Abstract
Focal adhesions (FAs) regulate cell migration. Vinculin, with its many potential binding partners, can interconnect signals in FAs. Despite the well-characterized structure of vinculin, the molecular mechanisms underlying its action have remained unclear. Here, using vinculin mutants, we separate the vinculin head and tail regions into distinct functional domains. We show that the vinculin head regulates integrin dynamics and clustering and the tail regulates the link to the mechanotransduction force machinery. The expression of vinculin constructs with unmasked binding sites in the head and tail regions induces dramatic FA growth, which is mediated by their direct interaction with talin. This interaction leads to clustering of activated integrin and an increase in integrin residency time in FAs. Surprisingly, paxillin recruitment, induced by active vinculin constructs, occurs independently of its potential binding site in the vinculin tail. The vinculin tail, however, is responsible for the functional link of FAs to the actin cytoskeleton. We propose a new model that explains how vinculin orchestrates FAs.
Introduction
Cell migration is coordinated by a complex of proteins that localizes to sites of cell–matrix interaction, the focal adhesions (FAs; Critchley, 2000; Geiger et al., 2001). The adaptor protein vinculin is a key regulator of FAs (Jockusch and Rudiger, 1996; Zamir and Geiger, 2001; Ziegler et al., 2006), and cells depleted of vinculin have reduced adhesion to a variety of ECM proteins, increased migration rates, and fewer and smaller adhesions compared with wild-type cells (Coll et al., 1995; Volberg et al., 1995; Xu et al., 1998; Saunders et al., 2006). Despite the profound role of vinculin in cell adhesion and motility, the molecular mechanisms by which vinculin exerts these distinct effects are poorly understood.
Structurally, vinculin comprises three major domains: an N-terminal head, a flexible proline-rich hinge (neck) region, and a C-terminal tail domain (Eimer et al., 1993; Winkler et al., 1996). Vinculin activation results from conformational rearrangements of these domains. Intramolecular associations between the head and tail domains constrain vinculin in an inactive conformation (Bakolitsa et al., 2004), causing it to be located within the cytoplasm (Chen et al., 2005). Upon recruitment to FAs, the structure of vinculin switches to an open, active conformation. This process of activation is crucial to allow the full access and direct interaction of talin and α-actinin to the head; ponsin, vinexin, vasodilator-stimulated phosphoprotein, and Arp2/3 to the neck; and actin, phosphatidylinositol (4,5)-bisphosphate (PIP2), and paxillin to the tail (Zamir and Geiger, 2001; Ziegler et al., 2006). However, because most of the studies characterizing interaction sites on vinculin rely on biochemical assays using purified proteins, it is not clear what relevance these potential interactions have for FA formation.
We have therefore taken a novel approach to the study of vinculin, focusing on its localization and function in cells. We found that in order for vinculin to drive the formation of FAs, it needs to interact with talin. The interaction of the opened, and therefore activated, form of vinculin with talin has a direct effect on integrins, clustering them in an active conformation and leading to FA enlargement. Subsequent to the appearance of vinculin in FAs, paxillin becomes recruited, though this is independent of the paxillin-binding site present in the vinculin tail (vinT) domain. Our data also suggest that vinT represents the major link between FAs and the actin network. Overall, we propose a new model that places vinculin in a key position regulating FA formation and turnover.
Results
Active vinculin induces an increase in FA size via its head domain
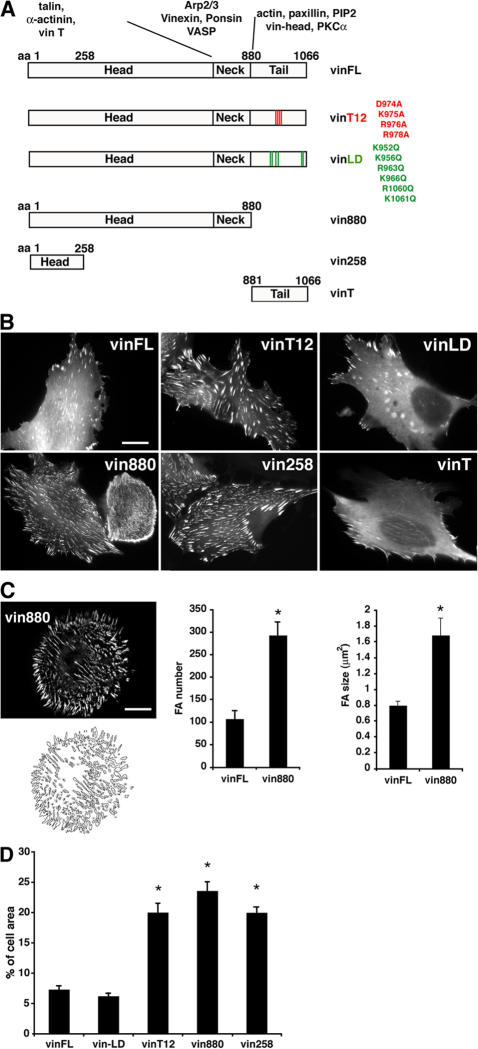
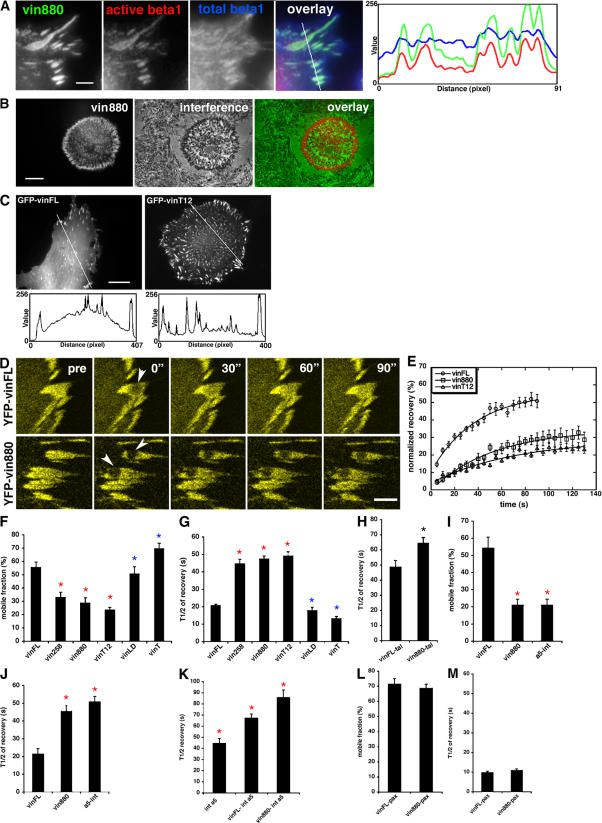
To investigate the role of different regions of vinculin in regulating FA number and size, a variety of vinculin constructs tagged to GFP or YFP were expressed in NIH3T3 cells (Fig. 1 A) and their effects on FA formation were compared. Among the tested constructs were full-length vinculin (vinFL); vinculin T12 (vinT12), a constitutively active form of vinculin bearing mutations that inhibit head–tail association (Cohen et al., 2005); vinculin LD (vinLD), which contains mutations that inhibit PIP2 binding (Chandrasekar et al., 2005); constructs that comprise the N-terminal 880 or 258 amino acids, thus lacking the tail (vin880 and vin258, respectively); and a vinT construct (comprising amino acids 881–1066; Fig. 1 A). All expressed constructs localized to FAs in a variety of cell types such as murine NIH3T3 fibroblasts, B16-F1 melanoma, and HeLa cells (Fig. 1 B and not depicted). Additionally, besides localizing to FAs, vinT also colocalized with actin filaments (see Fig. 7 A and not depicted). Notably, vinT12, vin880, and vin258 induced a dramatic increase in the size and number of FAs (Fig. 1, B and C), and the area of the cell surface that contained FAs was approximately three- to fourfold larger than that observed for vinFL and vinLD (Fig. 1 D). Thus, although all vinculin constructs locate to adhesion sites, the size and number of these adhesions dramatically increase by preventing head–tail associations. By using vinculin fragments, this property was shown to reside within the N-terminal 258 amino acid.
Figure 1.
Effect of wild-type and mutant forms of vinculin on FA growth. (A) Vinculin constructs that were expressed as fusion constructs to GFP derivatives in NIH3T3 cells: vinFL; vinculin comprising head and neck domains, amino acids 1–880 (vin880); vinculin head domain 1, amino acids 1–258 (vin 258); vinT, amino acids 880–1066; vinT12 (Cohen et al., 2005), with point mutations that render the molecule constitutively active; and vinLD (Chandrasekar et al., 2005), with point mutations that inhibit PIP2 binding. (B) NIH3T3 cells expressing indicated GFP-tagged constructs. Note the dramatic increase of FAs expressing vinT12, vin880, and vin258 compared to cells expressing vinFL or vinLD. vinT localizes to FAs and filamentous structures. (C) Quantification of FA size and number. FAs were masked, and their sizes and numbers where calculated. Asterisks indicate statistical significance (P < 0.001, t test). (D) Quantification of FA areas. FA area was calculated as a percentage of the total cell area. Asterisks indicate significant differences of values compared with vinFL (P < 0.0001, HSD test). There was no statistical difference in FA values between vinFL and vinLD. Error bars indicate ± SEM. Bars, 10 μm.
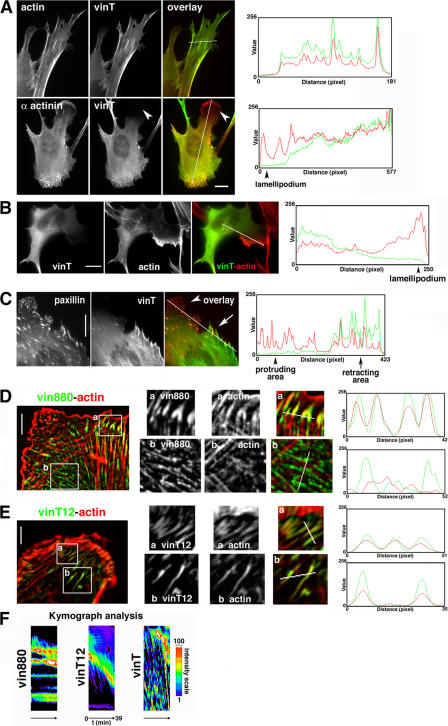
Figure 7.
vinT colocalization correlates with a subset of actin but not paxillin. (A) Cells expressing vinT were either labeled for actin or cotransfected with CFP–α-actinin (bottom). The intensity profiles on the right are from the area covered by the line in the overlay images. Note the high correlation of actin stress fibres with YFP-vinT. However, protrusive areas (white arrowheads) positive for α-actinin remain free of YFP-vinT. Bar, 10 μm. (B) In vin−/− MEF cells, there is little YFP-vinT in protruding lamellipodia when compared to F-actin labeled with rhodamine-phalloidine. Note the strong correlation of the intensity profiles (right) on the left side and the lack of correlation on the right side (lamellipodium). (C) NIH3T3 cell expressing CFP-paxillin and YFP-vinT. The protruding area of cell shows many FAs in the protruding area positive for paxillin but devoid of vinT. To the right are line profiles of fluorescence intensities of the line in the overlay image (compare with Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200703036/DC1). (D) NIH3T3 cell expressing YFP-vin880 and colabeled for actin. (a) FAs at the cell periphery with similar intensity profiles, indicating that these FAs are linked to actin stress fibers. (b) FAs away from the periphery of the cell. Intensity profiles show that intensity peaks of vin880 do not correlate with the actin intensities. (E) NIH3T3 cell expressing vinT12 and colabeled for actin. Intensity profiles taken from lines in inserts of peripheral and inner FA demonstrate a significant overlap of vinT12 and actin fluorescence intensity peaks, suggesting a link of these FAs with the actin cytoskeleton. (F) Kymograph analysis of NIH3T3 cells expressing YFP-vin880, GFP-vinT12, and YFP-vinT. Kymograph analysis shown in a spectral fluorescence intensity scale derived from a one-pixel line perpendicular to the cell edge of time-lapse recordings presented in Videos 2–4. Note that high intensity areas in vin880 cells remain stable with time and therefore are visualized as straight lines in this type of analysis, whereas those of vinT12 and vinT follow a retrograde flow. Bars (A–C) 10 μm; (D and E) 8 μm.
Talin and paxillin localize identically to vinculin-enlarged FAs
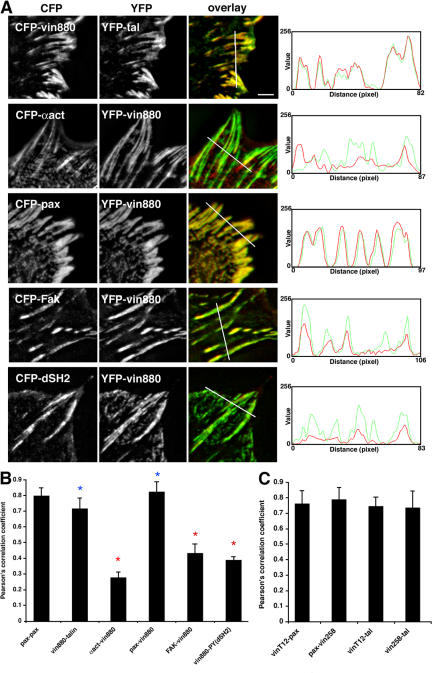
To identify associated molecules involved in the induction of enlarged FAs by active vinculin (vinT12) and C-terminally truncated constructs such as vin880 and vin258, image correlation analysis (ICA), which is a pixel-to-pixel comparison based on the Pearson's correlation coefficient (r), was used. In initial experiments on cells, coexpression of CFP- and YFP-paxillin revealed r values of ∼0.8 (Figs. 2 B and S1, available at http://www.jcb.org/cgi/content/full/jcb.200703036/DC1). These values reflect the virtually identical localization of two components.
Figure 2.
ICA of vin880 with other FA proteins. (A) Sections of NIH3T3 cells expressing CFP- and YFP-tagged proteins as indicated. Fluorescence intensity profiles depict the area of the line drawn in image overlays. Potential direct interacting proteins talin and α-actinin show different correlations with vin880. In contrast to talin, which colocalizes identically with vin880, α-actinin colocalizes only poorly with pixels positive for vin880. vin880 colocalization with potentially indirectly associating proteins paxillin and FAK and correlation with phosphotyrosine in FA is shown. Paxillin colocalizes identically with vin880 in oversized FAs, whereas pixels positive for FAK and a probe that recognizes phosphotyrosine in FA (dSH2) correlate less well with vin880 localization. Bar, 2 μm. (B) Quantification of colocalization by ICA based on the Pearson's correlation coefficient (a perfect linear correlation is shown at +1; see Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200703036/DC1). Note the high correlation of vin880 with talin and paxillin but lower correlation with α-actinin, FAK, and dSH2. Red asterisks indicate significant differences of correlation values compared with pax-pax control (P < 0.0001, HSD test). Blue asterisks indicate no statistical differences. (C) Quantification of colocalization in FAs by ICA based on the Pearson's correlation coefficient of GFP-vinT12/RFP-paxillin, CFP-paxillin/YFP-vin258, GFP-vinT12/RFP-talin, and YFP-vin258/RFP-talin. The correlation between indicated pairs was as equally high as CFP-paxillin/YFP-paxillin, demonstrating identical colocalization. Error bars indicate ± SEM.
CFP- or YFP-vin880 was then coexpressed pairwise with other prominent FA regulators fused to YFP or CFP and correlation coefficients were calculated. The Pearson's correlation coefficient measures colocalization in 2D (Fig. 2 B). To produce a more visual illustration of the degree of correlation between pairs of components, fluorescence intensities in 1D line profiles drawn over FA areas were also compared (Fig. 2 A, right). Although talin and paxillin showed essentially identical colocalization with vin880 (r = 0.72 and 0.82, respectively), the correlations of α-actinin (r = 0.27), FAK (r = 0.42), and a reporter for tyrosine-phosphorylated SH2-binding sites (dSH2; r = 0.39; Kirchner et al., 2003; Ballestrem et al., 2006) with vin880 in FAs were low (Fig. 2, A and B). The paxillin result was particularly surprising because vin880 lacks the reported paxillin binding site in vinculin. The high degree of colocalization of vin880 with paxillin and talin was also observed when endogenous paxillin and talin were detected with antibodies (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200703036/DC1; and not depicted). Thus, of the previously reported direct vinculin interaction partners, talin but not α-actinin is efficiently recruited to vin880-enlarged FAs. Also, the identical colocalization of vin880 with paxillin demonstrates that vinculin can trigger downstream events, resulting in the recruitment of paxillin independent of its interaction site located in the tail domain (also observed in vinculin null cells, see Fig. 6 A). Moreover, the absence of FAK and dSH2 from most of the vin880-induced FAs close to the nuclei (unpublished data) suggests that FAK and tyrosine phosphorylation are not likely to play a role in the recruitment of paxillin to vin880-induced hypertrophic FAs.
Figure 6.
Vinculin-induced FA hypertrophy and paxillin recruitment to FAs is not due to the potential dimerization of tailless vinculin constructs with endogenous vinculin. (A) CFP-paxillin was coexpressed with YFP-vinFL or -vin258 in vin−/− MEF cells. Overlay images are from insert areas outlined in the vinFL and vin258 labeled images. The line profiles taken from the lines indicated on the left outline the nearly identical localization of the respective coexpressed proteins in FAs. Bar, 8 μm. (B) Quantification of FA sizes in vin−/− MEF cells expressing the indicated YFP fusion proteins. Area of FAs was calculated as a percentage of the total cell area. Red asterisks indicate significant differences of correlation values compared with vinFL (P < 0.0001, HSD test). Blue asterisks indicate no statistical differences to vinFL. (C) YFP-vin880 t 1/2 of recovery is increased compared to YFP-vinFL when expressed in vin−/− MEF cells. Asterisk indicates statistical significance (P < 0.001, t test).
Analysis of intensity profiles of vin880 cotransfected with FAK or dSH2 constructs revealed differences compared with the relationship between vin880 and α-actinin. Although all high intensity peaks of FAK or dSH2 correlated well with intensity peaks of vin880, high α-actinin intensity often showed no clear correlation with the intensity profile of vin880 (Fig. 2 A). Conversely, α-actinin–positive structures showed a strong correlation with actin (unpublished data). Together, these data suggest that α-actinin is unlikely to have a key role in vinculin-induced FA growth.
To assess whether vin258 and vinT12 signal via different mechanisms, their presence in FAs was correlated with the same series of proteins. Again, high correlation coefficients were only obtained between talin and paxillin (Fig. 2 C and not depicted). These data suggest that talin and/or paxillin may play key roles in the formation of the enlarged FAs induced by active vinculin.
Vinculin regulates the clustering of integrins in FA
To determine the relationship between FA formation and receptor distribution, the colocalization of integrins with vin880 was examined in cells spread on fibronectin, the major ligand for α5β1 integrin. A YFP–α5 integrin construct colocalized with CFP-vin880 in all visible FAs (unpublished data). Integrins adopt different conformations, which can be reported by mAbs. A striking colocalization of active β1 with vin880 in FAs was observed (Fig. 3 A), which was distinct from the colocalization of total β1 with vin880. This is most obviously seen in the fluorescence intensity profile from the area of the line drawn in the image overlay (Fig. 3 A). Although the total β1 staining labeled integrins in FAs and the cell membrane, active β1 was found almost exclusively in FAs correlating highly with vin880 (r = 0.79). To examine whether these FAs link the cell to the ECM, interference reflection microscopy was used to visualize regions of the cell membrane proximal to the substratum. Indeed, FAs induced by vin880 were detected by interference reflection microscopy, demonstrating a physical association with the ECM (Fig. 3 B).
Figure 3.
Vinculin regulates integrin clustering and dynamics. (A) HeLa cells overexpressing YFP-vin880 were costained with an antibody recognizing active β1 (9EG7; Bazzoni et al., 1995) and the total pool of β1 integrin (TS2/16). FAs positive for YFP-vin880 are also positive for β1 integrins. The intensity profiles in red and green outline the high correlation of YFP-vin880 and active β1 integrins. The blue intensity profile shows the high levels of β1 integrins in the cell membrane and FAs. Fluorescence intensity profiles depict the area of the lines drawn in image overlays. (B) FAs induced by YFP-vin880 (left) are readily visible by interference reflection microscopy (middle), indicating that they participate in adhesion to ECM proteins. Overlay fluorescence and interference are shown on the right. (C) Images and line profiles drawn over the area of NIH3T3 cells expressing indicated constructs demonstrate that the cytoplasmic pool of GFP-vinFL is higher than that of GFP-vinT12. (D) To assess the turnover of indicated proteins, circular areas of 1.5-μm diameter were bleached, and recovery was measured. Note the slower fluorescence recovery of bleached areas in YFP-vin880–expressing cells compared to areas in YFP-vinFL–expressing cells. (E) Normalized recovery of vinFL, vin880, and vinT12 in FAs, with lines indicating the single exponential fit of the data. MF (F) and t 1/2 of recovery (G) of indicated YFP-fusion proteins in FAs of NIH3T3 cells. Red asterisks indicate significant differences compared to vinFL (P < 0.005, HSD test). Blue asterisks indicate no statistical differences. (H) Talin t 1/2 of recovery is longer when coexpressed with vin880 in comparison with vinFL. Asterisk indicates statistical significance (P < 0.05, t test). Comparison of MFs (I) and t 1/2 (J) of YFP-vinFL, -vin880, and −α5 integrin in FA. Note the striking similarity of vin880 and α5 integrin dynamics. Red asterisks indicate significant differences compared to vinFL (P < 0.001, HSD test). (K) Coexpression of vinFL or vin880 leads to a longer t 1/2 of recovery of YFP–α5 integrin in FAs of vin−/− MEF. Red asterisks indicate significant differences between the samples (P < 0.01, HSD test). (L and M) Expression of vin880 changes neither the MF (L) nor the t 1/2 of recovery (M) of paxillin in FAs. Error bars indicate ± SEM. Bars: (A) 2 μm; (B and C) 10 μm; (D) 2 μm.
Vinculin constructs that induce FA enlargement have increased residency time in FAs and form tight complexes with talin and integrins
During FA size measurements, it was observed that YFP-vin258, YFP-vin880, and GFP-vinT12, all of which induced enlarged FAs, had a clearer FA pattern with a reduced cytoplasmic pool compared with GFP-vinFL (Fig. 3 C and not depicted). The reduction of the cytoplasmic pool suggests differences in mobilities and affinities of the proteins, leading to an enhanced recruitment to FAs. To study mobilities of the different vinculin mutants in FAs, FRAP experiments were performed (Fig. 3, D and E; Lippincott-Schwartz et al., 2001). The observed FRAP recoveries presented in Fig. 3 E appear to be slightly biphasic, as was previously observed for vinculin (Lele et al., 2006). The possibility that this might be caused by the ability of vinculin to bind multiple binding partners or the enhanced fast reversible photobleaching for YFP-tagged probes used in the majority of these experiments needs further investigation. However, to avoid overinterpretation of our results, single exponential fits were used as reported previously by others with similar probes (Cohen et al., 2006). Using such fits provided estimates of the mobile fractions (MFs) and t 1/2 of recoveries. A striking twofold decrease in the MFs (Fig. 3, E and F) and a twofold increase in t 1/2 (Fig. 3 G) of vinT12, vin880, and vin258 were found compared with vinFL, vinLD, and vinT. This indicates that by switching to an active conformation or exposing binding sites within N-terminal domains, vinculin changes its affinity for binding partners, resulting in an increased stability of vinculin within FAs.
Because talin binds to integrins and provides an early mechanical link to ECM proteins (Giannone et al., 2003; Zaidel-Bar et al., 2003), it was hypothesized that the reduction of vinT12, vin880, and vin258 mobility may be caused by the formation of a stable complex with talin and integrins. A recent paper demonstrated the delayed turnover of talin in FAs upon coexpression with constitutively active vinT12 (Cohen et al., 2006), thus indicating their tight association in cells. Similar results were obtained for talin turnover when coexpressed with vin880 (talin t 1/2 increased ∼25% compared with cells coexpressing vinFL; Fig. 3 H).
If integrins were also part of such a tight complex, we predicted that integrin subunits would turn over at similar rates to vinT12, vin880, or vin258. Indeed the MF and t 1/2 of the GFP–α5 integrin chain were almost identical to YFP-vin880 (Fig. 3, I and J). Interestingly, YFP-paxillin, which colocalizes precisely with vin880 (Fig. 2, A and B), was considerably more mobile, with a t 1/2 of 11 s and a MF of 70%, and did not change upon coexpression of vin880 (Fig. 3, L and M). These data suggest that in cells cultured on fibronectin, talin, α5β1 integrin, and active vinculin form a tight complex at points of cell–ECM contact, whereas paxillin only transiently associates with this complex.
To test whether integrin dynamics change in the presence of vinculin in FAs, YFP–α5 integrin was expressed alone or in combination with CFP-vinFL or -vin880 in vin−/− mouse embryonic fibroblasts (MEFs; Saunders et al., 2006) and integrin turnover rates were measured using FRAP. A 40% increase in the t 1/2 of α5 integrin in cells coexpressing vinFL compared with α5 alone was observed. A further 30% increase of t 1/2 was observed in cells coexpressing vin880, demonstrating that integrin turnover within FAs is regulated by vinculin activity or exposure of N-terminal binding domains within vinculin (Fig. 3 K).
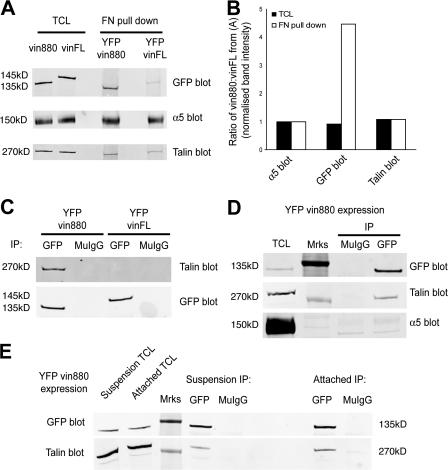
To confirm the apparent tight association between integrins, talin, and vin880, their interactions were tested biochemically. FN-coated beads were added to cells expressing YFP-vinFL or -vin880 and bead-bound fractions were isolated after cross-linking and detergent extraction. α5 integrin and talin were identified in all bead-bound cell fractions (Fig. 4 A). Despite the similar total levels of vin880 and vinFL expression, within the bead-bound fraction, vin880 was enriched by approximately fourfold, unlike talin, when normalized to the respective α5 integrin band intensity (Fig. 4 B). Moreover, coimmunoprecipitation experiments from cells expressing YFP-vin880 or -vinFL demonstrated the association of talin with vin880 but not vinFL (Fig. 4 C). Performing the assay after incubation with a chemical cross-linker resulted in the additional coimmunoprecipitation of the α5 integrin subunit with vin880 (1.85 ± 0.49–fold increase over control; P = 0.03, Fisher Sign test, n = 5; Fig. 4 D). The weak α5 integrin signal observed in the coimmunoprecipitation is partly caused by the low stoichiometry of the interaction, i.e., the vast majority of talin and vinculin within the cell is not complexed with integrin at any one time, partly because of the lability of the complex and the inaccessibility of a transmembrane receptor associated with poorly soluble cytoskeletal and matrix components. This latter point means that stringent detergent conditions are required to solubilize the individual components of the complex; these conditions subsequently lead to the dissociation of the complex.
Figure 4.
YFP-vin880 is enriched in FN– integrin complexes and coimmunoprecipitates talin and integrin. (A) FN–bead bound complexes isolated from NIH3T3 cells expressing either YFP-vin880 or -vinFL and immunoblotted for GFP, α5 integrin, or talin. (B) Quantification of A expressed as a ratio of vin880/vinFL after immunoblot band signal intensity normalization to the respective α5 integrin signal intensity. Open bars represent ratios of indicated proteins from pull-down experiments with pFN-coated beads; shaded bars represent protein ratios detected in total cell lysate (TCL) of cells expressing YFP-vin880 or -vinFL. (C) Immunoprecipitations using anti-GFP or control mouse IgG (MuIgG) from NIH3T3 cells expressing either YFP-vin880 or -vinFL. (D and E) Immunoprecipitations using anti-GFP or control MuIgG from NIH3T3 cells expressing YFP-vin880 after treatment with a chemical cross-linker (D) or from cells in suspension for 15 min (E) versus cells left attached to tissue culture dishes. All blots are representative of more than two independent transfections. Mrks denotes the position of molecular mass standards (250 kD for talin blots and 150 kD for GFP and α5 integrin blots), which are visible by Western blotting using the infrared imaging system.
Interestingly, talin coimmunoprecipitated with vin880 regardless of whether cells were in suspension or attached to the ECM (Fig. 4 E), and neither vin880 nor vinFL coimmunoprecipitated α-actinin, paxillin, or FAK (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200703036/DC1). Overall, these biochemical data support the FRAP and immunofluorescence data (Figs. 1–3 ) and indicate that the N-terminal domains of vinculin, e.g., vin880 without the C-terminal tail region, constitute a vinculin construct that forms a tight complex with talin and α5β1 integrin.
The talin–vinculin interaction is required for the vinculin-induced FA enlargement
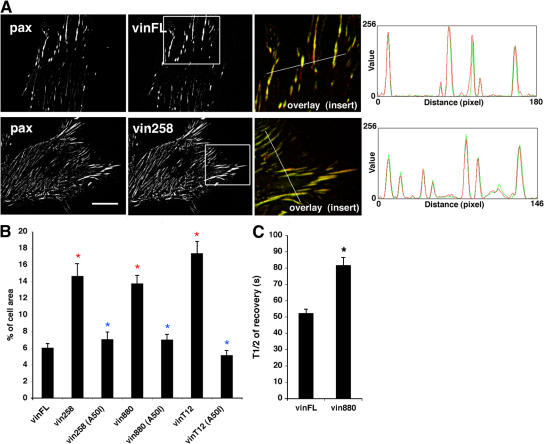
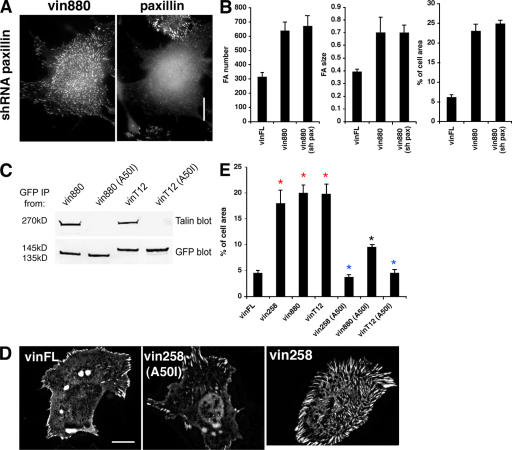
It was observed previously that talin and paxillin but not FAK, α-actinin, or phosphotyrosine correlated highly with localization of vin880 in FAs, suggesting that the former might be involved in FA enlargement or formation. To test this possibility, paxillin and talin (talin1) were deleted by small hairpin RNA knockdown. YFP-vin880 expressed in paxillin-deficient B16 cells was still able to induce FAs of a similar size and number as those in wild-type cells (Fig. 5, A and B). For the knockdown of talin1, the interpretation of data was difficult, primarily because many of the talin knockdown cells rounded up and were therefore unsuitable for FA measurements (unpublished data). Therefore, to test directly the role of the talin–vinculin interaction in the formation of enlarged FAs, an A50I mutation was introduced into vin258, vin880, and vinT12. This mutation is known to reduce talin binding to vinculin in vitro (Bakolitsa et al., 2004). Immunoprecipitations demonstrated that the A50I mutation in vin880 and vinT12 abrogated the coimmunoprecipitation of talin (Fig. 5 C). Expression of these constructs in NIH3T3 cells and subsequent analysis of FAs showed that vin258 (A50I) and vinT12 (A50I) no longer induced FA growth, whereas vin880 (A50I) exhibited greatly reduced activity (Fig. 5, D and E). Collectively, these experiments indicate not only that “activated” vinculin, or vinculin with exposed binding sites within its N-terminal domains (vin880 and vin258), binds to talin, but that this interaction is required for the formation of enlarged FAs. In contrast, paxillin is not required for the formation of FAs and is likely to be recruited downstream (or independently) of vinculin.
Figure 5.
FA size is regulated by vinculin interaction with talin. (A) Paxillin was knocked-down using small hairpin RNA in B16 melanoma cells expressing vin880. Despite the absence of paxillin, vin880 still induced FA growth. (B) Quantification of FA number, size, and area fraction of indicated constructs in B16 cells with or without (sh pax) paxillin. (C) Immunoprecipitations from cell lysates of HeLa cells expressing either YFP-vin880, -vin880 (A50I), -vinT12, or -vinT12 (A50I) with anti-GFP antibodies. Note talin coimmunoprecipitates with vin880 and vinT12 but not with A50I mutants. (D) NIH3T3 cells expressing YFP-vinFL and -vin258 (A50I) show no significant differences in FA size in contrast to cells expressing YFP-vin258. (E) Quantification of FA size of cells expressing indicated constructs. Area of FAs was calculated as a percentage of the total cell area. Red asterisks indicate significant differences compared with vinFL (P < 0.0001, HSD test). Blue asterisks indicate no statistical differences. Black asterisk indicates statistical difference with vinFL (P < 0.001, t test). Error bars indicate ± SEM. Bars: (A) 10 μm; (D) 10 μm.
Induction of oversized FAs and underlying signaling mechanisms are independent of endogenous vinculin
The possibility exists that both FA growth and paxillin recruitment to active vinculin could have been the result of an interaction of paxillin with endogenous vinculin that had dimerized, via an intermolecular head–tail interaction, with the expressed vinculin constructs. To test this possibility, vinculin constructs were expressed in vin−/− MEFs. Expression of tailless vinculin forms and active vinT12 in the absence of endogenous wild-type vinculin induced a two- to threefold increase of FAs (Fig. 6, A and B), which was abolished by the A50I mutation (Fig. 6 B). Interestingly, the induction of FAs in vin−/− MEFs by a vin880 (A50I) mutant was abrogated (compare Fig. 6 B with Fig. 5 E), suggesting that vin880 induced the activation of endogenous vinculin to a small extent. In further experiments, the colocalization of paxillin and vin258 was analyzed in enlarged FAs of vin−/− MEFs. As outlined in Fig. 6 A, YFP-vin258 colocalized identically with CFP-paxillin, demonstrating that the highly efficient paxillin recruitment induced by tailless vinculin forms is independent of the putative dimerization of endogenous vinculin with the expressed constructs.
The similar behavior of the vinculin expression constructs in cells with and without endogenous vinculin was examined using FRAP to measure t 1/2 of recovery of YFP-vinFL and YFP-vin880 (Fig. 6 C). As in cells with endogenous vinculin, the t 1/2 for vin880 increased ∼50%. This is in accordance with a previous paper using a vinculin head domain and active vinculin (Cohen et al., 2006). These experiments suggest that there is little contribution of endogenous vinculin to the data presented here using C-terminal truncations or activated vinculin constructs.
vinT colocalizes with a subset of filamentous actin but not with paxillin
Paxillin and actin have been shown to bind to the vinT region (Menkel et al., 1994; Wood et al., 1994; Huttelmaier et al., 1997). To elucidate possible associations of these two proteins with vinT, YFP-vinT was expressed in NIH3T3 cells and its colocalization with actin and paxillin was analyzed. The localization of vinT correlated well with actin stress fibers (r = 0.7) but was diminished or absent in large protruding lamellipodia that were positive for α-actinin (Fig. 7 A). This strong reduction of vinT in protruding areas was not caused by the potential competition with endogenous vinculin because the phenomenon also occurred in vinculin null cells (Fig. 7 B). Furthermore, time-lapse experiments showed that it was only when large lamellipodia collapsed and started to retract that vinT became strongly associated with these structures (Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200703036/DC1), suggesting that vinT only binds a subset of actin filaments. In contrast, CFP-paxillin was abundant in FAs of protruding cell areas (Fig. 7 C) and only colocalized with vinT in retracting areas, albeit with a low correlation between their intensity profiles. Thus, paxillin localization correlates with the head region of vinculin, and there is little, if any, interaction of paxillin with its tail. vinT, however, appears to be the major domain involved in actin binding.
vinT links adhesion sites to the actin cytoskeleton
The major factor implicated in the growth of FAs has, until now, been intracellular tension mediated by the actomyosin contractile machinery (Burridge and Chrzanowska-Wodnicka, 1996; Balaban et al., 2001). Our data indicate that the vinculin head but not the tail induces FA growth (Fig. 1 A), suggesting that a link to actin via the C terminus of vinculin may not be required for FA growth. To examine whether the vinculin–actin interaction has the potential to modulate FAs, the association of FA growth–promoting vinculin mutants that either lacked (vin880) or retained (vinT12) the tail domain were examined in relation to actin. Approximately 75% of the internal nucleoproximal FAs (defined in this paper as >10 μm from the cell edge) induced by overexpression of vinculin mutants without tail domains were not linked to actin stress fibers (Fig. 7 D). In contrast, overexpression of vinT12, which retains its tail domain, resulted in >80% of FAs that were streaklike and linked to actin filaments (Fig. 7 E). Furthermore, in ∼30% of these cells, long ropelike structures positive for both vinT12 and actin were apparently clustered together. These data suggested that the tail of vinculin forms a crucial link between FAs and the actin cytoskeleton. Because actin filaments in cells have been shown to undergo retrograde flow (Ponti et al., 2004; Vallotton et al., 2004; Gupton and Waterman-Storer, 2006; Hu et al., 2007), the possibility that vinculin dynamics are influenced by the link to actin was tested using live cell time-lapse analysis of vinT12 and the tailless vin880 (Fig. 7 F and Videos 2 and 3, available at http://www.jcb.org/cgi/content/full/jcb.200703036/DC1). Although vin880 was stably localized in FAs, vinT12 was transported in a retrograde manner with a mean velocity of 0.366 ± 0.112 μm/min toward the cell center upon retraction. Furthermore, vinT, which lacks a head domain, followed a retrograde flow with a mean velocity of 0.679 ± 0.295 μm/min from peripheral FAs toward the cell center (Fig. 7 F and Video 4). F-actin retrograde flow in similar velocity ranges has been observed previously in newt lung and PtK1 epithelial cells (Ponti et al., 2004). Thus, it is the vinT domain that links vinculin to the actin cytoskeleton, which may in turn exert forces on vinculin resulting in its relocalization outside of FAs.
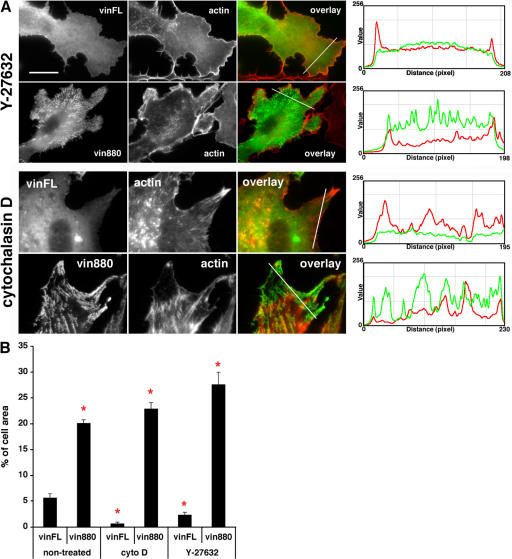
The vinculin head stabilizes adhesion sites despite inhibition of actomyosin-mediated tension
Because the vinculin head region is able to form large FAs in the absence of the actin-binding tail, we reasoned that the vinculin head might be able to initiate or stabilize cell–matrix adhesions independently of the actomyosin machinery. Inhibition of Rho-kinase (ROCK) leads to the release of actomyosin-mediated tension and the disruption of actin stress fibers. As a consequence of this perturbation of the actin cytoskeleton, FAs dissolve and only focal complexes of a transient nature remain visible. The effect of the ROCK inhibitor Y-27632 in NIH3T3 cells expressing vinFL and vin880 was therefore tested. Although Y-27632 treatment resulted in the loss of essentially all adhesion sites in vinFL-expressing cells (except a few complexes at the cell periphery), a large number of adhesion sites were still apparent in cells expressing vin880 (Fig. 8, A and B). This was the case even when Y-27632 was used at concentrations up to 300 μM, which leads to the complete distortion of the cell morphology. Similar observations were made when vinFL- and vin880-expressing cells were treated with the actin-disrupting agent cytochalasin D (Fig. 8, A and B). Although quantification of adhesion sites in vin880-expressing cells revealed no change in adhesion area when cells were treated with actin-perturbing reagents, the morphology of the remaining FA structures did appear to be altered (compare Fig. 8 A with Fig. 1 B). They were less streaklike and resembled those of β3 integrin clustering induced by switching to an active conformation through the addition of manganese, integrin-activating mutations, or talin head overexpression (Cluzel et al., 2005). Although the clusters observed by Cluzel et al. (2005) contained talin, they were not linked to the actomyosin machinery. This is in keeping with our model of N-terminal vinculin domains controlling the clustering of activated integrin via talin independently of linkages with the actin cytoskeleton. These results suggest that vinculin without the actin-binding tail domain is able to initiate FA formation in the absence of actomyosin-mediated forces.
Figure 8.
Vin880 stabilizes adhesion sites in cells despite the inhibition of actomyosin function or disruption of actin filaments. (A, top) NIH3T3 cells expressing YFP-vinFL or -vin880 treated with 100 μM Y-27632 for 60 min. Although only small dotlike adhesions (focal complexes) can be seen at the cell periphery in vinFL-expressing cells, a large number of adhesion sites are still apparent in cells expressing vin880, despite the absence of stress fibers. (bottom) Cells treated with 1 μM cytochalasin D for 30 min. Although essentially no adhesions can be seen in YFP-vinFL–expressing cells, many adhesion sites are still visible in YFP-vin880–expressing cells. Fluorescence intensity profiles on the right are from the area of the line drawn in image overlays. Note that there is little if any correlation of high vinFL or vin880 intensity peaks with those of actin. Bar, 12 μm. (B) Quantification of FA area. FA area was calculated as a percentage of the total cell area. Note the significant loss in FA area in vinFL-expressing cells when treated with cytochalasin D (cyto D) or Y-27632. No loss of adhesion area was found in vin880 cells treated with actin-perturbing reagents. Asterisks indicate significant differences of values compared with nontreated vinFL (P < 0.001, HSD test). The apparent slight increase of vin880 FA area in vin880-expressing cells treated with cytochalasin D or Y-27632 compared to nontreated cells was caused by a decrease in total cell area (in the case of cytochalasin D) or increased de novo formation of adhesions at the cell periphery (in the case of Y-27632).
Together, these findings separate the vinculin head and tail regions into two distinct functional domains: a head region that binds to talin and is involved in the growth of cell–matrix adhesions associated with clustering of active integrins and a tail domain that is involved in binding actin and coupling with the mechanotransduction force machinery.
Discussion
FAs are composed of >100 components (Geiger et al., 2001). Because vinculin depletion in cells leads to dramatic changes in cell adhesion motility and FA sizes (Coll et al., 1995; Volberg et al., 1995; Xu et al., 1998; Saunders et al., 2006), it has been proposed that vinculin is a key player in the regulation of cell adhesion. Although the structure of vinculin and its binding sites for 11 binding partners have been well characterized in vitro, the mechanisms underlying its stabilizing function on FAs in cells has, until now, been unclear. We sought to investigate how vinculin controls FAs within a cellular context and reevaluate current models of its action in light of this information. Our major findings are that (a) the interaction of the N-terminal head of vinculin with talin drives the clustering of integrins in cell–matrix adhesions, possibly by maintaining integrins in an activated state; (b) the vinculin–talin interaction leads to the highly efficient recruitment of paxillin independently of the paxillin-binding site located in the tail region of vinculin; and (c) vinculin, via the interaction of its tail with actin, is the major link of the FA core to the actin cytoskeleton.
Vinculin interaction with talin clusters integrins in an active conformation leading to FA growth
Previously, the interaction of vinculin with talin was reported to be crucial for the process of vinculin activation (Ziegler et al., 2006). It is now well established that this activation process leads to structural rearrangements that allow the access of a large number of vinculin binding partners whose role in FA stability was not clear. Our data demonstrate that the 258 N-terminal amino acids, the D1 domain of vinculin, is sufficient to induce FA enlargement and that its interaction with talin is crucial to maintain this function. Notably, tensile forces mediated by actomyosin are not required because (a) enlarged adhesion sites induced by tailless vinculin constructs are not necessarily linked to actin stress fibers and (b) a large number of adhesion sites remain present in vin880-expressing cells when intracellular tension is perturbed by the disruption of the actin cytoskeleton with cytochalasin D or blocking of actomyosin function with the ROCK inhibitor Y-27632. Thus, we now propose that vinculin activation, by an unknown mechanism, leads to enhanced integrin clustering and, consequently, the formation of FAs and that this occurs independently of, or in cooperation with, tensile forces exerted by the actomyosin machinery.
Using FRAP, we demonstrated that integrin turnover is influenced by vinculin activity. Recently, talin turnover was shown to be dependent on vinculin activity (Cohen et al., 2006), and β3 integrin clustering was found to require the presence of talin (Cluzel et al., 2005). It has been found that mutations leading to integrin activation increased integrin residency in FAs (Cluzel et al., 2005). Together with the observation that vin880 was enriched to, and isolated with, integrins bound to FN-coated beads and that α5 integrin and talin coimmunoprecipitate with vin880, our paper reveals that active vinculin, talin, and integrin form a tight complex. The colocalization of vin880 with active β1 integrins suggests that the integrin–talin–vinculin ternary complex alters the dynamics of integrins by clustering integrins in an active conformation, which in turn leads to FA growth.
It is now well established that talin is a key molecule regulating integrin activation (Ginsberg et al., 2005). Reducing the expression of talin by RNAi leads to the down-regulation of integrin activation levels (Tadokoro et al., 2003), and overexpression of the F2,3 domain of talin leads to integrin activation (Wegener et al., 2007). In our paper, the overexpression of full-length talin in all the cells tested was not able to enhance FA growth (Fig. S2 B and not depicted). Indeed, because FA growth was only achieved by coexpressing C-terminally truncated vinculin head constructs or active vinculin (Fig. 1 and not depicted), we would argue that vinculin activity may be the essential driving force for FA growth. It has been shown recently that talin has 11 potential binding sites for vinculin, some of which may be of low affinity or even cryptic (Fillingham et al., 2005; Patel et al., 2006). Moreover, the binding of a vinculin head construct to talin leads to a conformational change of talin that in turn may lead to the activation of the cryptic or low-affinity vinculin-binding sites (Fillingham et al., 2005). Therefore, an intriguing hypothesis is that active vinculin locks talin in FAs in an active conformation, which then induces further recruitment of vinculin and talin molecules, resulting in the growth of the adhesion site, thus providing an ideal platform for the recruitment other FA components and a link to the actin cytoskeleton.
Vinculin recruits paxillin to FAs independently of its binding site in the tail region
In contrast to α-actinin, paxillin colocalized identically with all constructs that induced FA enlargement. This was unexpected because the binding site of paxillin on vinT (Turner et al., 1990; Wood et al., 1994) is absent in the vin258 and 880 constructs. In fact, the observation that the vinT domain does not colocalize with paxillin in cell protrusions suggest there is little, if any, direct association between vinculin and paxillin in cells. Thus active vinculin induces paxillin recruitment either via a so far unidentified paxillin-binding site in the first 258 amino acids of vinculin or in an indirect manner through another protein. Interestingly, this protein cannot be FAK, which interacts with paxillin, as neither FAK nor dSH2 correlated with vinculin or paxillin. Other possible candidates involved in the direct recruitment of paxillin to FAs might be β1 integrin cytoplasmic domains (Schaller et al., 1995; Tanaka et al., 1996) or talin (Salgia et al., 1995), both of which colocalize in vin880-enlarged adhesion sites. Despite the colocalization, depleting paxillin via siRNA knockdown demonstrated that paxillin is not required for the induction of FA growth. However, whether indirect paxillin recruitment via vinculin activation has further important roles, such as balancing Rac activity (Brown and Turner, 2004) or controlling survival and motility by the regulation of paxillin–FAK interactions (Subauste et al., 2004), remains to be determined.
Vinculin provides the major connection of adhesion sites to the actin cytoskeleton
A recent study compared the motion of actin with FA components (Hu et al., 2007). It demonstrated that integrins as well as “core” proteins without direct interaction sites for actin (e.g., paxillin, zyxin, and FAK) have a low correlation with actin flow. In contrast, α-actinin mimicked actin kinematics, whereas vinculin and talin showed partial coupling to the actin flow. This differential coupling of FA components suggests a model of molecular hierarchies differentially linked to the actomyosin force machinery (Hu et al., 2007). It was speculated that the partial coupling of talin and vinculin to actin motions reflects different roles, whereby they spend part of the time bound to moving actin or to the less mobile FA component.
Our data shed more light onto such observations. First, we show by immunofluorescence, FRAP, and biochemistry that active and tailless forms of vinculin are linked to integrins via talin, which explains their slow mobility in FAs. Second, because only vinculin constructs that comprise the actin-binding vinT follow a retrograde flow, we suggest that vinculin is exposed to forces exerted by the actomyosin machinery. Third, our observation that talin, which bears at least two actin-binding sites (Hemmings et al., 1996; Lee et al., 2004) and colocalizes precisely with vin880 in FAs, does not provide an efficient link for many vin880-induced FAs to F-actin suggests that vinculin acts as the major link of the FA core to actin filaments. Thus, vinculin may represent the major transmitter of forces mediated by the actomyosin machinery in FAs. The apparent selective binding of the vinT to actin in contractile areas but not in protruding lamellipodia possibly contributes to specifically localized transmission of forces in different regions of the cell.
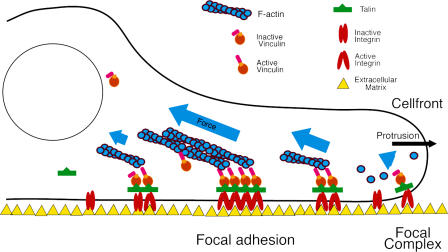
Model of vinculin action
Taking our findings together, we propose a new model for vinculin as a major regulator of FAs (Fig. 9). Vinculin is recruited via low-affinity binding to talin or neck-binding proteins to focal complexes at the cell front (Chen et al., 2005). Low-affinity interactions of vinculin with talin in initial adhesion complexes at the leading edge keeps vinculin in place for possible associations with PIP2 or actin, which subsequently leads to its activation (Huttelmaier et al., 1998; Bakolitsa et al., 1999; Bakolitsa et al., 2004; Chen et al., 2006; Janssen et al., 2006). If vinculin does not become activated at this stage, adhesion complexes turn over rapidly. However, once vinculin becomes activated, the conformational changes leading to a switch from low- to high-affinity binding of vinculin to talin stabilizes an active conformation of integrins in FAs, resulting in reduced FA turnover and growth. Similar changes of vinculin affinities to the invasin IpaA were reported during Shigella entry to cells, whereby IpaA mimics vinculin-binding sites on talin (Izard et al., 2006). Moreover, this initial phase of the model is in line with the observed conformational switch of vinculin during the transition of initial adhesions to later actin-bound stages (Cohen et al., 2005). In addition, vinculin activity regulates paxillin recruitment, which, depending on cosignals, may lead to additional modifications of FAs and cell migration (Turner, 2000; Zaidel-Bar et al., 2007). The activation process subsequently links vinculin, via its tail domain, to the contractile actomyosin machinery (Chen et al., 2006), which in turn allows the effective transmission of forces and thus bidirectional information between the inside and the outside of the cell. Ultimately, all this information is used to modulate a variety of cellular functions, including cell motility or active remodeling processes. To complete the cycle, FAs may be destabilized via further retrograde flow, leading to the removal of vinculin from FAs, or, alternatively, if actomyosin-mediated forces play a role in maintaining vinculin activity, via the transition of FA to areas of low actomyosin activity, with vinculin adopting a low affinity for talin upon refolding to an inactive state.
Figure 9.
Model of vinculin action in cells. See text in Discussion.
Materials and methods
Plasmids and cloning
vinFL, YFP-vin880, and YFP-vinT constructs, as well as CFP- and YFP-paxillin were provided by B. Geiger (Weizmann Institute of Science, Rehovot, Israel); vinT12 (Cohen et al., 2005) was provided by S. Craig (John Hopkins School of Medicine, Baltimore, MD); the plasmid for vinLD was a gift of W. Ziegler (University of Leizpig, Leipzig, Germany); YFP-vin258 was recloned from a bacterial expression vector from S. Craig into pcDNA3; YFP/CFP–α-actinin was recloned into pEYFP/pECFP vectors (Invitrogen) from GFP–α-actinin obtained by C. Otey (University of North Carolina at Chapel Hill, Chapel Hill, NC); α5 integrin and YFP-FAK were obtained from A.R. Horwitz (University of Virginia, Charlottesville, VA). The CFP/YFP-talin constructs were provided by K. Yamada and K. Matsumoto (National Institute of Dental and Craniofacial Research, Bethesda, MD). Cloning of the talin constructs was performed by introducing full-length talin (available from GenBank/EMBL/DDBJ under accession no. X56123; obtained from R. Hynes, Massachusetts Institute of Technology, Cambridge, MA) into pEYFPC1 and pECFPC1 vectors (Invitrogen) that contained a modified multiple cloning sequence with Not1 and EcoR1 restriction sites. Cloning of full-length talin into these sites resulted in a linker sequence with the base pairs TCCGGACTCAGATCTCGAGCTGCGGCCGCC.
The QuickChange site-directed mutagenesis kit (Stratagene) was used to generate the vinculin A50I mutants. Plasmids for the knockdown of paxillin and talin1 were constructed in pSuper (Oligoengine). The hairpin target sequences were paxillin (AGAGAAGCCAAAGCGAAAT) and talin (GAAGCACAGAGCCGATTGA). Their efficiency for protein knockdown was assessed by FACS selection of GFP cotransfected cells after 72-h expression followed by immunoblotting.
Cells and transfections
NIH3T3 mouse fibroblasts, B16F1 mouse melanoma cells, and HeLa cells were cultured in DME (Sigma-Aldrich), supplemented with penicillin/streptomycin, 10% FCS, and l-glutamine (Invitrogen). MEFs deficient of vinculin were provided by D. Critchley (University of Leicester, Leicester, UK; Saunders et al., 2006). For the culture of MEFs, nonessential vitamins and β-mercaptoethanol (Sigma-Aldrich) were added.
For transient transfections, Lipofectamine Plus (Invitrogen) was used according to the manufacturer's instructions. Cells were replated at 3 h after transfection in glass-bottom dishes (MatTek Corporation) coated with 10 μg/ml bovine plasma fibronectin (pFN; Sigma-Aldrich).
For perturbation of the actin cytoskeleton, cells expressing indicated vinculin constructs were treated for 60 min with 100 μM Y-27632 or for 30 min with 1 μM cytochalasin D (both obtained from Sigma-Aldrich) before fixation with 4% PFA.
Antibodies
Primary antibodies used for immunolabeling were anti-paxillin (clone 349; BD Biosciences), anti-talin 8d4 (binds only talin1; Sigma-Aldrich), talin C20 (recognizes talin1 and 2; Santa Cruz Biotechnology, Inc.), and anti–human vinculin (Sigma-Aldrich). 9EG7 and TS2/16 recognizing the active conformation and total pool of human β1 integrin (Bazzoni et al., 1995), respectively, were provided by D. Vestweber (University of Münster, Münster, Germany) and A. Sonnenberg (Netherlands Cancer Institute, Amsterdam, Netherlands), respectively.
Immunofluorescence and video microscopy
For ICA, cells expressing fluorophor-tagged constructs were plated on glass-bottom dishes and fixed with 3% PFA at 24–36 h after transfection. Cells were then imaged using an inverted microscope (IX70; Olympus) controlled by a Deltavision system (Applied Precision). For immunolabeling, cells plated on glass coverslips were fixed with 3% PFA for 15 min, permeabilized for 5 min with 0.5% Triton X-100 (Sigma-Aldrich), and subsequently incubated for 45 min with primary antibodies directed against indicated proteins. After three washes with PBS, cells were incubated in the presence of secondary antibodies conjugated to Cy2, 3, or 5 (Jackson ImmunoResearch Laboratories). In the case of colabeling for actin, TRITC-phalloidin (Invitrogen) was added to cells together with the secondary antibody. After three more washes, coverslips were mounted on slides using elvanol (Monwiol 4-88; Sigma-Aldrich). Mounted cells were imaged using the Deltavision microscope system.
Ham's F12 medium was used for time-lapse imaging of cells using an inverted microscope (Axiovert 200M; Carl Zeiss, Inc.) driven by software (IP Lab; BD Biosciences) and equipped with an incubation chamber (37°C). Images were taken with a 100× α Plan-Fluar objective (Carl Zeiss, Inc.) at the indicated time intervals.
For velocity measurements of YFP-vinT and GFP-vinT12, time-lapse sequences of cells expressing these constructs were 2D high-pass filtered to better visualize moving spots. Spots were then tracked using the ImageJ manual particle tracker. The dynamics of each construct were assessed by measuring 20–30 tracks in three different cells.
Image processing and Pearson's correlation coefficient
Images, unless stated otherwise, were processed using ImageJ version 1.32j. For calculation of the Pearson's correlation coefficient, captured CFP and YFP images were background subtracted (Zamir et al., 1999), an overlay image was created, a threshold was set to restrict analysis to FAs, and Pearson's correlation was calculated using the Image Correlator Plus plug-in for ImageJ. The Pearson's correlation coefficient reflects the degree of linear relationship between two variables; in this case, the fluorescence intensities of two fluorescently tagged proteins. ImageJ was also used to create fluorescence intensity line profiles over FAs of CFP and YFP merged images. Time-lapse images captured by IP Lab were exported to ImageJ and transformed into AVI time-lapse movies (ImageJ).
Photoshop 7 and Illustrator 9 (both from Adobe) were used for the assembly of figures for publication.
FRAP
A confocal microscope (TCS SP2; Leica) and an inverted microscope (IX70) equipped with a 488-nm laser lines (Olympus) under the control of software (DeltaVisionRT) were used for FRAP experiments. Cells plated on glass-bottom dishes and expressing indicated YFP- or GFP-tagged constructs were imaged at 37°C in Ham's F12 medium.
FRAP with the confocal microscope was performed similarly to the procedure described previously (Ballestrem et al., 2001). Initial fluorescence intensity was measured at low laser powers (5%) followed by photobleaching of a 1.5-μm-diam area in FAs at 100% laser power for 10 iterations. The fluorescence recovery was then followed with low laser powers at 5-s intervals until the fluorescence intensities recovered to a plateau. Images were transferred into ImageJ, background subtracted, and corrected for fluorescence loss caused by photobleaching. Corrected recovery fluorescence intensities were normalized to prebleach intensity. The MF was calculated according to MF = 100 × (Finfl − F(0))/(Fpre − F(0)), where Fpre is the prebleach intensity of bleached area, Finf is the postbleach intensity at the plateau, and F(0) is the postbleach intensity at time 0 in the bleached area. For determination of t 1/2 of recovery, the normalized recovery data were fitted to the single exponential equation F(t) = MF × (1 − eτt) and the t 1/2 of recovery was calculated by t 1/2 = ln 0.5/−τ.
For FRAP using the Deltavision system, 1.5-μm-diam regions of interest were selected. MF and t 1/2 were calculated using softWoRx FRAP analysis software (see application notes at http://www.api.com/lifescience/dv-pubsandapps.html#appnotes).
Immunoprecipitation
Where indicated, cells were detached with 0.05% (wt/vol) trypsin and 0.02% EDTA or lysed in situ. Lysis was performed at 4°C for 30 min in 150 mM NaCl, 20 mM Tris, 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 10 mM EDTA, pH 7.4, and 1% Triton X-100. Where indicated, cells were treated for 10 min at room temperature with the cross-linking agent dimethyl 3,3′-dithiobispropionimidate (Thermo Fisher Scientific) before lysis. Lysates were passed 10 times through a narrow bore tip before centrifugation (800 g for 10 min at 4°C) and protein G–Sepharose (GE Healthcare) was subsequently added to the supernatant for 30–60 min at 4°C. After centrifugation, immunoprecipitating mAbs were added to the lysate (1 μg/ml final concentration) together with protein G–Sepharose for 16 h at 4°C. Protein G–Sepharose was then collected and washed four times in lysis buffer by centrifugation. Immunoprecipitated complexes were eluted at 70°C for 5 min in sample buffer (80 mM Tris, 2.8% [wt/vol] SDS, 12% [vol/vol] glycerol, and 0.01% [wt/vol] bromophenol blue containing 2% [vol/vol] β-mercaptoethanol). Samples were then subjected to SDS-PAGE and Western blotting using an infrared imaging system (Odyssey; LI-COR Biosciences).
Biochemical isolation of plasma membrane fractions enriched for integrin-cytoskeletal/signaling complexes
Isolation of plasma membrane fractions enriched for integrin membrane complexes was performed according to a modified protocol of Plopper and Ingber (1993). In brief, NIH3T3 cells (2 × 107) were incubated for 60 min at 37°C with rotation in the presence of 107 pFN-coated beads (4.5-μm-diam tosyl-activated paramagnetic beads; Invitrogen) and dimethyl 3,3′-dithiobispropionimidate. Bead–cell complexes were isolated using a magnetic particle concentrator (Invitrogen) and lysed with sonication (VibraCell; Jencons) in ice-cold cytoskeletal stabilization buffer (50 mM NaCl, 150 mM sucrose, 3 mM MgCl2, 0.5 mM 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride, 1 mM NaVO4, 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 10 mM Pipes, pH 6.0, containing 0.5% [wt/vol] Triton X-100). Beads were washed with lysis buffer by repeated magnetic pelleting and bead-bound material was eluted in sample buffer and processed for Western blotting as described in Immunoprecipitation section. For quantification, band intensities were background subtracted and normalized to the α5 signal intensity before being expressed as a ratio of vin880 signal to vinFL signal.
Statistical analysis
t test or the Fisher Sign test (where indicated) were used to test statistical significances between two groups of data and analysis of variance (ANOVA) for comparison of multiple groups. Data found to be significant for ANOVA were tested post hoc by Tukey's honest significant difference (HSD) test. KaleidaGraph software (Synergy Software) was used for all statistical analysis.
Online supplemental material
Fig. S1 adds information of how we quantified the localization of two molecules using the Pearson's correlation coefficient. Fig. S2 shows that endogenous talin colocalizes with vin880 in enlarged FAs and that overexpression of YFP-talin does not lead to FA increase. Fig. S3 shows that paxillin, FAK, and α-actinin do not coimmunoprecipitate with vin880, supporting our view that they only transiently associate with the tight integrin–talin–vinculin complex.
Video 1 complements data presented in Fig. 7 (A–C) showing that large protrusions have reduced levels of vinT. Videos 2–4 correspond to data presented in Fig. 7 F outlining the differences in dynamics between vin880, vinT12, and vinT. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200703036/DC1.
Acknowledgments
We thank Susan Craig for the vinT12 and vin258 constructs; W. Ziegler for the vinLD constructs; David Critchley for the MEF vinculin-null cells; Ken Yamada for the talin constructs; Rick Horwitz for GFP/YFP–α5 integrin and CFP/YFP-FAK; Andrew Gilmore, Andreas Prokop, and Richard Kammerer for critical reading of the manuscript; Peter March and the members of the Faculty of Life Sciences Bioimaging core facility for the help with various microscopes; and Barbara Ciani and David Reynolds for assistance with statistical analysis.
C. Ballestrem and M. Humphries were supported by Wellcome Trust grants 077100 and 074941, respectively.
Abbreviations used in this paper: FA, focal adhesion; HSD, honest significant difference; ICA, image correlation analysis; MEF, mouse embryonic fibroblast; MF, mobile fraction; pFN, plasma fibronectin; PIP2, phosphatidylinositol (4,5)-bisphosphate; ROCK, Rho-kinase; vinFL, full-length vinculin; vinT, vinculin tail.
References
- Bakolitsa, C., J.M. de Pereda, C.R. Bagshaw, D.R. Critchley, and R.C. Liddington. 1999. Crystal structure of the vinculin tail suggests a pathway for activation. Cell. 99:603–613. [DOI] [PubMed] [Google Scholar]
- Bakolitsa, C., D.M. Cohen, L.A. Bankston, A.A. Bobkov, G.W. Cadwell, L. Jennings, D.R. Critchley, S.W. Craig, and R.C. Liddington. 2004. Structural basis for vinculin activation at sites of cell adhesion. Nature. 430:583–586. [DOI] [PubMed] [Google Scholar]
- Balaban, N.Q., U.S. Schwarz, D. Riveline, P. Goichberg, G. Tzur, I. Sabanay, D. Mahalu, S. Safran, A. Bershadsky, L. Addadi, and B. Geiger. 2001. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3:466–472. [DOI] [PubMed] [Google Scholar]
- Ballestrem, C., B. Hinz, B.A. Imhof, and B. Wehrle-Haller. 2001. Marching at the front and dragging behind: differential αVβ3-integrin turnover regulates focal adhesion behavior. J. Cell Biol. 155:1319–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballestrem, C., N. Erez, J. Kirchner, Z. Kam, A. Bershadsky, and B. Geiger. 2006. Molecular mapping of tyrosine-phosphorylated proteins in focal adhesions using fluorescence resonance energy transfer. J. Cell Sci. 119:866–875. [DOI] [PubMed] [Google Scholar]
- Bazzoni, G., D.T. Shih, C.A. Buck, and M.E. Hemler. 1995. Monoclonal antibody 9EG7 defines a novel beta 1 integrin epitope induced by soluble ligand and manganese, but inhibited by calcium. J. Biol. Chem. 270:25570–25577. [DOI] [PubMed] [Google Scholar]
- Brown, M.C., and C.E. Turner. 2004. Paxillin: adapting to change. Physiol. Rev. 84:1315–1339. [DOI] [PubMed] [Google Scholar]
- Burridge, K., and M. Chrzanowska-Wodnicka. 1996. Focal adhesions, contractility, and signaling. Annu. Rev. Cell Dev. Biol. 12:463–518. [DOI] [PubMed] [Google Scholar]
- Chandrasekar, I., T.E. Stradal, M.R. Holt, F. Entschladen, B.M. Jockusch, and W.H. Ziegler. 2005. Vinculin acts as a sensor in lipid regulation of adhesion-site turnover. J. Cell Sci. 118:1461–1472. [DOI] [PubMed] [Google Scholar]
- Chen, H., D.M. Cohen, D.M. Choudhury, N. Kioka, and S.W. Craig. 2005. Spatial distribution and functional significance of activated vinculin in living cells. J. Cell Biol. 169:459–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., D.M. Choudhury, and S.W. Craig. 2006. Coincidence of actin filaments and talin is required to activate vinculin. J. Biol. Chem. 281:40389–40398. [DOI] [PubMed] [Google Scholar]
- Cluzel, C., F. Saltel, J. Lussi, F. Paulhe, B.A. Imhof, and B. Wehrle-Haller. 2005. The mechanisms and dynamics of αvβ3 integrin clustering in living cells. J. Cell Biol. 171:383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, D.M., H. Chen, R.P. Johnson, B. Choudhury, and S.W. Craig. 2005. Two distinct head-tail interfaces cooperate to suppress activation of vinculin by talin. J. Biol. Chem. 280:17109–17117. [DOI] [PubMed] [Google Scholar]
- Cohen, D.M., B. Kutscher, H. Chen, D.B. Murphy, and S.W. Craig. 2006. A conformational switch in vinculin drives formation and dynamics of a talin-vinculin complex at focal adhesions. J. Biol. Chem. 281:16006–16015. [DOI] [PubMed] [Google Scholar]
- Coll, J.L., A. Ben-Ze'ev, R.M. Ezzell, J.L. Rodriguez Fernandez, H. Baribault, R.G. Oshima, and E.D. Adamson. 1995. Targeted disruption of vinculin genes in F9 and embryonic stem cells changes cell morphology, adhesion, and locomotion. Proc. Natl. Acad. Sci. USA. 92:9161–9165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Critchley, D.R. 2000. Focal adhesions - the cytoskeletal connection. Curr. Opin. Cell Biol. 12:133–139. [DOI] [PubMed] [Google Scholar]
- Eimer, W., M. Niermann, M.A. Eppe, and B.M. Jockusch. 1993. Molecular shape of vinculin in aqueous solution. J. Mol. Biol. 229:146–152. [DOI] [PubMed] [Google Scholar]
- Fillingham, I., A.R. Gingras, E. Papagrigoriou, B. Patel, J. Emsley, D.R. Critchley, G.C. Roberts, and I.L. Barsukov. 2005. A vinculin binding domain from the talin rod unfolds to form a complex with the vinculin head. Structure. 13:65–74. [DOI] [PubMed] [Google Scholar]
- Geiger, B., A. Bershadsky, R. Pankov, and K.M. Yamada. 2001. Transmembrane crosstalk between the extracellular matrix–cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2:793–805. [DOI] [PubMed] [Google Scholar]
- Giannone, G., G. Jiang, D.H. Sutton, D.R. Critchley, and M.P. Sheetz. 2003. Talin1 is critical for force-dependent reinforcement of initial integrin– cytoskeleton bonds but not tyrosine kinase activation. J. Cell Biol. 163:409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg, M.H., A. Partridge, and S.J. Shattil. 2005. Integrin regulation. Curr. Opin. Cell Biol. 17:509–516. [DOI] [PubMed] [Google Scholar]
- Gupton, S.L., and C.M. Waterman-Storer. 2006. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration. Cell. 125:1361–1374. [DOI] [PubMed] [Google Scholar]
- Hemmings, L., D.J. Rees, V. Ohanian, S.J. Bolton, A.P. Gilmore, B. Patel, H. Priddle, J.E. Trevithick, R.O. Hynes, and D.R. Critchley. 1996. Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J. Cell Sci. 109:2715–2726. [DOI] [PubMed] [Google Scholar]
- Hu, K., L. Ji, K.T. Applegate, G. Danuser, and C.M. Waterman-Storer. 2007. Differential transmission of actin motion within focal adhesions. Science. 315:111–115. [DOI] [PubMed] [Google Scholar]
- Huttelmaier, S., P. Bubeck, M. Rudiger, and B.M. Jockusch. 1997. Characterization of two F-actin-binding and oligomerization sites in the cell-contact protein vinculin. Eur. J. Biochem. 247:1136–1142. [DOI] [PubMed] [Google Scholar]
- Huttelmaier, S., O. Mayboroda, B. Harbeck, T. Jarchau, B.M. Jockusch, and M. Rudiger. 1998. The interaction of the cell-contact proteins VASP and vinculin is regulated by phosphatidylinositol-4,5-bisphosphate. Curr. Biol. 8:479–488. [DOI] [PubMed] [Google Scholar]
- Izard, T., G. Tran Van Nhieu, and P.R. Bois. 2006. Shigella applies molecular mimicry to subvert vinculin and invade host cells. J. Cell Biol. 175:465–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen, M.E., E. Kim, H. Liu, L.M. Fujimoto, A. Bobkov, N. Volkmann, and D. Hanein. 2006. Three-dimensional structure of vinculin bound to actin filaments. Mol. Cell. 21:271–281. [DOI] [PubMed] [Google Scholar]
- Jockusch, B.M., and M. Rudiger. 1996. Crosstalk between cell adhesion molecules: vinculin as a paradigm for regulation by conformation. Trends Cell Biol. 6:311–315. [DOI] [PubMed] [Google Scholar]
- Kirchner, J., Z. Kam, G. Tzur, A.D. Bershadsky, and B. Geiger. 2003. Live-cell monitoring of tyrosine phosphorylation in focal adhesions following microtubule disruption. J. Cell Sci. 116:975–986. [DOI] [PubMed] [Google Scholar]
- Lee, H.S., R.M. Bellin, D.L. Walker, B. Patel, P. Powers, H. Liu, B. Garcia-Alvarez, J.M. de Pereda, R.C. Liddington, N. Volkmann, et al. 2004. Characterization of an actin-binding site within the talin FERM domain. J. Mol. Biol. 343:771–784. [DOI] [PubMed] [Google Scholar]
- Lele, T.P., J. Pendse, S. Kumar, M. Salanga, J. Karavitis, and D.E. Ingber. 2006. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J. Cell. Physiol. 207:187–194. [DOI] [PubMed] [Google Scholar]
- Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2:444–456. [DOI] [PubMed] [Google Scholar]
- Menkel, A.R., M. Kroemker, P. Bubeck, M. Ronsiek, G. Nikolai, and B.M. Jockusch. 1994. Characterization of an F-actin–binding domain in the cytoskeletal protein vinculin. J. Cell Biol. 126:1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel, B., A.R. Gingras, A.A. Bobkov, L.M. Fujimoto, M. Zhang, R.C. Liddington, D. Mazzeo, J. Emsley, G.C. Roberts, I.L. Barsukov, and D.R. Critchley. 2006. The activity of the vinculin binding sites in talin is influenced by the stability of the helical bundles that make up the talin rod. J. Biol. Chem. 281:7458–7467. [DOI] [PubMed] [Google Scholar]
- Plopper, G., and D.E. Ingber. Rapid induction and isolation of focal adhesion complexes. 1993. Biochem. Biophys. Res. Commun. 193:571–578. [DOI] [PubMed] [Google Scholar]
- Ponti, A., M. Machacek, S.L. Gupton, C.M. Waterman-Storer, and G. Danuser. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science. 305:1782–1786. [DOI] [PubMed] [Google Scholar]
- Salgia, R., J.L. Li, S.H. Lo, B. Brunkhorst, G.S. Kansas, E.S. Sobhany, Y. Sun, E. Pisick, M. Hallek, T. Ernst, et al. 1995. Molecular cloning of human paxillin, a focal adhesion protein phosphorylated by P210BCR/ABL. J. Biol. Chem. 270:5039–5047. [DOI] [PubMed] [Google Scholar]
- Saunders, R.M., M.R. Holt, L. Jennings, D.H. Sutton, I.L. Barsukov, A. Bobkov, R.C. Liddington, E.A. Adamson, G.A. Dunn, and D.R. Critchley. 2006. Role of vinculin in regulating focal adhesion turnover. Eur. J. Cell Biol. 85:487–500. [DOI] [PubMed] [Google Scholar]
- Schaller, M.D., C.A. Otey, J.D. Hildebrand, and J.T. Parsons. 1995. Focal adhesion kinase and paxillin bind to peptides mimicking β integrin cytoplasmic domains. J. Cell Biol. 130:1181–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subauste, M.C., O. Pertz, E.D. Adamson, C.E. Turner, S. Junger, and K.M. Hahn. 2004. Vinculin modulation of paxillin–FAK interactions regulates ERK to control survival and motility. J. Cell Biol. 165:371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro, S., S.J. Shattil, K. Eto, V. Tai, R.C. Liddington, J.M. de Pereda, M.H. Ginsberg, and D.A. Calderwood. 2003. Talin binding to integrin beta tails: a final common step in integrin activation. Science. 302:103–106. [DOI] [PubMed] [Google Scholar]
- Tanaka, T., R. Yamaguchi, H. Sabe, K. Sekiguchi, and J.M. Healy. 1996. Paxillin association in vitro with integrin cytoplasmic domain peptides. FEBS Lett. 399:53–58. [DOI] [PubMed] [Google Scholar]
- Turner, C.E. 2000. Paxillin and focal adhesion signalling. Nat. Cell Biol. 2:E231–E236. [DOI] [PubMed] [Google Scholar]
- Turner, C.E., J.R. Glenney Jr., and K. Burridge. 1990. Paxillin: a new vinculin-binding protein present in focal adhesions. J. Cell Biol. 111:1059–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallotton, P., S.L. Gupton, C.M. Waterman-Storer, and G. Danuser. 2004. Simultaneous mapping of filamentous actin flow and turnover in migrating cells by quantitative fluorescent speckle microscopy. Proc. Natl. Acad. Sci. USA. 101:9660–9665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volberg, T., B. Geiger, Z. Kam, R. Pankov, I. Simcha, H. Sabanay, J.L. Coll, E. Adamson, and A. Ben-Ze'ev. 1995. Focal adhesion formation by F9 embryonal carcinoma cells after vinculin gene disruption. J. Cell Sci. 108:2253–2260. [DOI] [PubMed] [Google Scholar]
- Wegener, K.L., A.W. Partridge, J. Han, A.R. Pickford, R.C. Liddington, M.H. Ginsberg, and I.D. Campbell. 2007. Structural basis of integrin activation by talin. Cell. 128:171–182. [DOI] [PubMed] [Google Scholar]
- Winkler, J., H. Lunsdorf, and B.M. Jockusch. 1996. The ultrastructure of chicken gizzard vinculin as visualized by high-resolution electron microscopy. J. Struct. Biol. 116:270–277. [DOI] [PubMed] [Google Scholar]
- Wood, C.K., C.E. Turner, P. Jackson, and D.R. Critchley. 1994. Characterisation of the paxillin-binding site and the C-terminal focal adhesion targeting sequence in vinculin. J. Cell Sci. 107:709–717. [PubMed] [Google Scholar]
- Xu, W., H. Baribault, and E.D. Adamson. 1998. Vinculin knockout results in heart and brain defects during embryonic development. Development. 125:327–337. [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar, R., C. Ballestrem, Z. Kam, and B. Geiger. 2003. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116:4605–4613. [DOI] [PubMed] [Google Scholar]
- Zaidel-Bar, R., R. Milo, Z. Kam, and B. Geiger. 2007. A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 120:137–148. [DOI] [PubMed] [Google Scholar]
- Zamir, E., and B. Geiger. 2001. Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 114:3583–3590. [DOI] [PubMed] [Google Scholar]
- Zamir, E., B.Z. Katz, S. Aota, K.M. Yamada, B. Geiger, and Z. Kam. 1999. Molecular diversity of cell-matrix adhesions. J. Cell Sci. 112:1655–1669. [DOI] [PubMed] [Google Scholar]
- Ziegler, W.H., R.C. Liddington, and D.R. Critchley. 2006. The structure and regulation of vinculin. Trends Cell Biol. 16:453–460. [DOI] [PubMed] [Google Scholar]