Abstract
Filamin A (FLNa) can effect orthogonal branching of F-actin and bind many cellular constituents. FLNa dimeric subunits have N-terminal spectrin family F-actin binding domains (ABDs) and an elongated flexible segment of 24 immunoglobulin (Ig) repeats. We generated a library of FLNa fragments to examine their F-actin binding to define the structural properties of FLNa that enable its various functions. We find that Ig repeats 9–15 contain an F-actin–binding domain necessary for high avidity F-actin binding. Ig repeats 16–24, where most FLNa-binding partners interact, do not bind F-actin, and thus F-actin does not compete with Ig repeat 23 ligand, FilGAP. Ig repeats 16–24 have a compact structure that suggests their unfolding may accommodate pre-stress–mediated stiffening of F-actin networks, partner binding, mechanosensing, and mechanoprotection properties of FLNa. Our results also establish the orientation of FLNa dimers in F-actin branching. Dimerization, mediated by FLNa Ig repeat 24, accounts for rigid high-angle FLNa/F-actin branching resistant to bending by thermal forces, and high avidity F-actin binding and cross-linking.
Introduction
The dynamic remodeling of actin filaments (F-actin) concentrated at the cell periphery is responsible for cell translocation, for cell shape changes, and for cellular resistance to potentially disruptive mechanical stresses. These mechanical tasks depend in large measure on the coherence of three-dimensional (3D) F-actin gel networks (Discher et al., 2005), and cross-linking agents confer this coherence on intracellular F-actin (Matsudaira, 1994).
The most potent among many F-actin cross-linking agents is the first recognized nonmuscle F-actin-binding protein, now known as filamin A (FLNa). FLNa expression is essential for mammalian development (Feng et al., 2006; Ferland et al., 2006; Hart et al., 2006) and even small FLNa deletions or point mutations lead to diverse congenital anomalies (Robertson et al., 2003; Robertson, 2005; Kyndt et al., 2007). Cultured cells lacking FLNa protein expression exhibit unstable surfaces, are incapable of locomotion, and have impaired mechanical resistance (Flanagan et al., 2001; Kainulainen et al., 2002). FLNa confers elastic properties on F-actin networks subjected to prestress in vitro, and the network rigidities achieved simulate values observed for prestressed living cells (Gardel et al., 2006).
The power of FLNa as an F-actin gelation promoter resides in its efficiency in recruiting F-actin into extended networks, and the source of this efficiency is its ability to orient each cross-linked rod-like actin filament at right angles, thereby minimizing redundant cross-linking (Hartwig et al., 1980; Hartwig and Shevlin, 1986). In addition, the mechanical properties of F-actin/FLNa networks depend on FLNa's capacity to cross-link F-actin with high avidity while permitting sufficient interfilament flexibility for networks to exhibit fully reversible elastic deformation in response to high stresses without rupturing (Gardel et al., 2006).
FLNa also binds numerous cellular components other than F-actin, including membrane receptors, enzymes, channels, signaling intermediates, and transcription factors, and it modulates the functional activities of these binding partners (Stossel et al., 2001; Feng and Walsh, 2004; Popowicz et al., 2006). Because many of these binding partners regulate actin assembly and disassembly, FLNa resides at the center of a complex feedback system in which signaling around it organizes actin architecture that, in turn, regulates signaling. A comprehension of the fine structure of FLNa is essential to understand how this molecule can execute diverse and complex functions and to relate specific arrangements of these functions to a growing catalogue of biological and clinical abnormalities ascribable to FLNa.
FLNa is a homodimer with conserved F-actin–binding domains (ABDs) consisting of two calponin homology (CH) sequences (CH1 & CH2) at the amino termini (N-T) of its 280.7-kD, 80-nm-long subunits. The amino acid sequence of FLNa's ABD is representative of ABDs of the α-actinin or spectrin superfamily (Hartwig, 1995), with the exception that the FLNa ABD has a unique calmodulin-binding site positioned in the CH1, and calcium-activated calmodulin (holocalmodulin) competes at this site for F-actin binding (Nakamura et al., 2005). 24 β pleated sheet repeat (Ig) segments separate the ABDs from a carboxyl-terminal (C-T) subunit self-association site, with two intervening calpain-sensitive “hinge” sequences separating repeats 15 and 16 (hinge 1) and repeats 23 and 24 (hinge 2), hinge 1 contributes to the high elasticity of prestressed FLNa/F-actin gels (Gardel et al., 2006). The series of repeats proximal and distal to hinge 1 are designated “rods 1 and 2” (Gorlin et al., 1990). Most FLNa binding partners interact with rod 2 and the molecular interfaces mediating some of these interactions at the atomic level are known (Kiema et al., 2006; Nakamura et al., 2006). Despite all of this information, how FLNa binds and architecturally organizes F-actin and serves as a functional platform for multiple cellular constituents is completely obscure.
We have therefore generated an extensive library of FLNa fragments and examined their individual and combined contributions to F-actin binding, F-actin branching, and interactions with a non-F-actin binding partner. The results inform a plausible model for how FLNa orthogonally cross-links F-actin with high avidity while simultaneously accommodating sufficient flexibility to account for pre-stress–mediated elasticity and binding to other ligands. The identification of specific FLNa domains responsible for these diverse functions sets the stage for the engineering of targeted reagents for modifying these functions and sorting out their roles at the cell and organism level.
Results
Expression and purification of human FLNa C-T and N-T truncates
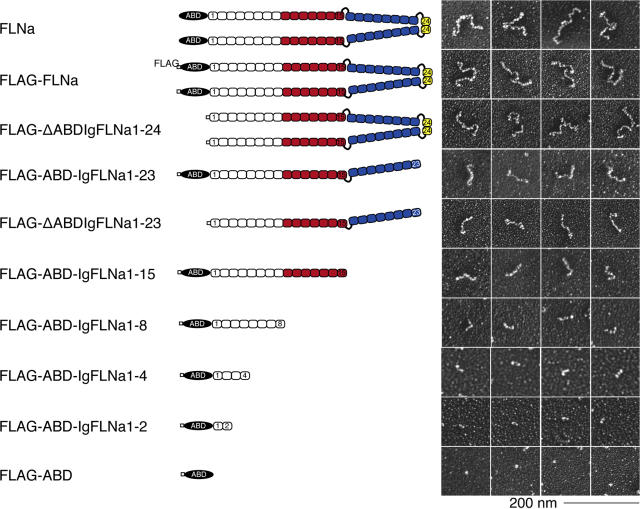
We engineered FLNa cDNAs encoding unmodified or FLAG-tagged full-length FLNa dimers or FLNa truncates encompassing defined sections of FLNa subunits, and expressed the recombinant proteins in Sf9 insect cells (Fig. 1).Truncations of the last C-T repeat, the dimerization site IgFLNa24, result in monomeric fragments, and further truncations between Ig repeat junctions lead to sequentially shorter constructs and eventually only the ABD. We also generated FLNa N-T truncates containing Ig repeats 1–23, Ig 1–24 lacking the ABD (ΔABD), and full-length FLNa dimers lacking the hinge 1 or hinge 2 sequences. We isolated full-length FLNa by chromatography as previously described (Nakamura et al., 2002) and purified constructs tagged at their N-T with the FLAG epitope using anti-FLAG mAb agarose affinity chromatography followed by gel filtration. Fig. S1 C shows that all recombinant proteins evaluated by SDS-PAGE consisted of single polypeptides of predicted molecular weight, and Fig. S1 D depicts the inferred isoelectric points of FLNa Ig repeats (Fig. S1 is available at http://www.jcb.org/cgi/content/full/jcb.200707073/DC1). In addition to the sequentially truncated FLNa subunit constructs, we prepared and purified His (hexahistidine)-tagged monomeric truncates encompassing 8 Ig-like repeats from the FLNa subunit (His-IgFLNa1-8, His-IgFLNa8-15, His-IgFLNa16-23; Fig. 2 A), and versions of these segments containing hinge 2 plus Ig repeat 24 at the C-T with (His-ABD-IgFLNa1-8+24, His-ABD-IgFLNa8-15+24 and His-ABD-IgFLNa16-24; Fig. S5, A and D) or without the N-T ABD (His-IgFLNa1-8+24, His-IgFLNa8-15+24 and His-IgFLNa16-24; Fig. 2, A and B). The constructs were of predicted sizes, and addition of hinge 2 and repeat 24 resulted in dimeric molecules (Fig. 2, A and B; Fig. S2).
Figure 1.
Recombinant FLNa constructs used in this study. Diagram of human FLNa and its recombinant fragments fused to a FLAG-tag. Color indicates the actin-binding domain (black), the secondary actin-binding site found in this study (red), the rod 2 segment (blue), and the dimerization domain (yellow). The right panel shows electron micrographs of rotary-shadowed molecules corresponding to the diagram on the left.
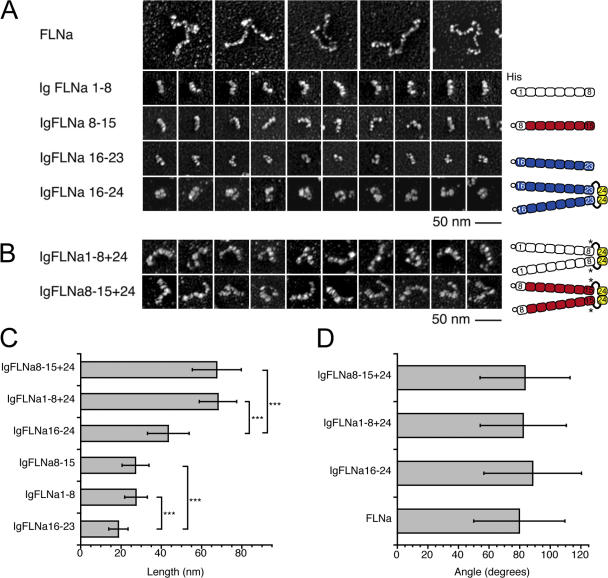
Figure 2.
Structures and topological properties of FLNa constructs. (A) Electron micrographs of rotary shadowed FLNa and truncated molecules fused to a His (hexahistidine)-tag. The 15 Ig repeats in the rod 1 domain are similar in structural composition as can be seen by comparing IgFLNa1-8 with IgFLNa8-15. The rod 2 domain has a more globular and compact appearance. The dimerized IgFLNa16-24 is also compact and consists of globular subdomains. (B) Addition of hinge 2 and dimerization domain Ig repeat 24 to IgFLNa1-8 or IgFLNa8-15 duplicates the size (C) and retains the rod-like structures. Asterisk indicates insertion of Ala residue derived from restriction site in the N-terminal of hinge 2 (see Materials and methods). (C) Contour length of FLNa truncates. The rod 2 segments are significantly shorter than the constructs that have the same number of Ig-like repeats but are derived from the rod 1 segment (n > 1,000 molecules). P < 0.0001 (***) (t test). (D) Angles at the dimerization sites of full-length FLNa and FLNa constructs (n > 200 molecules). The average angle of the C-terminal domain of FLNa is ∼80°. The dimerized constructs have similar angle and angle distribution as full-length FLNa (see Fig. S3, B and C). Data are means ± SD.
Structure of FLNa molecules: role of the subdomains
Fig. S1 A compares the ultrastructures of intact FLNa molecules to the recombinant C-T and N-T truncated proteins. FLAG-tagged full-length FLNa molecules appear identical to native FLNa purified from rabbit lung macrophages (Hartwig and Stossel, 1981) and to untagged recombinant human FLNa (Fig. 1 and Fig. S1 A) (Nakamura et al., 2002). All of the constructs were monodisperse, and the lengths of truncates from the rod 1 segment are directly proportional to the number of repeats expressed, as predicted (Fig. S1 B). The contour lengths of rod 2 segments are significantly shorter (19.2 ± 6 nm) than subsegments of rod 1 containing equivalent numbers of FLNa Ig repeats (28.5 ± 7 and 28.8 ± 8 nm for His-IgFLNa1-8 and His-IgFLNa8-15, respectively; n > 1,000, P < 0.0001, Fig. 2, A and C). Rod 2 segments appear more compact in electron micrographs than the elongated rod-like structures of rod 1 segments, although they are somewhat flexible as they take on variable shapes. Rod 2 dimers appear as three globular units that branch to form an L-shaped structure.
Although individual FLNa subunits exhibit flexibility along their contour lengths, which accounts for their variable shape in electron micrographs, they diverge from their self-association site at one end with fairly fixed high angles. Therefore, the N-T two-thirds of each subunit accounts for most of its flexibility, which is similar along the subunit contour length with the exception of a hot spot for bending at a distance of ∼60 nm from the free end (Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200707073/DC1). This location corresponds to the site of hinge 1 in the FLNa subunit sequence, and a subunit construct missing hinge 1 lacks this flexible hot spot. The C-T segments of FLNa dimers diverge at nearly right angles irrespective of whether the subunits contain hinge 2 (79.8 ± 30° versus 85.1 ± 34; Fig. S3 B). Dimeric rod fragments consisting of 8 Ig repeat segments derived from the FLNa subunits branch at similar angles as full-length FLNa (averaging ∼80–90°) irrespective of the subunit location from which the branches originate (Fig. 2 D; Fig. S3 C).
F-actin binding of FLNa and FLNa fragments reveals two F-actin binding sites on FLNa subunits and the importance of dimerization for high avidity F-actin binding
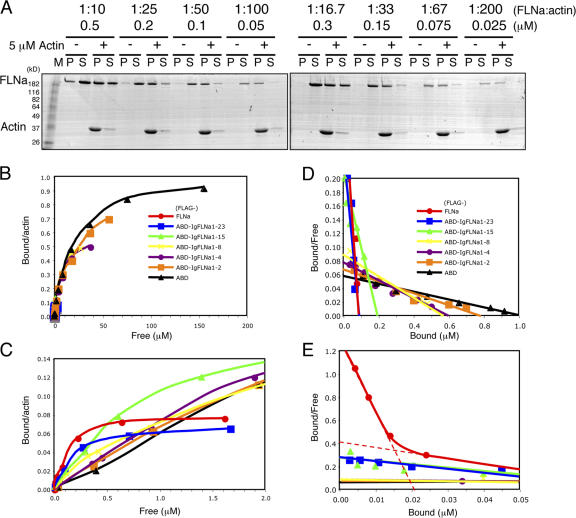
We evaluated the actin-binding properties of purified FLNa truncates by cosedimentation with 5 μM F-actin using either 1:10 (0.5 μM) or 1:100 (0.05 μM) concentrations of the recombinant FLNa proteins. Fig. 3 A shows Coomassie blue–stained SDS-PAGE gels of supernatant and pellet fractions prepared by centrifugation of FLNa after incubation with or without 5 μM F-actin.Because a portion of the recombinant protein sedimented in the absence of actin, we estimated specific actin-bound protein after subtracting the nonspecific component and expressed it as the percentage bound versus total input (Fig. 3 B). At a 1:10 ratio of FLNa to actin, 56.3 ± 6.3% (n = 3) of FLNa bound, whereas a significantly higher percentage (83.6 ± 21%, n = 3) of FLNa bound to F-actin when the ratio of FLNa to actin was 1:100. Recombinant FLNa proteins bound progressively less F-actin in inverse proportion to the number of rod Ig domains expressed, and only ∼10% (8.7 ± 1.9% and 11.7 ± 2.6% at 1:10 or 1:100 actin, respectively) of the ABD domains lacking attached Ig repeats bound F-actin.
Figure 3.
Binding of FLNa and FLNa truncates to F-actin. (A) Purified recombinant protein (0.5 or 0.05 μM) was incubated with or without 5 μM rabbit skeletal muscle actin in a polymerization buffer for 1 h at 25°C. F-actin (pellet) was separated from soluble actin (supernatant) by centrifugation. The supernatant (S) and pellet (P) fractions were diluted to equivalent volumes in SDS-PAGE sample buffer and equal volumes of each were analyzed by SDS-PAGE. The slower migrating polypeptide (arrow) is the recombinant FLNa construct. The migration of actin is indicated. (B) The amount of free and bound recombinant protein was determined by quantitative densitometric analysis from the slab gels (A) and is plotted as bound/total (%) for each of the FLNa recombinant proteins. Data are means ± SD for three separate measurements. (C) ABD deficient FLNa dimers and IgFLNa8-15+24 align F-actin into bundles. 2 μM F-actin was incubated in the absence (top left) or presence of 0.4 μM ΔABDIgFLNa1-23 (monomer: top middle) or 0.4 μM ΔABDIgFLNa1-24 (dimer: top right). Dimeric truncated molecules consisting each of 2 eight-domain monomers dimerized by repeat 24 were also incubated with F-actin (bottom). Bar, 200 nm.
Deletion of the ABD from FLNa subunits (ΔABDIgFLNa1-24) greatly diminished F-actin binding, but did not completely abolish it (Fig. 3 B). This binding was dependent on FLNa subunit dimer formation, because a construct lacking the self-association repeat, IgFLNa24 (ΔABDIgFLNa1-23) hardly interacted with F-actin (Fig. 3 B). These data suggest that FLNa can interact with F-actin in the absence of the ABD, and negative staining of F-actin incubated with ΔABDIgFLNa1-24 at high ratios revealed the formation of actin filament bundles. IgFLNa8-15+24 could also induce bundling. This bundling was dependent on dimer formation, as ΔABDIgFLNa1-23 and IgFLNa8-15 did not have this effect (Fig. 3 C).
We also determined the binding affinities (Kapp) and stoichiometries of the various FLNa truncates for F-actin. Fig. 4 A and Fig. S4 A show a representative binding assay of Coomassie blue–stained SDS-PAGE gels of supernatant and pellet fractions prepared by centrifugation of FLNa and its recombinant truncates after incubation with or without 5 μM F-actin (Fig. S4 is available at http://www.jcb.org/cgi/content/full/jcb.200707073/DC1). Binding of FLNa and its truncates to F-actin was specific and saturable.Fig. 4, B and C (note that Fig. 4 C is an enlarged view of Fig. 4 B), plot bound FLNa against free (supernatant) FLNa, and Fig. 4, D and E, show these data in the form of a Scatchard plot. The nonlinearity of binding of full-length FLNa indicates either multiple binding sites or negative cooperativity (Klotz and Hunston, 1971; Wilkinson, 2004). A nonlinear regression analysis of the data reveals a high affinity site of Kapp = 0.017 μM and a low affinity site of Kapp = 0.22 μM, assuming a two independent site model justified by additional findings described below (Fig. 4 E and Table I).The binding parameters reveal limited high affinity sites for the binding of full-length FLNa protein to actin (∼1 FLNa dimer per 92 actin subunits in F-actin or 1 FLNa molecule for every 255 nm of F-actin contour length).
Figure 4.
Binding parameters of the different recombinant FLNa constructs for F-actin. (A) Representative cosedimentation experiment used to determine the apparent dissociation constant of FLNa for F-actin. The concentration of FLNa was varied in a fixed 5 μM concentration of F-actin. (B) Amount of free and bound FLNa construct determined from panel A is plotted as indicated. The hyperbolic binding behavior displayed by intact FLNa indicates multiple bindings or negative cooperativity. Klotz plots are provided in Fig. S4. (C) Enlargement of critical binding data between the X-axis values of 0 and 2.0 μM of free FLNa. (D) Scatchard plot of binding data. (E) Enlargement of critical binding region on X-axis of 0 to 0.05 μM for bound FLNa. Each graph plots the data resulting from a set of representative experiments. Each of these experiments was repeated at least three times.
Table I.
Dissociation constants and stoichiometries for the filamin A constructs
| Constructs | Dissociation constant [μM] |
Stoichiometry [ratio to actin, mol/mol] |
|---|---|---|
| FLNa (high avidity) | 0.017 (0.013–0.021)a | 46b (39–54)a |
| FLNa (low affinity) | 0.22 (0.15–0.28) | 10.5b (9.5–11.4) |
| ABD-IgFLNa1-23 | 0.31 (0.28–0.34) | 12 (11.8–12.5) |
| ABD-IgFLNa1-15 | 0.67 (0.38–0.97) | 5.4 (4.8–6.5) |
| ABD-IgFLNa1-8 | 7.1 (6.0–8.1) | 1.8 (1.7–1.9) |
| ABD-IgFLNa1-4 | 7.5 (6.2–8.9) | 1.7 (1.6–1.8) |
| ABD-IgFLNa1-2 | 11.8 (8.1–15.6) | 1.3 (1.2–1.5) |
| ABD | 17 (14.5–20.4) | 1 (0.95–1.05) |
95% confidence interval.
Actin bound mol per FLNa subunit.
Table I summarizes the binding affinities and stoichiometries of the different FLNa truncates. These data reveal single classes of binding sites for F-actin on monomeric FLNa constructs containing the N-T ABD. FLNa's ABD alone binds F-actin at a 1:1 ratio (mol/mol to each actin subunit in filaments), although it has a Kapp that is 1,000-fold less (Kapp= 17 μM) than intact FLNa. Addition of the first half of rod domain 1 (IgFLNa1-8) to the ABD did not greatly increase its binding parameters. However, further extension of the rod 1 domain with the addition of IgFLNa9-15 increased its Kapp for actin ∼25-fold, implicating an additional F-actin binding region in the distal half of rod segment 1. Addition of rod 2 resulted in only a small enhancement in affinity for F-actin compared with the ABD-rod 1 construct.
We could not determine the affinity and stoichiometry of monomeric or dimeric FLNa constructs containing N-terminal ABDs and 8 Ig repeats encompassing proximal or distal halves of rod segment 1, because we could not prepare these proteins in sufficient concentrations to enable formal affinity measurements. However, we were able to take advantage of the effect of dimerization to increase binding avidity and use the low construct concentrations in F-actin gelation assays to demonstrate unequivocally the substantive contributions of the two ABDs in FLNa subunits.
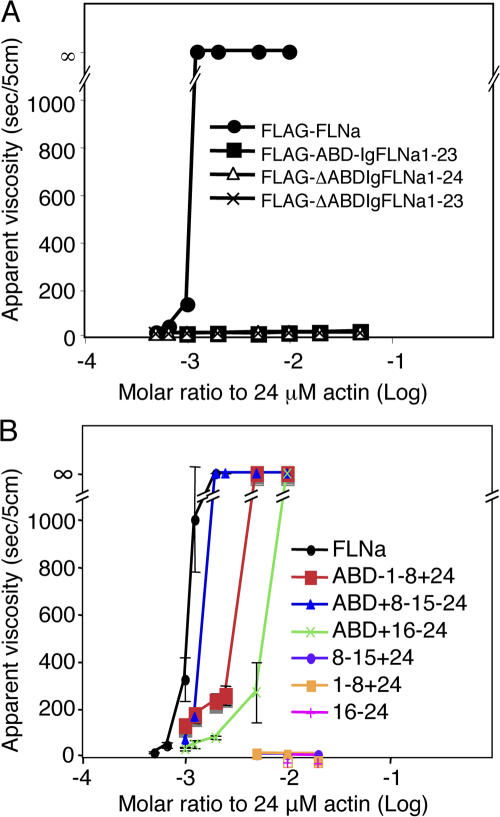
Fig. 5 A shows the apparent viscosity of 24 μM actin polymerized with various concentrations of FLAG-tagged FLNa or recombinant FLNa deletion constructs, for 1 h at room temperature.A molar ratio of one FLAG-tagged full-length FLNa dimer per 800 actin monomers in F-actin was sufficient to induce an abrupt and large viscosity increase indicative of gelation. This ratio is identical to that seen with non-FLAG-tagged recombinant human FLNa or of FLNa purified from human tissues (Nakamura et al., 2002), indicating that the FLAG-tag at the N-T of FLNa does not interfere with actin-binding activity. Deletion of the N-T ABD or the IgFLNa24 dimerization domain, however, completely abolished gelation activity.
Figure 5.
Gel points for actin polymerized with various recombinant FLNa constructs. Apparent viscosity of 24 μM actin after polymerization in the presence of various concentrations of recombinant FLNa proteins as indicated in A and B. Error bars represent SD (n = 3). Error bars are smaller than the symbols in (A).
Fig. 5 B shows that a dimeric construct containing the N-T ABD and Ig repeats 8–15 of rod segment 1 was nearly as effective as full-length FLNa in promoting F-actin gelation. Constructs with N-T ABDs and 8 Ig repeat cassettes from other regions than the distal half of rod segment 1 of FLNa subunits were less efficient. As established by previous work, the more potent gelation activity of the mini-FLNa dimers containing both ABDs is reflected in their ability to promote orthogonal F-actin branching, whereas the other constructs led to more F-actin bundle formation (Fig. S5 C, available at http://www.jcb.org/cgi/content/full/jcb.200707073/DC1). Presumably reduced F-actin binding permits actin filaments to dissociate from the FLNa template and to rotate into bundle configurations.
Orientation of FLNa-actin filament cross-links in vitro
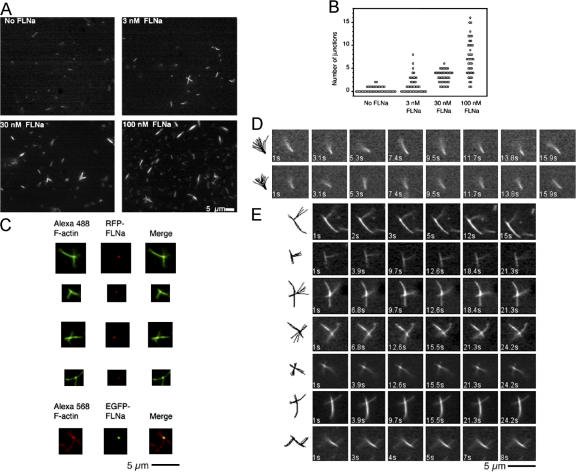
To observe the dynamic behavior of FLNa/F-actin cross-links in real time, we polymerized actin in the presence of gelsolin, which nucleates actin in the slow-growing (pointed) direction and caps the fast-growing (barbed) filament ends (Yin et al., 1981), and attached these gelsolin-capped F-actins stabilized with Alexa488 phallodin to glass coverslips coated with anti-gelsolin IgG. We then added FLNa and a second population of gelsolin-capped, Alexa488 phalloidin–coated F-actin, and observed the mixture by fluorescence microscopy. In the absence of FLNa, only individual actin filaments, tethered at one end, attached to the surface. These filaments rotated rapidly around their attachment points (Fig. 6 D; Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200707073/DC1). Addition of FLNa, however, in a dose-dependent fashion, generated a secondary population of filaments cross-linked to the glass-attached filaments (Fig. 6, A–C).These filaments had a striking perpendicular orientation relative to the first actin filaments. Time-lapse photography revealed that these perpendicular branches were extremely rigid in contrast to the rapid oscillations of F-actin bound to the substrate (Fig. 6 E; Video 2).
Figure 6.
Orthogonal branching of F-actin by FLNa. (A) Relationship between FLNa concentration and filament branching. Treatment of actin filaments tethered to a surface by gelsolin with FLNa and free F-actin results in the formation of orthogonal actin filament crossings and branches. An increase in filament junctions is observed with the addition of increasing FLNa concentrations. (B) Plot of branch number versus FLNa concentration added. For each experiment, junctions in 40 randomly selected areas in >5 coverslips (3 experiments) were analyzed. (C) Fluorescence microscopy of F-actin-FLNa junctions. The presence of FLNa at the junctions is detected using RFP or EGFP labeled FLNa and Alexa 488- or Alexa 568 phalloidin-labeled F-actin, respectively. (D) Representative time-lapse images that record the movement of actin filaments bound to anti-gelsolin IgGs (see Video 1). Note that the gelsolin-tethered end of the filament is immobile while the free end oscillates rapidly. The superimposed tracings to the left track the movement of the filament with time. (E) Behavior of actin filaments cross-linked by FLNa molecules. FLNa creates stable orthogonal filament cross-links that are not deflected by thermal motion. Filament junctions are at nearly perpendicular angles. Note that the addition of filament stabilizes it in the vicinity of the cross-link. The images on the left record the filament movements (see Video 2).
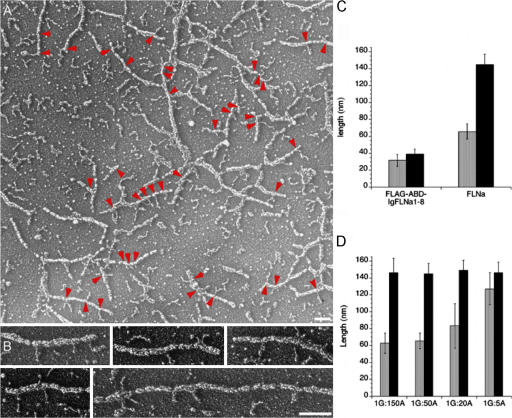
Low angle rotary shadowed images of actin filaments decorated with FLNa provide a closer picture of the interactions between FLNa subunits and F-actin. Fig. 7 A shows a representative field of F-actin with multiple FLNa molecules attached along the actin filaments and a few cross-linked actin filaments. Filamentous structures 65 ± 13 nm in length (approximately half the contour length of a 158 ± 12 nm FLNa dimer; Fig. 2) protrude at right angles from the actin filaments (Fig. 7, B and C).In contrast, FLNa truncates containing only the ABD and the first 8 Ig repeats (ABD-IgFLNa1-8) attach mainly through their N-T ABD (Fig. 7 C), and the rod segments separate from the F-actin.
Figure 7.
Decoration of F-actin with FLNa molecules. Mixtures of F-actin in the presence or absence of gelsolin were stabilized with phallacidin and incubated with recombinant human FLNa and visualized by low angle metal casting. (A) Representative field of actin filaments both cross-linked and coated with FLNa molecules. The arrowheads identify FLNa molecules attached by one subunit. Bar, 100 nm. (B) Selected images of filaments showing the FLNa–actin interaction. Bar, 100 nm. (C) Quantification of the unbound portion of FLNa associated with the actin filament. The unattached length is approximately equivalent to one subunit (n = 23). Identical experiments with the FLNa truncate FLAG-ABD-IgFLNa1-8 reveal that it attaches only by its ABD (n = 38). (D) Relationship of filament length to the amount of the FLNa molecule associated. Actin filaments of lengths ranging from ∼400 nm to 14 nm were prepared by assembling actin in the presence of gelsolin and calcium (G = gelsolin, A = Actin). As the length of the actin filament becomes limiting, FLNa molecules attach only with one of their ABDs, P < 0.0001 (t test; n = 11–45).
We evaluated the effect of actin filament length on the amount of FLNa associated with F-actin (Fig. 7 D). We polymerized actin in the presence of increasing amounts of gelsolin, thereby generating populations of filaments of decreasing lengths. The results show that as the average length decreased below 50 nm, the portion of FLNa molecules not associated with actin increases, consistent with the role of the FLNa subunit rod 1 segments in F-actin binding.
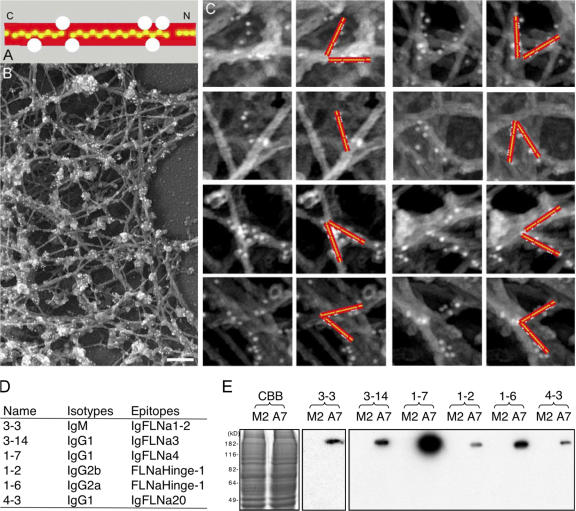
Topography of FLNa binding to F-actin in situ
We labeled detergent-extracted actin cytoskeletons of A7 melanoma cells with a mixture of monoclonal anti-FLNa antibodies reactive against different FLNa epitopes (catalogued in Fig. 8). We visualized the antibodies bound in the electron microscope using 10 nm anti–mouse IgG-coated gold particles. As shown in Fig. 8, B and C, this labeling reveals the contours of the FLNa molecules at orthogonal F-actin junctions in 3D reconstructions of stereo-paired micrographs.The immunogold labeling pattern shows that the rod domains of FLNa associate with the sides of the actin filaments forming the vertices of the junctions. These observations are consistent with the in vitro binding studies and demonstrate that FLNa molecules bind F-actin by their ABD and the rod segment in situ.
Figure 8.
Visualization of the path made by FLNa molecules at F-actin cross-links in the actin cytoskeletons from A7 cells. (A) Location of the binding epitopes of anti-FLNa mouse monoclonal antibodies on the FLNa molecule. Detailed data mapping these sites is provided in D and E. (B) Anti-FLNa immunogold electron microscopy in the cortex of a cytoskeleton from a representative A7 melanoma cell. Bar, 200 nm. (C) Larger anti-FLNa immunogold electron microscopy image of an A7 melanoma cell cytoskeleton at the cortex. The columns are paired: the left column in each pair shows original images while the right shows the images with schematic gold-labeled filamin arms superimposed. The red arms each measure 80 nm and have been placed following stereo three-dimensional analysis. Control treatment of M2 cells with this monoclonal cocktail did not result in labeling (not depicted). (D) Isotypes and epitopes of the FLNa mAbs. The epitopes were mapped by ELISA using various domains of recombinant FLNa. (E) Coomassie brilliant blue (CBB) stain of 7.5% SDS-PAGE of M2 and A7 cell lysates (left) and Western blotting of the corresponding samples with the FLNa mAbs.
Effect of F-actin binding on FLNa binding partner interactions
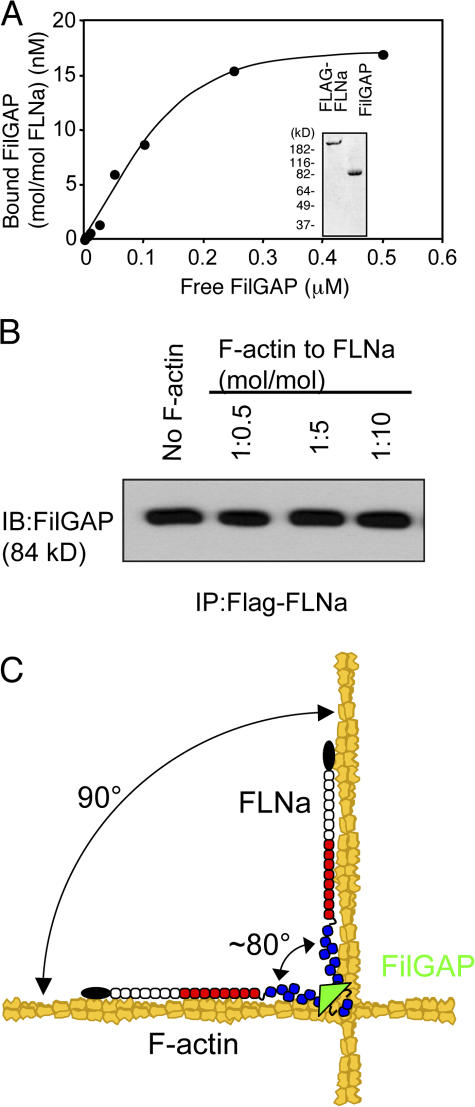
To test whether the binding of the rod segments along F-actin affects FLNa partner interactions, we used full-length recombinant human FilGAP, shown to bind IgFLNa23 in the rod 2 segment (Ohta et al., 2006). Fig. 9 A demonstrates that purified FLAG-FLNa stoichiometrically interacts with full-length FilGAP in vitro. Addition of F-actin up to a 1:10 molar ratio to FLNa did not affect FLNa-FilGAP binding (Fig. 9 B).
Figure 9.
F-actin-binding to FLNa does not affect the FLNa-FilGAP interaction. (A) Binding of full-length FilGAP to FLNa. Increasing amounts of FilGAP (2.5–500 nM) were incubated with 20 nM FLAG-FLNa. The complex was pulled down with anti-FLAG M2 agarose beads. After washing the beads, bound proteins were separated by SDS-PAGE and the amount of bound FilGAP was determined densitometrically from immunoblotting using rabbit polyclonal anti-FilGAP antibodies. Graph plots the data resulting from a set of representative experiments. Each of these experiments was repeated three times. Inlet shows purified FLAG-FLNa and FilGAP stained with Coomassie blue after 8–16% gradient SDS-PAGE. (B) Effect of F-actin on the FLNa-FilGAP binding was assessed by pull-down assays followed by immunoblotting using anti-FilGAP antibodies. Gelsolin shortened (∼120 nm) F-actin was added at 1:0.5, 1:5, or 1:10 (FLNa:F-actin, mol/mol) ratio (N = 3). (C) The model showing the coherent interaction of FLNa with actin filaments. The Ig9-15 rod 1 segments of FLNa (red) facilitate F-actin-binding and FLNa favors junctions of actin filaments because of its high avidity. The rod 2 segment (blue) accommodates “breathing space” for partner interactions (FilGAP, green, binds IgFLNa repeat 23).
Discussion
We have collected internally consistent and mutually supportive kinetic and morphologic data informing as to how FLNa subunits bind F-actin, how FLNa promotes perpendicular F-actin branching, how F-actin branching by FLNa dimers achieves high avidity binding to F-actin, and how FLNa provides interacting domains for its partners. Contributing importantly to these diverse properties are previously unrecognized structural and functional differences between FLNa's rod segments 1 and 2 and between subsegments of rod 1.
Mechanism of FLNa subunit/F-actin binding
The previously characterized ABD represents a key aspect of FLNa subunit attachment to F-actin, as FLNa subunits without it lack gelation activity. This finding is consistent with the effectiveness of calcium-calmodulin binding to this domain in dissociating FLNa/F-actin complexes in vitro (Nakamura et al., 2005). However, the affinity of the ABD for F-actin is relatively low if not attached to Ig repeats 9–15 in rod segment 1. Although in the absence of the ABD, this rod subsegment does not induce gelation, binding is detectable by its F-actin bundling activity. Addition of the N-T ABD to the Ig8-15+24 segments, but not to the Ig1-8+24 or the Ig16-24 segments, reconstituted low stoichiometry orthogonal F-actin gelation, supporting the conclusion that the distal half of rod 1 augments F-actin binding. The restriction of binding to this segment of rod 1 suggests that specifics of the Ig repeats contribute to F-actin binding activity. Utrophin, a member of the same protein family as FLNa, binds F-actin through electrostatic interactions between a basic spectrin-repeat and acidic residues on actin (Amann et al., 1998). The isoelectric points of repeats IgFLNa 9 to 15 are acidic, however (Fig. S1 D), suggesting a different type of binding mechanism.
The additive contributions of the ABD and Ig repeats 9–15 to F-actin binding and the flexibility of the N-T portion of the FLNa subunit suggest that when the ABD engages an actin monomer in F-actin the flexibility of Ig repeats 1–8 enables repeats 9–15 to find the proper alignments of actin monomers in F-actin downstream from the initial binding site. Accordingly, electron micrographs revealed that constructs containing the ABD and Ig repeats 1–8 only have a single point attachment to F-actin, whereas full-length subunits align along F-actin, implying multiple attachments. Conversely, limiting the ability of Ig repeats 9–15 to bind F-actin by presenting FLNa subunits with shortened actin filaments also resulted in single point attachments. Immunohistochemical labeling of FLNa subunits showed that they align along actin filaments at F-actin junctions of cell cytoskeletons in situ.
The overall linearity and flexibility of rod segment 1 can accommodate the twist in the helix groove of F-actin to facilitate such binding. Previous work has shown that proteins known to bind in the helix groove compete with FLNs for F-actin binding (Nomura et al., 1987). The participation of two F-actin interaction sites, involving the N-T two-thirds of the FLNa subunit contour length, accounts for the relatively tight binding of subunits to F-actin and the observation that FLNa/F-actin gels withstand much higher applied stresses than α-actinin/F-actin gels (Gardel et al., 2006).
Mechanism of FLNa dimerization and F-actin branching
Perpendicular branching of F-actin by FLNa, initially inferred from its potent gelation activity, has been observed in static electron micrographs of FLNa/F-actin complexes and FLNa/F-actin networks prepared in vitro (Hartwig et al., 1980; Hartwig and Stossel, 1981; Niederman et al., 1983), and FLNa molecules reside at orthogonal F-actin branch points in the cell periphery in situ (Hartwig and Shevlin, 1986). We have now shown such branching by video microscopy and that the F- actin branches are sufficiently rigid to withstand deformation in the absence of imposed stress.
The F-actin branching activity of FLNa resides at its C-T, consistent with the engagement of the N-T two-thirds of the subunits with F-actin binding. In contrast to the extended flexible structure of the FLNa rod 1 segment, the rod 2 segment in full-length or truncated FLNa dimers has a more compact configuration with three globular units. Moreover, the subunits diverge at fixed high angles to form L-shaped structures. Although previous biochemical, morphological and structural studies localized the filamin dimerization site to its C-terminus and have implicated the self-association of Ig repeat 24 as essential for this function (Gorlin et al., 1990; Pudas et al., 2005), our systematic analysis of hybrid dimeric constructs revealed for the first time that Ig repeat 24 is sufficient to confer this branching morphology and that hinge 2 does not contribute to it. A recently reported X-ray analysis of repeat 23–24 dimers of filamin C (FLNc) is consistent with our findings. The Ig-24 repeats diverge with the same L-shape (∼90 degree angle) observed in our electron micrographs, and interactions with repeat 23 did not appear to contribute to this configuration (Sjekloca et al., 2007).
Mechanism of F-actin cross-linking
Our FLNa/F-actin binding studies revealed that dimeric FLNa has 2 classes of apparent binding affinities for F-actin (Table I). One class has a high affinity, with a Kapp in the nM range, and a limited stoichiometry (one FLNa subunit per ∼50 monomers in F-actin); the other has a lower affinity, with a μM Kapp, and ∼10-times more FLNa binding sites on F-actin. The simplest explanation for this differential binding behavior is that the tightness of association depends on whether one or both FLNa dimer subunits engage F-actin. It stipulates that each of the FLNa subunits can only bind to different filaments, and that the second actin filament be positioned in the correct orientation to the first to permit binding. Hence, when only one FLNa subunit binds F-actin, its cumulative binding affinity is μM (0.2 μM), and the free subunit extends perpendicularly as observed in Fig. 7, B and C. If a second actin filament is available in the correct perpendicular orientation, the unbound subunit engages it, and the apparent affinity (avidity) of the interaction increases markedly, a phenomenon well exemplified by IgG antibody molecules with two antigen binding sites that bind at least 100 times more strongly than monovalent Fab subfragments to a polyvalent antigen (Morris, 1995). The higher affinity of dimeric ABD-IgFLNa1-8+24 compared with monomeric ABD-IgFLNa1-8 for F-actin also supports the model that high-avidity binding of FLNa occurs only when both FLNa subunits engage F-actin (Fig. S5 B). Previous reports of binding affinities between FLNa and F-actin documenting a Kapp in the μM range (Goldmann and Isenberg, 1993) used high FLNa to F-actin ratios, which dominantly report the low-affinity and not the high affinity interactions.
Estimates of relative actin and FLNa concentrations in cells, assuming average F-actin lengths of 1 μm (Hartwig and Shevlin, 1986; Podolski and Steck, 1990), are in the stoichiometric range of the high-avidity binding interaction we measured in vitro. In addition, morphometric analysis of orthogonal F-actin cellular networks reveals an average inter-branch spacing of 100 nanometers, approximately the contour length of individual FLNa subunits (Hartwig and Shevlin, 1986). Therefore, FLNa dimers may act as templates for F-actin networks in the cell.
Cross-link flexibility and unfolding is an explanation offered to account for prestress-mediated elasticity increases of FLNa/F-actin networks (Gardel et al., 2006; DiDonna and Levine, 2007). The lack of binding of FLNa subunit rod 2 to F-actin renders nearly a third of the FLNa subunit contour length available for unfolding, and the observable flexibility of rod 2 in electron micrographs is consistent with the capacity to unfold. Moreover, we showed that hinge 1, a requirement for the elasticity associated with prestress of FLNa/F-actin gels (Gardel et al., 2006), truly is a hinge, a site of greater than average flexibility within the FLNa subunit.
Large but not small externally imposed strains increase FLNa/F-actin network elasticity, implying that unfolding requires high mechanical energy input. This idea fits the invariant perpendicular branching we observed for unstressed FLNa/F-actin complexes by video microscopy.
Differential arrangement of IgFLNa domains in rod segment 2
The dimensions of the individual Ig domains that have been solved for FLN family proteins are 4.0–4.8 nm × 2.0–2.6 nm × 1.5–1.8 nm (Gorlin et al., 1990; Fucini et al., 1997; Kiema et al., 2006; Nakamura et al., 2006). Structural studies of three tandem repeats from dictyostelium FLN (IgddFLN4-6) showed neighboring Ig domains to array in a zigzag configuration (Popowicz et al., 2004) with an average spacing of ∼3.7 nm per repeat. This spacing along the long axis of the rod is in good agreement with our measurements of rod segment 1 (IgFLNa1-8: 28.5 nm/8 ∼3.6 nm per IgFLNa, IgFLNa8-15: 28.8 nm/8 ∼3.6 nm per IgFLNa), suggesting that IgFLNa repeats in rod 1 of FLNa arrange in an overlapping configuration. The repeat spacing in rod 2, however, is 33% shorter (19.2 nm/8 ∼2.4 nm per IgFLNa), suggesting a more packed configuration of Ig domains. Recently the atomic structure of FLNa repeats 19–21 has been solved demonstrating a nonlinear packing of these Ig repeats (Lad et al., 2007).
Differential roles of FLNa rod segments for partner interactions
Since rod segment 2 does not contribute to F-actin binding or branching, it is potentially free to interact with other partner proteins, explaining why most partner interactions occur at this site in the FLNa subunit. Although the ability to cosediment FLNa with some of its partners such as Gp1bα or β-integrins with F-actin is consistent with this idea (Sharma et al., 1995; Kovacsovics and Hartwig, 1996), it has not been formally tested. We now have shown that FilGAP maintains interactivity with rod 2 segments even in the presence of F-actin (Fig. 9).
The same principle of bivalency underlying the high avidity interaction between FLNa and F-actin applies to the connections between FLNa and its other binding partners, such as the von Willebrand factor receptor and integrins, that make for tight connections between extracellular matrix proteins and the actin cytoskeleton. Two Gp1bα subunits of the von Willebrand factor receptor complex and two β chain cytoplasmic tails of clustered integrins each bind sites on Ig repeat 17 or Ig repeat 21, respectively on two FLNa dimer subunits (Pfaff et al., 1998; Kiema et al., 2006; Nakamura et al., 2006). Whereas the widely separated N-T ABD and rod 1 segments of FLNa subunits can reach out to numerous submembrane actin filaments, separations between membrane receptor tails are small. Therefore, in addition to avoiding competition with F-actin binding, the placement of binding sites for these ligands in rod segment 2 near the C-termini of the FLNa subunits accommodates to the topography of membrane proteins.
Many FLNa-binding partners may constitutively interact with the rod 2 domain of FLNa bound to F-actin, but mechanical stretching of the rod may, in some circumstances, regulate the availability of binding interfaces, possibly explaining the role of FLNa in mechanotransduction (Kainulainen et al., 2002). The nonlinear alignment of repeats IgFLNa19-21 provides a mechanism by which paired domains can dissociate to reveal cryptic binding sites (Lad et al., 2007). F-actin binding to FLNa, however, may regulate by competition the binding of certain partners thought to interact with rod segment 1, such as furin (Liu et al., 1997), cyclin B1 (Cukier et al., 2007), the cystic fibrosis transmembrane conductance regulator (Thelin et al., 2007), CD28 (Tavano et al., 2006), and protein kinase Cα (Tigges et al., 2003).
Materials and methods
Recombinant FLNa constructs
cDNA for human FLNa was expressed using a Baculovirus Expression System (Invitrogen) in Sf9 insect cells and recombinant protein was purified as previously described (Nakamura et al., 2002). Full-length, and various N-T and C-T truncates of FLNa (Fig. 1), were fused at their N-T to the FLAG epitope in the pFASTBAC-FLAG vector. pFASTBAC-FLAG-FLNa was prepared by PCR. A cDNA fragment encoding the N-T of FLNa was amplified using pFASTBAC FLNa (Nakamura et al., 2002) as the template, with the forward primer GGAATTCCATATGAGTAGCTCCCACTCTC containing a NdeI site, and with the reverse primer GTCCAGATGAGGCCCAGGATCAG. The amplified fragment was purified, NdeII/SalI-digested, and ligated into NdeI/SalI sites in the pFASTBAC FLAG vector to generate pFASTBAC FLAG-FLNa/SalI, which was confirmed by sequencing. The 3′-site in FLNa cDNA was prepared by cutting pFASTBAC FLNa with SalI and HindIII and ligating it into the pFASTBAC FLAG-FLNa/SalI opened with SalI and HindIII, thereby generating pFASTBAC FLAG-FLNa. Deletion constructs illustrated in Fig. 1 A were made by PCR with the appropriate primers.
The His-tagged constructs were made by PCR using modified pET-23a(+)-HT(a or b) plasmids that were generated by inserting NdeI/HindIII fragment of PCR product amplified from pFASTBAC-HT(a or b) (Invitrogen) with two primers, GGAATTCCATATGTCGTACTACCATCAC and CTAGTACTTCTCGACAAGCTTGG, into NdeI/HindIII sites of pET-23a(+) (Novagen). pET-23a(+)-HT-IgFLNa1-8, 8–15, 16–23, or 16–24 were generated by PCR. Dimerized constructs of IgFLNa1-8+24, 8–15+24 or 16–23+24 were made inserting NheI/Not fragment of PCR product amplified from cDNA of hinge 2 and IgFLNa24 (NheI/NotI). These dimerized constructs have, therefore, an additional alanine residue just before the hinge 2 domain. cDNA of FLNa ABD (1–278 aa) were also added at their N-terminal by PCR.
Expression and purification of FLNa fusion proteins
FLAG-tagged fusion proteins were expressed in accordance with manufacturer's instructions in Sf9 cells (5 × 108 cells) and the expressing cells were harvested 72 h post-infection. After washing with PBS, the cells were lysed in 20 ml of 20 mM Tris-HCl, pH 7.4, 100 mM NaCl, 1% Triton X-100, 5 mM EGTA, 2 mM PMSF, 10 μg/ml aprotinin, and 10 μg/ml leupeptin. The supernatant of this lysis, separated from insoluble material by centrifugation at 20,000 g for 30 min at 4°C, was loaded onto a HitrapQ column (5 ml; GE Healthcare). Bound recombinant protein was eluted with a 100 ml linear gradient of 100–500 mM NaCl in 10 mM Tris-HCl, pH7.4, 0.01% Triton X-100, and 0.5 mM EGTA at a flow rate of 2 ml/min. Fractions containing the recombinant fusion protein were absorbed on a 2-ml anti-FLAG M2 agarose column (Sigma-Aldrich) and sequentially washed with 20 ml of 10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1 mM EGTA, and 0.1% Triton X-100 then 20 ml of TBS (10 mM Tris-HCl, pH 7.4 and 150 mM NaCl). Bound protein was eluted using TBS containing 100 μg/ml FLAG peptide (Sigma-Aldrich). Purified proteins were concentrated using an Amicon Ultra-15 (Millipore), with a molecular weight retention of >5,000 daltons, and gel-filtered on Superose 6 or 12 columns (GE Healthcare) to remove the FLAG peptide. Protein concentrations were measured by UV spectrophotometry at 280 nm (http://au.expasy.org/tools/protparam.html). Recombinant protein was stored at −80°C and thawed and pre-cleared by centrifugation before use. The typical yield of each FLAG-tagged protein was ∼5 mg. The purity of recombinant protein was assessed by Coomassie blue-staining of SDS-polyacrylamide gels and by immunoblot analysis with the anti-FLNa and anti-FLAG clone M2 monoclonal antibodies (Sigma-Aldrich).
His-tagged fusion proteins were expressed in BL21 Star (DE3) Escherichia coli (Invitrogen) at 37°C. The fusion proteins were extracted from the E. coli by sonicating in lysis solution (20 mM sodium phosphate, pH 8.0, 100 mM NaCl, 20 mM imidazole,1 mM β-mercaptoethanol, 2 mM PMSF, 10 μg/ml aprotinin, and 10 μg/ml leupeptin) at 4°C. The extracts were centrifuged at 20,000 g for 30 min at 4°C and loaded onto a Ni-NTA column (1 ml; QIAGEN). The column was washed with washing solution I (20 mM sodium phosphate, pH 8.0, 20 mM imidazole, 1 mM β-mercaptoethanol, 300 mM NaCl, 0.1% Triton X-100) followed by washing solution II (20 mM sodium phosphate, pH 8.0, 20 mM imidazole, 100 mM NaCl, 1 mM β-mercaptoethanol) and bound recombinant proteins were eluted with 20 mM sodium phosphate, pH 8.0, 200 mM imidazole, 1 mM β-mercaptoethanol. Purified proteins were concentrated using an Amicon Ultra-15 (Millipore) with a molecular weight retention of >5,000 daltons and gel-filtered on Superdex 200 pg (16/600; GE healthcare) column.
Prediction of isoelectric points
The isoelectric points of FLNa repeats were predicted using Iso-Electric Point Calculator (http://maths.sci.shu.ac.uk/aramakum/cgi-bin/respond.pl?hpdi=analysis).
Stokes radius
The Stokes' radii of recombinant proteins were determined by gel filtration chromatography on a Superdex 200 pg (16/600) column at a flow rate of 1 ml/min in 20 mM Tris-HCl, pH7.4, 150 mM NaCl, 0.5 mM EGTA, 0.5 mM β-mercaptoethanol, using ferritin, aldolase, ovalbumin, and ribonuclease A as a standard at 4°C.
Non-denaturing (native) and denaturing gel electrophoresis
Non-denaturing and denaturing gel electrophoresis were performed using Novex Tris-glycine gel (Invitrogen) in accordance with manufacturer's instruction and proteins were stained with SimplyBlue safe stain (Invitrogen).
MALDI-TOF mass spectroscopy
Molecular weights of recombinant proteins were determined by MALDI-TOF mass spectroscopy in linear mode with an accuracy of 0.1% in Tufts University core facility.
Mouse monoclonal antibodies
Anti-FLNa mouse mAbs were prepared as previously described. Their reactive epitopes were mapped by enzyme-linked immunosorbent assays or immunoblotting. The antibody subtypes were determined using a kit (Boehringer Mannheim). Monoclonal IgG or IgM antibodies were purified from hybridoma supernatants using GammaBind G-Sepharose (GE Healthcare) or by KAPTIV-M affinity chromatography (Genomics One International).
Rotary shadowing of FLNa
FLNa, FLNa truncates, or FLNa/F-actin mixtures containing 0.1 μM phallacidin were mixed in a final glycerol concentration of 50% and sprayed on the surface of freshly cleaved mica as previously described (Tyler et al., 1980). Samples were dried under high vacuum in a CFE-60 freeze-fracture apparatus (Cressington, Inc.) then metal cast with platinum at 5° with rotation and carbon at 90° without rotation. Metal films were separated from the mica in distilled water, picked up on 200 mesh carbon grids, and viewed and photographed in a JEOL 1200-EX transmission electron microscope. FLNa molecules were also monitored by stereo-pairing pictures taken from −10° and +10° angles upon metal cast with tantalum at 5° while rotating and carbon at 90° without rotation. This revealed that the molecule attaches flat to mica upon glycerol spraying. Therefore, angular measurements could be performed as well as length measurements.
Measurements of FLNa and truncates length and C-T angles
TEM pictures were analyzed using Scion Image 1.62a software. Length measurements of molecules were made by tracing along their contour from N-T to C-T. C-T angle measurements were done in Scion Image by using the angle tool. Since FLNa is a very flexible molecule, measurements were performed at the C-T (extending only ∼10–15 nm along each rod). Determination of the position of the dimerization site was done by tracing from the N-T along the molecule. For full-length FLNa, 80 nm from the N-T was measured and determined as the C-T. Shorter molecules were treated the same way but measurements to find the dimerization domain was adjusted to their shorter length.
Measurements of FLNa bending points
Full-length FLNa molecules containing or lacking hinge 1 were mixed with 50% glycerol and sprayed onto freshly cleaved mica at a concentration of 50 μg/ml. Proteins were rotary shadowed using Pt at 5° angle and replicas monitored by TEM. Pictures were analyzed using Scion Image 1.62a software. Measurements of molecules were made by tracing their contour. Before determination of bending points, the entire length of the molecule (N-T to N-T) was measured to ensure full-length and dimeric state. Bending points were acquired by tracing the molecule from the N-T and measuring the length from the N-T at which sharp bending occurred. Both strands of dimeric proteins were measured for a total of 55 FLNa molecules.
Actin cosedimentation assay
Recombinant FLNa or FLNa truncates were mixed with or without 5 μM G-actin in polymerization buffer (solution F: 20 mM Tris-HCl, pH 7.4, 0.5 mM Na2ATP, 5 mM MgCl2, 120 mM NaCl, and 0.2 mM DTT) and incubated for 1 h at 25°C. F-actin was collected by centrifugation at 200,000 g (70,000 rpm; Beckman TLA-100) for 30 min at 25°C. Proteins remaining in the resultant supernatant or in the pellet were solubilized in SDS gel sample buffer and displayed by SDS-PAGE. Polypeptides were visualized by Coomassie-staining and were scanned and quantified using the software program NIH ImageJ 1.37v for Mac. The amount of the FLNa constructs bound to F-actin was fit to a single rectangular hyperbola using Prism (ver. 4; GraphPad Software) for Mac. Two independent site model was used for full-length FLNa (see text) and their apparent dissociation constants were independently calculated using Prism software. Klotz plots were generated as previously described (Wilkinson, 2004).
Falling ball viscometry
Gel point measurements were determined by falling ball viscometry as described previously (Nakamura et al. 2002).
Immunogold labeling of cells in the electron microscope
A7 and M2 melanoma cells, attached to 5 mm glass coverslips, were washed twice in PHEM buffer (Schliwa et al., 1981) and incubated for 2 min in PHEM buffer containing 0.75% Triton X-100, 1 μM phalloidin, protease inhibitors, and 0.36% formaldehyde. Without intermediate washing, the permeabilization buffer was removed and the detergent-insoluble residue was fixed for 10 min in PHEM buffer containing 3.6% formaldehyde. After fixation, the samples were blocked for a minimum of 2 h with PBS (pH 7.4) containing 1% BSA. After blocking, coverslips were incubated with 10 μl of solution containing a mixture of 6 monoclonal antibodies against FLNa (2 μg/ml of each) for 2 h at 37°C. After the incubation, the coverslips were washed three times with the blocking buffer to remove unbound antibody and then incubated with secondary anti–mouse 10-nm gold conjugates for 2 h at 37°C. The immunogold-labeled coverslips were washed three times in the PBS/BSA buffer to remove unbound reagents, five times in PBS to remove BSA, and then post-fixed by immersion in 1% glutaraldehyde in PBS for 10 min. Coverslips were washed three times in distilled, microfiltered water and stored at 4°C until they were processed by rapid-freezing, freeze-drying, and metal-casting with 1.2 nm of tantalum-tungsten at 45° and 2.5 nm of carbon at 90°. Stereo-paired micrographs were taken at ± 10° of tilt in a JEOL 1200-EX electron microscope. Actin filament junctions in the cytoskeletons from A7 cells were reconstructed in 3D by tracing the filament paths in stereo-paired micrographs (Niederman et al., 1983).
Visualization of FLNa/F-actin cross-links
Chambers, formed by mounting a glass coverslip onto a 35-mm-diameter cell culture dish with a 1-cm hole, were glow discharged in a Cressington 308R coating system, using an output current of <20 mA for 40 s. The coverslips were coated with 10 μg/ml of monoclonal 2C4 anti-gelsolin antibody in PBS for 12 h at 4°C. Coverslips were blocked with 3% BSA in PBS for 2 h at RT after five quick washes in PBS. F-actin, 10 μM was polymerized in buffer B (0.1 M KCl, 0.2 mM MgCl2, 1.5 mM CaCl2, 0.5 mM ATP, 10 mM Tris, 0.5 mM mercaptoethanol, pH 7.4) in the presence of human recombinant plasma gelsolin (Biogen Inc.) at a molar ratio of 1:1,200 gelsolin/actin and stabilized with 0.1 μM Alexa 488 phalloidin (Invitrogen). The F-actin-capped by gelsolin solution was diluted to 1 μM and incubated on the 2C4 antibody-coated coverslips for 2h at RT after washing them with PBS followed by buffer B. Unbound F-actin was removed by washing three times in buffer B containing 0.5% BSA and 0.1 μM Alexa 488 phalloidin (buffer C). Non-specific FLNa binding sites were blocked by incubating the coverslips with 1 μM of ΔABDIgFLNa1-23 for 15 min at RT. After three washes in buffer C, FLNa at a concentration of 30 nM was added for 10 min to the coverslip. A secondary layer of short (∼1–1.5 μm) F-actin, polymerized in buffer B in the presence of human recombinant plasma gelsolin at a molar ratio of 1:500 gelsolin/actin and stabilized with 0.1 μM Alexa488-conjugated phalloidin, was added for 2 min at RT. Specimens were washed once in buffer C and three times in buffer B. Stable F-actin -FLNa cross-links in 100 μl buffer B were visualized at 22°C by fluorescence microscopy using a Zeiss Axiovert 200M microscope with a Zeiss 100× (NA 1.4) DIC oil-immersion objective lens and captured with an Orca II ER cooled CCD camera (Hamamatsu) equipped with an electronic shutter. Shutter and image acquisition was controlled by Metamorph software version 5.0r7 (Universal Imaging Corporation). Movement of F-actin due to Brownian motion was captured at a rate of 0.3–2 frames per second with a 0.5-s exposure time. To monitor the localization of FLNa in the cross-linked F-actin, red-fluorescent protein- (RFP, DsRedmonomer) or EGFP-tagged FLNa was used in combination with F-actin labeled with Alexa 488 or Alexa 568 phalloidin, respectively. The exposure time was increased to 5–10 s to visualize single EGFP- or RFP-tagged FLNa molecules at the junctions.
Expression and purification of full-length human recombinant FilGAP
The cDNA of full-length human FilGAP was cloned into pFASTBAC vector (Invitrogen) using EcoRI/HindIII restriction sites. The FilGAP protein was expressed in Sf9 cells as described above and purified by a HitrapQ column followed by Superose 6 column.
FLAG-FLNa pull-down assay
The full-length FilGAP (2.5 - 500 nM) was incubated with 10 μl of anti-FLAG M2 agarose (50% slurry) in the presence or absence of 20 nM FLAG-FLNa and in binding/washing buffer (50 mM Tris-HCl, 150 mM NaCl, 0.1% Triton X-100, 0.1 mM b-mercaptoethanol, 0.1 mM EGTA, pH 7.4; 400 μl) containing 1% BSA for 1 h at 25°C. The beads were sedimented and washed four times in ice cold binding/washing buffer (600 μl). Proteins bound to the beads were solubilized in SDS sample buffer and separated by 9.5% SDS-PAGE followed by immunoblotting.
To investigate an effect of F-actin on the FLNa-FilGAP binding, G-actin (58 μM) was polymerized at 25°C in the presence of human recombinant plasma gelsolin (gift from Biogen Inc.) at a ratio of 1:40 gelsolin/G-actin in buffer B (0.1 M KCl, 0.2 mM MgCl2, 1.5 mM CaCl2, 0.5 mM ATP, 10 mM Tris, 0.5 mM mercaptoethanol, pH 7.4), stabilized with phalloidin (1:1, mol/mol). F-actin was added to the binding assay described above with the exception that FLAG-FLNa (20 nM) was preincubated with anti-FLAG M2 agarose and unbound FLNa washed away (4 washes in ice cold binding/washing buffer, 600 μl) before addition of F-actin and FilGAP. F-actin (10–200 nM) was added to the binding assay 30 min before addition of FilGAP (1 nM).
Online supplemental material
Fig. S1 shows electron micrographs of the purified recombinant FLNa constructs. Fig. S2 shows characterization of His-FLNa constructs. Fig. S3 shows topological properties of FLNa constructs. Fig. S4 shows binding of FLNa truncates to F-actin. Fig. S5 shows differential roles of FLNa rod segments for F-actin branching. Video 1 shows time-lapse capturing of actin filaments tethered to the bottom of a culture dish by gelsolin IgG, rotating freely around the axis of attachment. Video 2 shows time-lapse capturing of actin filaments cross-linked by FLNa. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200707073/DC1.
Acknowledgments
None of the authors have any financial interest related to this work.
This work was supported by National Institutes of Health grants HL19429 and HL56252 (J.H. Hartwig), an American Cancer Society Clinical Research Professorship and a gift from the Edwin S Webster Foundation to T.P. Stossel. T.M. Osborn was a recipient of the Göteborg University Jubileumsfond's scholarship.
F. Nakamura and T. M. Osborn contributed equally to this paper.
Abbreviations used in this paper: ABD, actin-binding domain; C-T, carboxy terminal; FLNa, filamin A; IgFLNa, immunoglobulin-like filamin A domain; Kapp, apparent dissociation constant: N-T, amino terminal.
References
- Amann, K.J., B.A. Renley, and J.M. Ervasti. 1998. A cluster of basic repeats in the dystrophin rod domain binds F-actin through an electrostatic interaction. J. Biol. Chem. 273:28419–28423. [DOI] [PubMed] [Google Scholar]
- Cukier, I.H., Y. Li, and J.M. Lee. 2007. Cyclin B1/Cdk1 binds and phosphorylates Filamin A and regulates its ability to cross-link actin. FEBS Lett. 581:1661–1672. [DOI] [PubMed] [Google Scholar]
- DiDonna, B.A., and A.J. Levine. 2007. Unfolding cross-linkers as rheology regulators in F-actin networks. Phys. R ev. E. Stat. Nonlin. Soft Matter Phys. 75:041909. [DOI] [PubMed] [Google Scholar]
- Discher, D.E., P. Janmey, and Y.L. Wang. 2005. Tissue cells feel and respond to the stiffness of their substrate. Science. 310:1139–1143. [DOI] [PubMed] [Google Scholar]
- Feng, Y., and C.A. Walsh. 2004. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat. Cell Biol. 6:1034–1038. [DOI] [PubMed] [Google Scholar]
- Feng, Y., M.H. Chen, I.P. Moskowitz, A.M. Mendonza, L. Vidali, F. Nakamura, D.J. Kwiatkowski, and C.A. Walsh. 2006. Filamin A (FLNA) is required for cell-cell contact in vascular development and cardiac morphogenesis. Proc. Natl. Acad. Sci. USA. 103:19836–19841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferland, R.J., J.N. Gaitanis, K. Apse, U. Tantravahi, C.A. Walsh, and V.L. Sheen. 2006. Periventricular nodular heterotopia and Williams syndrome. Am. J. Med. Genet. A. 140:1305–1311. [DOI] [PubMed] [Google Scholar]
- Flanagan, L.A., J. Chou, H. Falet, R. Neujahr, J.H. Hartwig, and T.P. Stossel. 2001. Filamin A, the Arp2/3 complex, and the morphology and function of cortical actin filaments in human melanoma cells. J. Cell Biol. 155:511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fucini, P., C. Renner, C. Herberhold, A.A. Noegel, and T.A. Holak. 1997. The repeating segments of the F-actin cross-linking gelation factor (ABP-120) have an immunoglobulin-like fold. Nat. Struct. Biol. 4:223–230. [DOI] [PubMed] [Google Scholar]
- Gardel, M.L., F. Nakamura, J.H. Hartwig, J.C. Crocker, T.P. Stossel, and D.A. Weitz. 2006. Prestressed F-actin networks cross-linked by hinged filamins replicate mechanical properties of cells. Proc. Natl. Acad. Sci. USA. 103:1762–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmann, W.H., and G. Isenberg. 1993. Analysis of filamin and alpha-actinin binding to actin by the stopped flow method. FEBS Lett. 336:408–410. [DOI] [PubMed] [Google Scholar]
- Gorlin, J.B., R. Yamin, S. Egan, M. Stewart, T.P. Stossel, D.J. Kwiatkowski, and J.H. Hartwig. 1990. Human endothelial actin-binding protein (ABP-280, nonmuscle filamin): a molecular leaf spring. J. Cell Biol. 111:1089–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, A.W., J.E. Morgan, J. Schneider, K. West, L. McKie, S. Bhattacharya, I.J. Jackson, and S.H. Cross. 2006. Cardiac malformations and midline skeletal defects in mice lacking filamin A. Hum. Mol. Genet. 15:2457–2467. [DOI] [PubMed] [Google Scholar]
- Hartwig, J.H. 1995. Actin-binding proteins. 1: Spectrin super family. Protein Profile. 2:703–800. [PubMed] [Google Scholar]
- Hartwig, J.H., and P. Shevlin. 1986. The architecture of actin filaments and the ultrastructural location of actin-binding protein in the periphery of lung macrophages. J. Cell Biol. 103:1007–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwig, J.H., and T.P. Stossel. 1981. Structure of macrophage actin-binding protein molecules in solution and interacting with actin filaments. J. Mol. Biol. 145:563–581. [DOI] [PubMed] [Google Scholar]
- Hartwig, J.H., J. Tyler, and T.P. Stossel. 1980. Actin-binding protein promotes the bipolar and perpendicular branching of actin filaments. J. Cell Biol. 87:841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainulainen, T., A. Pender, M. D'Addario, Y. Feng, P. Lekic, and C.A. McCulloch. 2002. Cell death and mechanoprotection by filamin a in connective tissues after challenge by applied tensile forces. J. Biol. Chem. 277:21998–22009. [DOI] [PubMed] [Google Scholar]
- Kiema, T., Y. Lad, P. Jiang, C.L. Oxley, M. Baldassarre, K.L. Wegener, I.D. Campbell, J. Ylanne, and D.A. Calderwood. 2006. The molecular basis of filamin binding to integrins and competition with talin. Mol. Cell. 21:337–347. [DOI] [PubMed] [Google Scholar]
- Klotz, I.M., and D.L. Hunston. 1971. Properties of graphical representations of multiple classes of binding sites. Biochemistry. 10:3065–3069. [DOI] [PubMed] [Google Scholar]
- Kovacsovics, T.J., and J.H. Hartwig. 1996. Thrombin-induced GPIb-IX centralization on the platelet surface requires actin assembly and myosin II activation. Blood. 87:618–629. [PubMed] [Google Scholar]
- Kyndt, F., J.P. Gueffet, V. Probst, P. Jaafar, A. Legendre, F. Le Bouffant, C. Toquet, E. Roy, L. McGregor, S.A. Lynch, et al. 2007. Mutations in the gene encoding filamin A as a cause for familial cardiac valvular dystrophy. Circulation. 115:40–49. [DOI] [PubMed] [Google Scholar]
- Lad, Y., T. Kiema, P. Jiang, O.T. Pentikainen, C.H. Coles, I.D. Campbell, D.A. Calderwood, and J. Ylanne. 2007. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO J. 26:3993–4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, G., L. Thomas, R.A. Warren, C.A. Enns, C.C. Cunningham, J.H. Hartwig, and G. Thomas. 1997. Cytoskeletal protein ABP-280 directs the intracellular trafficking of furin and modulates proprotein processing in the endocytic pathway. J. Cell Biol. 139:1719–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira, P. 1994. Actin crosslinking proteins at the leading edge. Semin. Cell Biol. 5:165–174. [DOI] [PubMed] [Google Scholar]
- Morris, R.J. 1995. Antigen-antibody interactions: how affinity and kinetics affect assay design and selection procedures. In Monoclonal Antibodies. M.A. Ritter and H.M. Ladyman, editors. Cambridge University Press, Cambridge. 34–57.
- Nakamura, F., E. Osborn, P.A. Janmey, and T.P. Stossel. 2002. Comparison of filamin A-induced cross-linking and Arp2/3 complex-mediated branching on the mechanics of actin filaments. J. Biol. Chem. 277:9148–9154. [DOI] [PubMed] [Google Scholar]
- Nakamura, F., J.H. Hartwig, T.P. Stossel, and P.T. Szymanski. 2005. Ca2+ and calmodulin regulate the binding of filamin A to actin filaments. J. Biol. Chem. 280:32426–32433. [DOI] [PubMed] [Google Scholar]
- Nakamura, F., R. Pudas, O. Heikkinen, P. Permi, I. Kilpelainen, A.D. Munday, J.H. Hartwig, T.P. Stossel, and J. Ylanne. 2006. The structure of the GPIb-filamin A complex. Blood. 107:1925–1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederman, R., P.C. Amrein, and J. Hartwig. 1983. Three-dimensional structure of actin filaments and of an actin gel made with actin-binding protein. J. Cell Biol. 96:1400–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, M., K. Yoshikawa, T. Tanaka, K. Sobue, and K. Maruyama. 1987. The role of tropomyosin in the interactions of F-actin with caldesmon and actin-binding protein (or filamin). Eur. J. Biochem. 163:467–471. [DOI] [PubMed] [Google Scholar]
- Ohta, Y., J.H. Hartwig, and T.P. Stossel. 2006. FilGAP, a Rho- and ROCK-regulated GAP for Rac binds filamin A to control actin remodelling. Nat. Cell Biol. 8:803–814. [DOI] [PubMed] [Google Scholar]
- Pfaff, M., S. Liu, D.J. Erle, and M.H. Ginsberg. 1998. Integrin beta cytoplasmic domains differentially bind to cytoskeletal proteins. J. Biol. Chem. 273:6104–6109. [DOI] [PubMed] [Google Scholar]
- Podolski, J.L., and T.L. Steck. 1990. Length distribution of F-actin in Dictyostelium discoideum. J. Biol. Chem. 265:1312–1318. [PubMed] [Google Scholar]
- Popowicz, G.M., R. Muller, A.A. Noegel, M. Schleicher, R. Huber, and T.A. Holak. 2004. Molecular structure of the rod domain of Dictyostelium filamin. J. Mol. Biol. 342:1637–1646. [DOI] [PubMed] [Google Scholar]
- Popowicz, G.M., M. Schleicher, A.A. Noegel, and T.A. Holak. 2006. Filamins: promiscuous organizers of the cytoskeleton. Trends Biochem. Sci. 31:411–419. [DOI] [PubMed] [Google Scholar]
- Pudas, R., T.R. Kiema, P.J. Butler, M. Stewart, and J. Ylanne. 2005. Structural basis for vertebrate filamin dimerization. Structure. 13:111–119. [DOI] [PubMed] [Google Scholar]
- Robertson, S.P. 2005. Filamin A: phenotypic diversity. Curr. Opin. Genet. Dev. 15:301–307. [DOI] [PubMed] [Google Scholar]
- Robertson, S.P., S.R. Twigg, A.J. Sutherland-Smith, V. Biancalana, R.J. Gorlin, D. Horn, S.J. Kenwrick, C.A. Kim, E. Morava, R. Newbury-Ecob, et al. 2003. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat. Genet. 33:487–491. [DOI] [PubMed] [Google Scholar]
- Schliwa, M., J. van Blerkom, and K.R. Porter. 1981. Stabilization and the cytoplasmic ground substance in detergent-opened cells and a structural and biochemical analysis of its composition. Proc. Natl. Acad. Sci. USA. 78:4329–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma, C.P., R.M. Ezzell, and M.A. Arnaout. 1995. Direct interaction of filamin (ABP-280) with the beta 2-integrin subunit CD18. J. Immunol. 154:3461–3470. [PubMed] [Google Scholar]
- Sjekloca, L., R. Pudas, B. Sjoblom, P. Konarev, O. Carugo, V. Rybin, T.R. Kiema, D. Svergun, J. Ylanne, and K.D. Carugo. 2007. Crystal structure of human filamin C domain 23 and small angle scattering model for filamin C 23-24 dimer. J. Mol. Biol. 368:1011–1023. [DOI] [PubMed] [Google Scholar]
- Stossel, T.P., J. Condeelis, L. Cooley, J.H. Hartwig, A. Noegel, M. Schleicher, and S.S. Shapiro. 2001. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2:138–145. [DOI] [PubMed] [Google Scholar]
- Tavano, R., R.L. Contento, S.J. Baranda, M. Soligo, L. Tuosto, S. Manes, and A. Viola. 2006. CD28 interaction with filamin-A controls lipid raft accumulation at the T-cell immunological synapse. Nat. Cell Biol. 8:1270–1276. [DOI] [PubMed] [Google Scholar]
- Thelin, W.R., Y. Chen, M. Gentzsch, S.M. Kreda, J.L. Sallee, C.O. Scarlett, C.H. Borchers, K. Jacobson, M.J. Stutts, and S.L. Milgram. 2007. Direct interaction with filamins modulates the stability and plasma membrane expression of CFTR. J. Clin. Invest. 117:364–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tigges, U., B. Koch, J. Wissing, B.M. Jockusch, and W.H. Ziegler. 2003. The F-actin cross-linking and focal adhesion protein filamin A is a ligand and in vivo substrate for protein kinase C alpha. J. Biol. Chem. 278:23561–23569. [DOI] [PubMed] [Google Scholar]
- Tyler, J.M., J.M. Anderson, and D. Branton. 1980. Structural comparison of several actin-binding macromolecules. J. Cell. Biol. 85:489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, K.D. 2004. Quantitative analysis of protein-protein interactions. Methods Mol. Biol. 261:15–32. [DOI] [PubMed] [Google Scholar]
- Yin, H.L., J.H. Hartwig, K. Maruyama, and T.P. Stossel. 1981. Ca2+ control of actin filament length. Effects of macrophage gelsolin on actin polymerization. J. Biol. Chem. 256:9693–9697. [PubMed] [Google Scholar]