separated DNA was blotted to Zetaprobe membrane (Bio-Rad) under denaturing conditions with a Vacugene-XL system. Analysis of VDE-DSB1 Δproduct and resection intermediates was undertaken as described [31]. For VDE-DSB2 the probe used to display SpeI- digested DNA after native separation is specific to Chromosome XV coordinates 566120–566811.
Quantification and calculations for Southern analyses.
Quantification was as described [31]; briefly, for VDE-DSB1 the amount of DNA in the VDE-DSB1 band and Δproduct were determined as a proportion of arg4-vde-containing chromatids by dividing the signal in each band by half of total signal (which represents both arg4-vde and arg4-bgl chromatids). For the denaturing gels, the amount of signal attributable to chromatids that contained parental arg4-vde insert was calculated taking into account the presence of signal from six other chromatids (two chromatids with arg4-bgl on the homologous Chromosome V and four chromatids with arg4-nsp,bgl at the natural ARG4 locus on both Chromosome VIII homologues) and the fact that chromatids repairing to Δproduct do not contribute signal.
If;
Δr = Recorded proportion of arg4-vde chromatids repaired by SSA
Sr = Signal recorded in lane on denaturing gel
Δm = Δproduct missing from lane on denaturing gel
Rr = Resection band recorded signal on denaturing gel
1/4 of total possible signal comes from arg4-vde chromatids
Then
Δm = 0.25ΔrSr/(1−0.25Δr)
Therefore signal calculated in lane attributable to arg4-vde chromatids;
Sc = 0.25(Δm+Sr)
Then proportion of arg4-vde chromatids in each resection band;
Rc = Rr/Sc
For VDE-DSB2 gels, the DNA in each band was quantified.
If
T = Total signal in lane
P = Signal in parent band
Δprox = signal in Δprox band
Δdistal = signal in Δdistal band
Then DNA repaired, as a proportion of breaks made equals (Δprox + Δdistal)/(T − P) and the proportion of DNA repaired to Δdistal equals Δdistal/(Δprox + Δdistal).
Rfa1 ChIP.
20 ml cells (4 × 109 cells) were treated with 1% fresh formaldehyde for 15 min at room temperature and 125 mM glycine for 5 min. ChIP was performed as described [67] using magnetic protein G beads (Dynal) and a polyclonal rabbit anti-Rfa1 antibody (supplied by S. Brill). Enrichment of DNA bound by RPA was estimated by quantitative PCR using an Applied Biosystems 7500 Real-Time PCR system with 0.4 μM primers, SYBR Green PCR master mix (Applied Biosystems), and the PCR program: 95 °C for 15 s; 60 °C for 1 min; 40 cycles. The following primers were used: YCR047c; 5′-TATGTCGTCCACCTGGTCGTCG-3′ and 5′-TCCTAAACAGCGGTTGATGAGG-3′ 10 bp from VDE-DSB1; 5′-GCGAATGAAAGACGTCTTGG-3′ and 5′-CGGCCCTCTTAATTAGAACTTC-3′ 5.2 kb from VDE-DSB1; 5′ -CGCACATTTCCCCGAAAA-3′ and 5′-TGAAGACGAAAGGGCCTCG-3′.
Primers close to the YCR047C promoter regions were used to measure recovery of Spo11 DSB-associated sequences. The approximate distance from the PCR product to the nearest DSB is 0.6 kb (Buhler et al., unpublished data). A dilution series of genomic DNA from dAG206 strain was used to establish a standard curve. The ChIP enrichment was calculated as percentage of the target locus present in the immunoprecipitated sample relative to the amount in the starting input material. Corrected ChIP enrichment (Figure 6) was calculated based on estimates of the maximum proportion of probed chromatids likely to receive a DSB (Pmax). At YCR047C this value is 0.12 [68]. For VDE-DSB1 the correction value accounts for knowledge that up to 95% (Figure 2B) of all arg4-vde chromatids can receive a VDE-DSB1, and the proportion of chromatids with homology to the PCR primers and containing the arg4-vde allele varies with primer site. For primers close to and far from VDE-DSB1, the respective proportions of input chromatids containing the arg4-vde allele are 0.25 and 0.50 creating Pmax values of 0.24 and 0.48. The corrected ChIP enrichment is the original ChIP enrichment value/Pmax and thus reports on the ChIP enrichment as a proportion of chromatids that will receive a DSB during the time course.
To determine the ratio between corrected ChIP enrichment and proportion of chromatids expected to be single-stranded at the primer site, ssDNA values close to the Spo11-DSB at YCR047C were taken to be equal to the proportion of DSBs visible by Southern analyses in wild-type and dmc1Δ strains [68] (unpublished data). For the 10 bp from VDE-DSB1, the proportion of chromatids with ssDNA close to the break site was taken as the total proportion of arg4-vde chromatids present in resection bands at the relevant time points (Figure 3A; unpublished data). For the 5.2 kb from VDE-DSB1, the proportion of chromatids with ssDNA 8.5 kb from the break site was calculated (Figure 3A; unpublished data) and used as a conservative estimate of the proportion of chromatids that would be single-stranded where the primers lie. The DNA used to determine the proportion of chromatids with ssDNA was derived from different time courses used in ChIP experiments.
Supporting Information
(21 KB XLS)
Accession Numbers
The accession numbers from the NCBI (http://www.ncbi.nlm.nih.gov) gene database for genes discussed in this paper are DMC1 (856926), HOP1 (854738), MEK1 (854533), NDT80 (−856524), RAD54 (852713), RFA1 (851266), RED1 (850968), SAE2 (852700), and SPO11 (856364).
Acknowledgments
We thank members of the Goldman lab, Enrique (Fadri) Martinez-Perez, anonymous referees, and the editor for comments that improved the manuscript; Douglas Bishop for strains and plasmids; and Steven Brill for anti-Rfa1 antibody.
Abbreviations
- ChIP
chromatin immunoprecipitation
- DSB
double-strand break
- RNR
ribonucleotide reductase
- RPA
replication protein A
- SSA
single-strand annealing
- ssDNA
single-strand DNA
- VDE
VMA1-derived endonuclease
Footnotes
¤a Current address: Helen L. and Martin S. Kimmel Center for Biology and Medicine, Skirball Institute for Biomolecular Medicine and Department of Pathology, New York University School of Medicine, New York, New York, United States of America
¤b Current address: Molecular Biology Program, Memorial Sloan-Kettering Cancer Center, New York, New York, United States of America
¤c Current address: Department of Nutritional Sciences and Toxicology, University of California Berkeley, Berkeley, California, United States of America
¤d Current address: Tumour Microcirculation Group, Royal Hallamshire Hospital, University of Sheffield, Sheffield, United Kingdom
A previous version of this article appeared as an Early Online Release on October 24, 2007 (doi:10.1371/journal.pgen.0030223.eor).
Author contributions. ASHG conceived and designed the experiments. RJ, VB, MJN, AB-B, and MN performed the experiments. SH performed the preliminary ChIP experiments. RJ, VB, MJN, AB-B, MN, and ASHG analyzed the data. AN contributed reagents/materials/analysis tools. ASHG wrote the paper with contributions from RJ, VB, MJN, and AN.
Funding. This work was funded by Yorkshire Cancer Research and BBSRC grants to ASHG and the Agence Nationale pour la Recherche (06-BLAN-0120–01) to AN.
Competing interests. The authors have declared that no competing interests exist.
References
- Petronczki M, Siomos MF, Nasmyth K. Un menage a quatre: the molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Sun H, Treco D, Schultes NP, Szostak JW. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338:87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: a meiosis-specific yeast homolog of E. coli recA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Niu H, Wan L, Baumgartner B, Schaefer D, Loidl J, et al. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol Biol Cell. 2005;16:5804–5818. doi: 10.1091/mbc.E05-05-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Weiner BM, Kleckner N. Meiotic cells monitor the status of the interhomolog recombination complex. Genes Dev. 1997;11:106–118. doi: 10.1101/gad.11.1.106. [DOI] [PubMed] [Google Scholar]
- Thompson DA, Stahl FW. Genetic control of recombination partner preference in yeast meiosis. Isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics. 1999;153:621–641. doi: 10.1093/genetics/153.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Nikolski Y, Oshiro J, Chon J, Shinohara M, et al. High copy number suppression of the meiotic arrest caused by a dmc1 mutation: REC114 imposes an early recombination block and RAD54 promotes a DMC1-independent DSB repair pathway. Genes Cells. 1999;4:425–444. doi: 10.1046/j.1365-2443.1999.00273.x. [DOI] [PubMed] [Google Scholar]
- Bailis JM, Roeder GS. Synaptonemal complex morphogenesis and sister-chromatid cohesion require Mek1-dependent phosphorylation of a meiotic chromosomal protein. Genes Dev. 1998;12:3551–3563. doi: 10.1101/gad.12.22.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AV, Roeder GS. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol. 1997;136:957–967. doi: 10.1083/jcb.136.5.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Byers B. HOP1: a yeast meiotic pairing gene. Genetics. 1989;121:445–462. doi: 10.1093/genetics/121.3.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. RED1: a yeast gene required for the segregation of chromosomes during the reductional division of meiosis. Proc Natl Acad Sci U S A. 1988;85:6057–6061. doi: 10.1073/pnas.85.16.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin FL, Sperle K, Sternberg N. Model for homologous recombination during transfer of DNA into mouse L cells: role for DNA ends in the recombination process. Mol Cell Biol. 1984;4:1020–1034. doi: 10.1128/mcb.4.6.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozenberger BA, Roeder GS. A unique pathway of double-strand break repair operates in tandemly repeated genes. Mol Cell Biol. 1991;11:1222–1231. doi: 10.1128/mcb.11.3.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Haber JE. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae . Mol Cell Biol. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, Fishman-Lobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae . Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Haber JE. Characterization of double-strand break-induced recombination: homology requirements and single-stranded DNA formation. Mol Cell Biol. 1992;12:563–575. doi: 10.1128/mcb.12.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Shinohara M, Ohta T, Matsuda S, Ogawa T. Rad52 forms ring structures and co-operates with RPA in single-strand DNA annealing. Genes Cells. 1998;3:145–156. doi: 10.1046/j.1365-2443.1998.00176.x. [DOI] [PubMed] [Google Scholar]
- Moreau S, Morgan EA, Symington LS. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1, and Rad27 nucleases in DNA metabolism. Genetics. 2001;159:1423–1433. doi: 10.1093/genetics/159.4.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Symington LS. The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double-strand breaks. Mol Cell Biol. 2004;24:9682–9694. doi: 10.1128/MCB.24.21.9682-9694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazanehdari KA, Borts RH. EXO1 and MSH4 differentially affect crossing over and segregation. Chromosoma. 2000;109:94–102. doi: 10.1007/s004120050416. [DOI] [PubMed] [Google Scholar]
- Umezu K, Sugawara N, Chen C, Haber JE, Kolodner RD. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wu TC, Lichten M. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 1995;14:4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta K, Shibata T, Nicolas A. Changes in chromatin structure at recombination initiation sites during yeast meiosis. EMBO J. 1994;13:5754–5763. doi: 10.1002/j.1460-2075.1994.tb06913.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto S, Mizutani R, Satow Y, Kawasaki M, Ohya Y, et al. Recognition and cleavage of double-stranded DNA by yeast VMA1-derived endonuclease. Nucleic Acids Symp Ser. 1999. pp. 197–198. [DOI] [PubMed]
- Nickoloff JA, Chen EY, Heffron F. A 24-bp DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc Natl Acad Sci U S A. 1986;83:7831–7835. doi: 10.1073/pnas.83.20.7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Nogami S, Ohya Y. VDE-initiated intein homing in Saccharomyces cerevisiae proceeds in a meiotic recombination-like manner. Genes Cells. 2003;8:587–602. doi: 10.1046/j.1365-2443.2003.00659.x. [DOI] [PubMed] [Google Scholar]
- Malkova A, Klein F, Leung WY, Haber JE. HO endonuclease-induced recombination in yeast meiosis resembles Spo11-induced events. Proc Natl Acad Sci U S A. 2000;97:14500–14505. doi: 10.1073/pnas.97.26.14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Ohya Y. Recruitment of RecA homologs Dmc1p and Rad51p to the double-strand break repair site initiated by meiosis-specific endonuclease VDE (PI-SceI) Mol Genet Genomics. 2006;275:204–214. doi: 10.1007/s00438-005-0078-4. [DOI] [PubMed] [Google Scholar]
- Neale MJ, Ramachandran M, Trelles-Sticken E, Scherthan H, Goldman ASH. Wild-type levels of Spo11-induced DSBs are required for normal single-strand resection during meiosis. Mol Cell. 2002;9:835–846. doi: 10.1016/s1097-2765(02)00498-7. [DOI] [PubMed] [Google Scholar]
- Clerici M, Mantiero D, Lucchini G, Longhese MP. The Saccharomyces cerevisiae Sae2 protein promotes resection and bridging of double-strand break ends. J Biol Chem. 2005;280:38631–38638. doi: 10.1074/jbc.M508339200. [DOI] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae . Mol Cell Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, et al. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- Lydall D, Nikolsky Y, Bishop DK, Weinert T. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature. 1996;383:840–843. doi: 10.1038/383840a0. [DOI] [PubMed] [Google Scholar]
- Diaz RL, Alcid AD, Berger JM, Keeney S. Identification of residues in yeast Spo11p critical for meiotic DNA double-strand break formation. Mol Cell Biol. 2002;22:1106–1115. doi: 10.1128/MCB.22.4.1106-1115.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson KA, Keeney S. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc Natl Acad Sci U S A. 2004;101:4519–4524. doi: 10.1073/pnas.0400843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbel A, Zenvirth D, Simchen G. Sister chromatid-based DNA repair is mediated by RAD54, not by DMC1 or TID1 . EMBO J. 1999;18:2648–2658. doi: 10.1093/emboj/18.9.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Shita-Yamaguchi E, Buerstedde JM, Shinagawa H, Ogawa H, et al. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics. 1997;147:1545–1556. doi: 10.1093/genetics/147.4.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick MA, Zgaga Z, Hieter P, Westmoreland J, Fogel S, et al. Recombinant repair of diverged DNAs: a study of homoeologous chromosomes and mammalian YACs in yeast. Mol Gen Genet. 1992;234:65–73. doi: 10.1007/BF00272346. [DOI] [PubMed] [Google Scholar]
- Nilsson-Tillgren T, Gjermansen C, Holmberg S, Peterson JGL, Kielland-Brandt MC. Analysis of Chromosome V and the ILV1 gene from Saccharomyces carlsebergensis . Carlsberg Res Commun. 1986;51:309–326. [Google Scholar]
- Kielland-Brandt MC, Gjermansen C, Nilsson-Tillgren T, Peterson JGL, Holmberg S. Genetic differences between Saccharomyces carlsebergensis and S. cerevisiae. Analysis of Chromosome III by single chromosome transfer. Carlsberg Res Commun. 1981;46:65–76. [Google Scholar]
- Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- Goldway M, Sherman A, Zenvirth D, Arbel T, Simchen G. A short chromosomal region with major roles in yeast Chromosome III meiotic disjunction, recombination, and double- strand breaks. Genetics. 1993;133:159–169. doi: 10.1093/genetics/133.2.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda T, Nagai Y, Ohya Y. Molecular mechanism of VDE-initiated intein homing in yeast nuclear genome. Adv Biophys. 2004;38:215–232. [PubMed] [Google Scholar]
- Borde V, Goldman ASH, Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290:806–809. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of joint molecules that form frequently between homologs but rarely between sister chromatids during yeast meiosis. Cell. 1994;76:51–63. doi: 10.1016/0092-8674(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Chabes A, Domkin V, Thelander L. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J Biol Chem. 1999;274:36679–36683. doi: 10.1074/jbc.274.51.36679. [DOI] [PubMed] [Google Scholar]
- Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci U S A. 2002;99:3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Chabes A, Domkin V, Thelander L, Rothstein R. The ribonucleotide reductase inhibitor Sml1 is a new target of the Mec1/Rad53 kinase cascade during growth and in response to DNA damage. EMBO J. 2001;20:3544–3553. doi: 10.1093/emboj/20.13.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc A, Merrill GF. Checkpoint deficient rad53–11 yeast cannot accumulate dNTPs in response to DNA damage. Biochem Biophys Res Commun. 2007;353:527–530. doi: 10.1016/j.bbrc.2006.12.049. [DOI] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- Brill SJ, Stillman B. Replication factor-A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev. 1991;5:1589–1600. doi: 10.1101/gad.5.9.1589. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Sung P. Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J Biol Chem. 1997;272:28194–28197. doi: 10.1074/jbc.272.45.28194. [DOI] [PubMed] [Google Scholar]
- Kim PM, Paffett KS, Solinger JA, Heyer WD, Nickoloff JA. Spontaneous and double-strand break-induced recombination, and gene conversion tract lengths, are differentially affected by overexpression of wild-type or ATPase-defective yeast Rad54. Nucleic Acids Res. 2002;30:2727–2735. doi: 10.1093/nar/gkf413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim PM, Allen C, Wagener BM, Shen Z, Nickoloff JA. Overexpression of human RAD51 and RAD52 reduces double-strand break-induced homologous recombination in mammalian cells. Nucleic Acids Res. 2001;29:4352–4360. doi: 10.1093/nar/29.21.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov S, Pellegrini M, Shuda K, Fernandez-Capetillo O, Liu Y, et al. RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J Cell Biol. 2007;176:581–592. doi: 10.1083/jcb.200608130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry SM, Adelman CA, Theunissen JW, Hassold TJ, Hunt PA, et al. The Mre11 complex influences DNA repair, synapsis, and crossing over in murine meiosis. Curr Biol. 2007;17:373–378. doi: 10.1016/j.cub.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS. Budding yeast Hed1 downregulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 2006;20:1766–1775. doi: 10.1101/gad.1422506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi H, Roeder GS. The importance of genetic recombination for fidelity of chromosome pairing in meiosis. Dev Cell. 2003;5:915–925. doi: 10.1016/s1534-5807(03)00357-5. [DOI] [PubMed] [Google Scholar]
- Wan L, Zhang C, Shokat KM, Hollingsworth NM. Chemical inactivation of Cdc7 kinase in budding yeast results in a reversible arrest that allows efficient cell synchronization prior to meiotic recombination. Genetics. 2006;174:1767–1774. doi: 10.1534/genetics.106.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin AE, McElver J, Sunjevaric I, Rothstein R, Bowen B, et al. Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell. 1999;11:809–824. doi: 10.1105/tpc.11.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski WP, Golubovskaya IN, Cande WZ. Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell. 2003;15:1807–1816. doi: 10.1105/tpc.012898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieler S, Penkner A, Borde V, Klein F. The control of Spo11′s interaction with meiotic recombination hotspots. Genes Dev. 2005;19:255–269. doi: 10.1101/gad.321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borde V, Lin W, Novikov E, Petrini JH, Lichten M, et al. Association of Mre11p with double-strand break sites during yeast meiosis. Mol Cell. 2004;13:389–401. doi: 10.1016/s1097-2765(04)00034-6. [DOI] [PubMed] [Google Scholar]
- Wu TC, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae . Genetics. 1995;140:55–66. doi: 10.1093/genetics/140.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(21 KB XLS)